Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
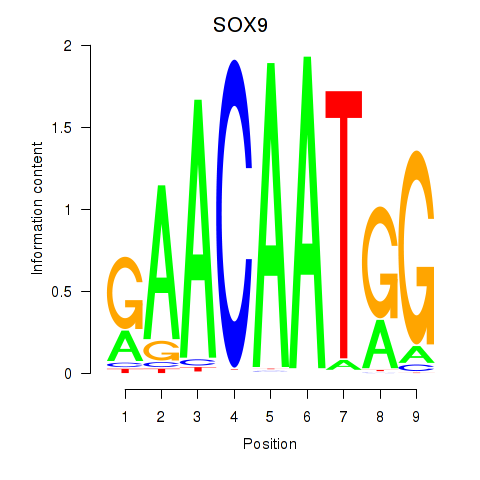
Results for SOX9
Z-value: 0.78
Transcription factors associated with SOX9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX9
|
ENSG00000125398.5 | SRY-box transcription factor 9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX9 | hg19_v2_chr17_+_70117153_70117174 | 0.32 | 1.3e-06 | Click! |
Activity profile of SOX9 motif
Sorted Z-values of SOX9 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX9
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.7 | 44.0 | GO:0051086 | chaperone mediated protein folding independent of cofactor(GO:0051086) |
| 11.2 | 55.8 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 5.9 | 53.2 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 4.6 | 13.7 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 3.5 | 10.4 | GO:1904647 | response to rotenone(GO:1904647) |
| 3.4 | 17.2 | GO:0042816 | vitamin B6 metabolic process(GO:0042816) |
| 3.2 | 38.6 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 2.8 | 19.4 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 2.4 | 9.4 | GO:0006238 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 2.3 | 6.9 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 2.3 | 11.3 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 2.1 | 12.9 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 2.1 | 21.2 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 2.1 | 10.6 | GO:0033490 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 2.0 | 6.0 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 2.0 | 19.7 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 1.9 | 24.8 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 1.7 | 35.9 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 1.7 | 25.5 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 1.7 | 6.7 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 1.6 | 9.5 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 1.6 | 4.7 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 1.4 | 17.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 1.4 | 11.2 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 1.4 | 6.9 | GO:0016128 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) phytosteroid metabolic process(GO:0016128) phytosteroid biosynthetic process(GO:0016129) |
| 1.4 | 5.4 | GO:0032072 | plasmacytoid dendritic cell activation(GO:0002270) T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) regulation of restriction endodeoxyribonuclease activity(GO:0032072) negative regulation of apoptotic cell clearance(GO:2000426) |
| 1.3 | 7.7 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 1.3 | 19.1 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 1.3 | 7.6 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 1.3 | 12.6 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 1.2 | 3.6 | GO:0045659 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 1.2 | 4.7 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 1.2 | 7.0 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 1.2 | 4.6 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 1.2 | 3.5 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 1.1 | 17.7 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 1.1 | 4.4 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 1.1 | 5.5 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 1.1 | 4.3 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 1.1 | 10.8 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 1.1 | 5.3 | GO:0030035 | microspike assembly(GO:0030035) |
| 1.1 | 15.9 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 1.1 | 4.2 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 1.0 | 10.4 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 1.0 | 3.1 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 1.0 | 6.8 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 1.0 | 8.6 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.9 | 2.8 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.9 | 2.7 | GO:0060926 | atrioventricular node development(GO:0003162) cardiac pacemaker cell differentiation(GO:0060920) cardiac pacemaker cell development(GO:0060926) |
| 0.9 | 4.5 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.8 | 5.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.8 | 8.5 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.8 | 3.8 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.7 | 4.4 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.7 | 3.7 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.7 | 2.0 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.7 | 8.6 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.6 | 5.1 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.6 | 15.5 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.6 | 26.9 | GO:2000816 | negative regulation of mitotic sister chromatid separation(GO:2000816) |
| 0.6 | 5.8 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.6 | 2.3 | GO:1903436 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.6 | 4.0 | GO:0009597 | detection of virus(GO:0009597) |
| 0.6 | 3.9 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.6 | 11.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.5 | 2.7 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.5 | 2.0 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.5 | 10.5 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.5 | 6.9 | GO:0090344 | negative regulation of cell aging(GO:0090344) |
| 0.5 | 2.4 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.5 | 6.1 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.5 | 2.3 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.5 | 1.8 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.4 | 4.4 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.4 | 2.8 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.4 | 6.1 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.4 | 4.1 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.4 | 4.8 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.4 | 6.6 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.4 | 9.7 | GO:0046653 | tetrahydrofolate metabolic process(GO:0046653) |
| 0.3 | 3.3 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.3 | 8.8 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.3 | 2.5 | GO:0001845 | phagolysosome assembly(GO:0001845) negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.3 | 55.7 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.3 | 0.9 | GO:0072708 | response to sorbitol(GO:0072708) cellular response to sorbitol(GO:0072709) |
| 0.3 | 3.2 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.3 | 3.5 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.3 | 9.8 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.3 | 2.0 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.3 | 10.6 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.3 | 4.7 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.3 | 2.7 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.3 | 6.3 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.3 | 7.9 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.3 | 9.4 | GO:0046949 | fatty-acyl-CoA biosynthetic process(GO:0046949) |
| 0.3 | 18.6 | GO:0031663 | lipopolysaccharide-mediated signaling pathway(GO:0031663) |
| 0.3 | 0.5 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.3 | 10.1 | GO:0042407 | cristae formation(GO:0042407) |
| 0.3 | 7.8 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.3 | 2.0 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.2 | 1.7 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.2 | 4.5 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.2 | 11.1 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.2 | 3.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.2 | 0.9 | GO:0021814 | cell motility involved in cerebral cortex radial glia guided migration(GO:0021814) negative regulation of glial cell migration(GO:1903976) |
| 0.2 | 3.2 | GO:0060965 | negative regulation of gene silencing by miRNA(GO:0060965) |
| 0.2 | 16.2 | GO:0055092 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.2 | 2.5 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.2 | 1.2 | GO:0009082 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.2 | 2.6 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.2 | 0.8 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.2 | 6.0 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.2 | 0.5 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.2 | 1.1 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.2 | 9.0 | GO:0071549 | cellular response to dexamethasone stimulus(GO:0071549) |
| 0.2 | 4.0 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.2 | 0.9 | GO:0030047 | actin modification(GO:0030047) |
| 0.2 | 6.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.2 | 4.3 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.2 | 6.1 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.2 | 1.3 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.2 | 5.8 | GO:0060706 | cell differentiation involved in embryonic placenta development(GO:0060706) |
| 0.2 | 2.0 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.2 | 3.3 | GO:0002643 | regulation of tolerance induction(GO:0002643) |
| 0.2 | 0.3 | GO:0070256 | negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 0.1 | 1.0 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.1 | 5.6 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.1 | 2.8 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.1 | 2.8 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 1.4 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.1 | 0.8 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.1 | 1.1 | GO:0001764 | neuron migration(GO:0001764) |
| 0.1 | 6.2 | GO:0051983 | regulation of chromosome segregation(GO:0051983) |
| 0.1 | 6.1 | GO:0043388 | positive regulation of DNA binding(GO:0043388) |
| 0.1 | 4.1 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.1 | 1.9 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.1 | 0.6 | GO:0070945 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 0.1 | 3.8 | GO:0097435 | fibril organization(GO:0097435) |
| 0.1 | 0.3 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.1 | 8.1 | GO:0071349 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.1 | 3.5 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.1 | 5.4 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 0.8 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.1 | 1.0 | GO:0006621 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.1 | 8.2 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.1 | 4.3 | GO:0000226 | microtubule cytoskeleton organization(GO:0000226) |
| 0.1 | 0.4 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 1.0 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.1 | 3.1 | GO:1901224 | positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.1 | 5.7 | GO:0070125 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 0.1 | 1.2 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.1 | 0.9 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.1 | 1.6 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.1 | 4.3 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.1 | 0.5 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.1 | 4.3 | GO:0008589 | regulation of smoothened signaling pathway(GO:0008589) |
| 0.1 | 0.4 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.1 | 3.9 | GO:0008542 | visual learning(GO:0008542) |
| 0.1 | 35.2 | GO:0002283 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.1 | 18.8 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.1 | 0.9 | GO:0021799 | cerebral cortex radially oriented cell migration(GO:0021799) |
| 0.1 | 1.8 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.1 | 4.6 | GO:0043647 | inositol phosphate metabolic process(GO:0043647) |
| 0.0 | 0.8 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:2000586 | regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) |
| 0.0 | 1.3 | GO:0010644 | cell communication by electrical coupling(GO:0010644) |
| 0.0 | 1.3 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 4.6 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 1.1 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.0 | 0.3 | GO:0021681 | cerebellar granular layer development(GO:0021681) |
| 0.0 | 0.3 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 3.9 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.0 | 0.6 | GO:0060065 | uterus development(GO:0060065) |
| 0.0 | 1.2 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 1.7 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 | 0.1 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.9 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 2.0 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.6 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.5 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 4.7 | GO:0007264 | small GTPase mediated signal transduction(GO:0007264) |
| 0.0 | 0.3 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 1.2 | GO:1903169 | regulation of calcium ion transmembrane transport(GO:1903169) |
| 0.0 | 0.3 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.0 | 0.0 | GO:0032972 | diaphragm contraction(GO:0002086) regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 0.3 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 1.4 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.9 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 44.0 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 3.2 | 12.9 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 2.7 | 53.2 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 2.7 | 21.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 2.6 | 10.5 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 2.6 | 7.7 | GO:0097447 | dendritic tree(GO:0097447) |
| 2.5 | 17.6 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 2.5 | 17.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 1.8 | 5.3 | GO:0044393 | microspike(GO:0044393) |
| 1.6 | 19.4 | GO:0005642 | annulate lamellae(GO:0005642) |
| 1.6 | 4.7 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 1.5 | 4.5 | GO:0031523 | Myb complex(GO:0031523) |
| 1.4 | 9.5 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 1.2 | 13.7 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 1.2 | 7.2 | GO:1990357 | terminal web(GO:1990357) |
| 1.1 | 16.7 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 1.1 | 5.6 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 1.1 | 25.4 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 1.1 | 4.4 | GO:0005694 | chromosome(GO:0005694) |
| 1.1 | 15.8 | GO:0030478 | actin cap(GO:0030478) |
| 1.0 | 3.0 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.9 | 4.3 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.8 | 6.7 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.8 | 5.5 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.8 | 36.1 | GO:0002102 | podosome(GO:0002102) |
| 0.7 | 7.5 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.7 | 2.8 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.6 | 4.2 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.6 | 11.3 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.6 | 5.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.6 | 16.9 | GO:1990752 | microtubule end(GO:1990752) |
| 0.5 | 2.7 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.5 | 8.6 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.5 | 3.5 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.5 | 7.8 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.4 | 13.7 | GO:0032420 | stereocilium(GO:0032420) |
| 0.4 | 17.3 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.4 | 38.9 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.4 | 175.5 | GO:0005874 | microtubule(GO:0005874) |
| 0.4 | 2.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.4 | 8.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.3 | 5.5 | GO:0010369 | chromocenter(GO:0010369) |
| 0.3 | 28.3 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.3 | 4.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.3 | 4.0 | GO:0042555 | MCM complex(GO:0042555) |
| 0.3 | 4.7 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.2 | 4.8 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.2 | 18.1 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.2 | 4.5 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.2 | 36.5 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.2 | 1.8 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.2 | 4.7 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.2 | 6.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 6.6 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.2 | 4.6 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.2 | 17.3 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.2 | 3.8 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.2 | 10.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.2 | 5.7 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 4.2 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 3.1 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.1 | 4.5 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.1 | 5.7 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 26.6 | GO:0031256 | leading edge membrane(GO:0031256) |
| 0.1 | 3.1 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 1.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 1.7 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 11.0 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.1 | 6.1 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 0.4 | GO:1903349 | omegasome membrane(GO:1903349) |
| 0.1 | 5.7 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.1 | 0.3 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.1 | 0.9 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.1 | 5.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 0.9 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 4.0 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 7.1 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 10.0 | GO:1904813 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.1 | 1.0 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 0.5 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.1 | 3.9 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 9.9 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 2.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 2.8 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.8 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) CNTFR-CLCF1 complex(GO:0097059) |
| 0.0 | 0.9 | GO:0015630 | microtubule cytoskeleton(GO:0015630) |
| 0.0 | 17.2 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 2.5 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 1.5 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 1.2 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 1.4 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.5 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.5 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.0 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 1.4 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 4.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.3 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 2.8 | GO:0012506 | vesicle membrane(GO:0012506) cytoplasmic vesicle membrane(GO:0030659) |
| 0.0 | 4.7 | GO:0005759 | mitochondrial matrix(GO:0005759) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 13.7 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 3.5 | 10.6 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 3.2 | 9.7 | GO:0004487 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 3.1 | 9.4 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 2.6 | 10.5 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 2.3 | 6.9 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 1.8 | 5.3 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 1.8 | 24.8 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 1.7 | 6.9 | GO:0004310 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 1.4 | 11.3 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 1.4 | 11.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 1.4 | 15.0 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 1.3 | 9.4 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 1.3 | 15.9 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 1.3 | 7.7 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 1.3 | 7.6 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 1.1 | 4.5 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 1.1 | 4.5 | GO:0038025 | glycoprotein transporter activity(GO:0034437) reelin receptor activity(GO:0038025) |
| 1.1 | 40.8 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.9 | 5.4 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.9 | 19.5 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.9 | 21.2 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.9 | 7.9 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.9 | 7.8 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.9 | 3.4 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.8 | 4.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.8 | 8.6 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.8 | 21.1 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.8 | 4.6 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.7 | 4.4 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.7 | 33.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.7 | 3.5 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.6 | 17.2 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.6 | 17.7 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.6 | 7.9 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.6 | 28.8 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.6 | 13.2 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.6 | 9.5 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.6 | 12.4 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.6 | 15.8 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.6 | 2.3 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.5 | 1.6 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.5 | 4.7 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.5 | 3.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.5 | 2.0 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.5 | 10.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.5 | 12.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.4 | 4.0 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.4 | 4.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.4 | 6.0 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.4 | 4.6 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.4 | 14.2 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.4 | 2.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.4 | 2.0 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.4 | 3.5 | GO:0015288 | porin activity(GO:0015288) |
| 0.4 | 126.6 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.3 | 17.4 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.3 | 1.0 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.3 | 10.9 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.3 | 4.0 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.3 | 6.1 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.3 | 14.5 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.3 | 12.7 | GO:1990939 | ATP-dependent microtubule motor activity(GO:1990939) |
| 0.2 | 5.3 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.2 | 17.5 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.2 | 99.1 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.2 | 19.7 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.2 | 3.6 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.2 | 0.8 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.2 | 2.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.2 | 4.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.2 | 0.8 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.2 | 1.3 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.2 | 2.8 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.2 | 3.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.2 | 4.7 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.2 | 1.7 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.2 | 0.5 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.2 | 0.5 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.2 | 5.8 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.2 | 6.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 37.4 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.2 | 0.9 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 39.0 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.1 | 11.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 5.6 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 13.3 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 2.0 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 25.2 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 1.0 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 3.1 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 3.5 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 0.3 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) heterotrimeric G-protein binding(GO:0032795) |
| 0.1 | 1.1 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 1.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 10.6 | GO:0005525 | GTP binding(GO:0005525) |
| 0.1 | 2.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 1.0 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 1.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 1.7 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 0.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 1.0 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 1.0 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 7.3 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 3.3 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 6.5 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.1 | 3.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 2.7 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 3.1 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.9 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 2.8 | GO:0019210 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.0 | 3.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.1 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 2.4 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 3.4 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.3 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.1 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.3 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.9 | GO:0004713 | protein tyrosine kinase activity(GO:0004713) |
| 0.0 | 3.1 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 2.4 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 3.2 | GO:0003779 | actin binding(GO:0003779) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 54.4 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.9 | 55.1 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.7 | 25.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.6 | 35.1 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.6 | 61.5 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.5 | 37.8 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.3 | 3.1 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.3 | 7.0 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.3 | 16.0 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.3 | 11.5 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.2 | 5.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 6.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.2 | 7.3 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.2 | 9.5 | PID ATR PATHWAY | ATR signaling pathway |
| 0.2 | 4.7 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 15.0 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.2 | 15.0 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.2 | 4.5 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.2 | 2.6 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.2 | 2.8 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.2 | 7.1 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.2 | 0.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.2 | 2.0 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.2 | 5.8 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 16.3 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 7.4 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 7.6 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.1 | 2.3 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 6.4 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 9.9 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 2.8 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 7.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 2.7 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 6.1 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 5.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 10.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 6.1 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 3.2 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 3.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 3.1 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 0.8 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 3.1 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 1.1 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 1.0 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.8 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 2.4 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.1 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.7 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.9 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.6 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.1 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.3 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.3 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 102.4 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 1.3 | 14.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 1.2 | 37.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 1.0 | 25.5 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 1.0 | 49.0 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.8 | 8.6 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.7 | 19.4 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.7 | 14.4 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.6 | 11.3 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.6 | 10.5 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.6 | 12.9 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.5 | 20.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.5 | 13.1 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.4 | 4.4 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.4 | 3.0 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.4 | 7.7 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.4 | 5.6 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.4 | 17.7 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.4 | 29.4 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.4 | 9.4 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.3 | 36.6 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.3 | 6.0 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.3 | 20.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.3 | 10.8 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.3 | 3.1 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.3 | 8.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.3 | 4.0 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.3 | 5.4 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.3 | 28.0 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF MITOTIC PROTEINS | Genes involved in APC/C:Cdc20 mediated degradation of mitotic proteins |
| 0.3 | 16.4 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.2 | 4.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.2 | 2.7 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.2 | 4.0 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.2 | 4.8 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 1.8 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.2 | 4.7 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.2 | 7.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 2.3 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 2.5 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 6.3 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 7.5 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 4.1 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 3.2 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.1 | 7.0 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 2.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 7.1 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 1.0 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 0.5 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 3.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 0.4 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.1 | 0.9 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.1 | 1.7 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 9.4 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 5.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 4.6 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 1.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 3.5 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.9 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 6.0 | REACTOME METABOLISM OF CARBOHYDRATES | Genes involved in Metabolism of carbohydrates |
| 0.0 | 2.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |