Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
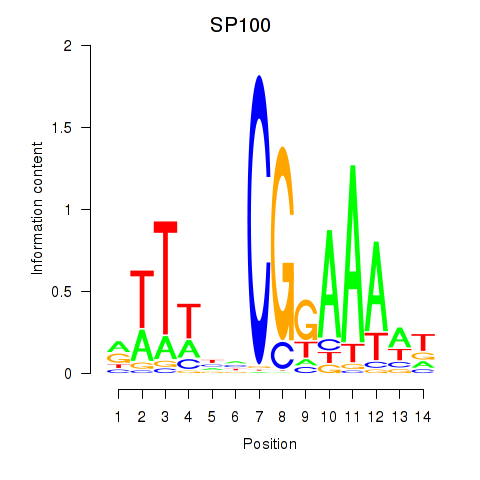
Results for SP100
Z-value: 0.77
Transcription factors associated with SP100
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SP100
|
ENSG00000067066.12 | SP100 nuclear antigen |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SP100 | hg19_v2_chr2_+_231280908_231280945 | 0.30 | 6.8e-06 | Click! |
Activity profile of SP100 motif
Sorted Z-values of SP100 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_137514687 | 33.53 |
ENST00000394894.3
|
KIF20A
|
kinesin family member 20A |
| chr5_+_72143988 | 24.14 |
ENST00000506351.2
|
TNPO1
|
transportin 1 |
| chr12_+_104682496 | 23.25 |
ENST00000378070.4
|
TXNRD1
|
thioredoxin reductase 1 |
| chr15_+_66797627 | 21.78 |
ENST00000565627.1
ENST00000564179.1 |
ZWILCH
|
zwilch kinetochore protein |
| chr15_+_66797455 | 21.78 |
ENST00000446801.2
|
ZWILCH
|
zwilch kinetochore protein |
| chr12_+_27863706 | 20.67 |
ENST00000081029.3
ENST00000538315.1 ENST00000542791.1 |
MRPS35
|
mitochondrial ribosomal protein S35 |
| chr5_+_33440802 | 19.69 |
ENST00000502553.1
ENST00000514259.1 ENST00000265112.3 |
TARS
|
threonyl-tRNA synthetase |
| chr16_-_69760409 | 18.73 |
ENST00000561500.1
ENST00000439109.2 ENST00000564043.1 ENST00000379046.2 ENST00000379047.3 |
NQO1
|
NAD(P)H dehydrogenase, quinone 1 |
| chr7_+_117824210 | 18.06 |
ENST00000422760.1
ENST00000411938.1 |
NAA38
|
N(alpha)-acetyltransferase 38, NatC auxiliary subunit |
| chr9_-_113018835 | 17.86 |
ENST00000374517.5
|
TXN
|
thioredoxin |
| chr5_+_33441053 | 17.69 |
ENST00000541634.1
ENST00000455217.2 ENST00000414361.2 |
TARS
|
threonyl-tRNA synthetase |
| chr11_+_35201826 | 17.53 |
ENST00000531873.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr1_+_214776516 | 16.32 |
ENST00000366955.3
|
CENPF
|
centromere protein F, 350/400kDa |
| chr5_+_159848807 | 16.17 |
ENST00000352433.5
|
PTTG1
|
pituitary tumor-transforming 1 |
| chr12_+_104324112 | 15.80 |
ENST00000299767.5
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1 |
| chr16_+_74330673 | 15.79 |
ENST00000219313.4
ENST00000540379.1 ENST00000567958.1 ENST00000568615.2 |
PSMD7
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 7 |
| chr9_+_33265011 | 15.45 |
ENST00000419016.2
|
CHMP5
|
charged multivesicular body protein 5 |
| chr7_+_117824086 | 15.15 |
ENST00000249299.2
ENST00000424702.1 |
NAA38
|
N(alpha)-acetyltransferase 38, NatC auxiliary subunit |
| chrX_+_77359671 | 14.31 |
ENST00000373316.4
|
PGK1
|
phosphoglycerate kinase 1 |
| chr1_-_87380002 | 14.14 |
ENST00000331835.5
|
SEP15
|
Homo sapiens 15 kDa selenoprotein (SEP15), transcript variant 2, mRNA. |
| chr1_-_63988846 | 14.02 |
ENST00000283568.8
ENST00000371092.3 ENST00000271002.10 |
ITGB3BP
|
integrin beta 3 binding protein (beta3-endonexin) |
| chr14_-_102605983 | 13.92 |
ENST00000334701.7
|
HSP90AA1
|
heat shock protein 90kDa alpha (cytosolic), class A member 1 |
| chr4_-_170679024 | 13.76 |
ENST00000393381.2
|
C4orf27
|
chromosome 4 open reading frame 27 |
| chr5_+_137514834 | 13.70 |
ENST00000508792.1
ENST00000504621.1 |
KIF20A
|
kinesin family member 20A |
| chr14_+_56127989 | 13.61 |
ENST00000555573.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr2_+_39005336 | 13.18 |
ENST00000409566.1
|
GEMIN6
|
gem (nuclear organelle) associated protein 6 |
| chr1_-_87379785 | 13.00 |
ENST00000401030.3
ENST00000370554.1 |
SEP15
|
Homo sapiens 15 kDa selenoprotein (SEP15), transcript variant 2, mRNA. |
| chr11_+_85955787 | 13.00 |
ENST00000528180.1
|
EED
|
embryonic ectoderm development |
| chr2_+_113403434 | 12.76 |
ENST00000272542.3
|
SLC20A1
|
solute carrier family 20 (phosphate transporter), member 1 |
| chrX_+_77359726 | 12.75 |
ENST00000442431.1
|
PGK1
|
phosphoglycerate kinase 1 |
| chr10_+_79793518 | 12.73 |
ENST00000440692.1
ENST00000435275.1 ENST00000372360.3 ENST00000360830.4 |
RPS24
|
ribosomal protein S24 |
| chr2_-_55496344 | 12.58 |
ENST00000403721.1
ENST00000263629.4 |
MTIF2
|
mitochondrial translational initiation factor 2 |
| chr13_+_43597269 | 12.48 |
ENST00000379221.2
|
DNAJC15
|
DnaJ (Hsp40) homolog, subfamily C, member 15 |
| chr2_+_201936707 | 12.45 |
ENST00000433898.1
ENST00000454214.1 |
NDUFB3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3, 12kDa |
| chr12_-_120884175 | 12.39 |
ENST00000546954.1
|
TRIAP1
|
TP53 regulated inhibitor of apoptosis 1 |
| chr3_+_159557637 | 12.38 |
ENST00000445224.2
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr10_-_27443294 | 12.30 |
ENST00000396296.3
ENST00000375972.3 ENST00000376016.3 ENST00000491542.2 |
YME1L1
|
YME1-like 1 ATPase |
| chr1_+_45205498 | 12.27 |
ENST00000372218.4
|
KIF2C
|
kinesin family member 2C |
| chr1_-_108735440 | 12.11 |
ENST00000370041.4
|
SLC25A24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr2_+_201170770 | 12.08 |
ENST00000409988.3
ENST00000409385.1 |
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr17_-_57784755 | 11.94 |
ENST00000537860.1
ENST00000393038.2 ENST00000409433.2 |
PTRH2
|
peptidyl-tRNA hydrolase 2 |
| chr1_-_75198681 | 11.87 |
ENST00000370872.3
ENST00000370871.3 ENST00000340866.5 ENST00000370870.1 |
CRYZ
|
crystallin, zeta (quinone reductase) |
| chr11_+_118958689 | 11.80 |
ENST00000535253.1
ENST00000392841.1 |
HMBS
|
hydroxymethylbilane synthase |
| chr9_-_139372141 | 11.71 |
ENST00000313050.7
|
SEC16A
|
SEC16 homolog A (S. cerevisiae) |
| chr11_-_6704513 | 11.64 |
ENST00000532203.1
ENST00000288937.6 |
MRPL17
|
mitochondrial ribosomal protein L17 |
| chr14_-_102552659 | 11.55 |
ENST00000441629.2
|
HSP90AA1
|
heat shock protein 90kDa alpha (cytosolic), class A member 1 |
| chrX_-_77225135 | 11.38 |
ENST00000458128.1
|
PGAM4
|
phosphoglycerate mutase family member 4 |
| chr3_+_62304648 | 11.34 |
ENST00000462069.1
ENST00000232519.5 ENST00000465142.1 |
C3orf14
|
chromosome 3 open reading frame 14 |
| chr2_+_39005325 | 11.30 |
ENST00000281950.3
|
GEMIN6
|
gem (nuclear organelle) associated protein 6 |
| chr2_+_201170596 | 11.23 |
ENST00000439084.1
ENST00000409718.1 |
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr3_+_62304712 | 11.19 |
ENST00000494481.1
|
C3orf14
|
chromosome 3 open reading frame 14 |
| chr3_+_180630090 | 11.19 |
ENST00000357559.4
ENST00000305586.7 |
FXR1
|
fragile X mental retardation, autosomal homolog 1 |
| chrX_+_118708493 | 11.14 |
ENST00000371558.2
|
UBE2A
|
ubiquitin-conjugating enzyme E2A |
| chr11_+_125496619 | 10.86 |
ENST00000532669.1
ENST00000278916.3 |
CHEK1
|
checkpoint kinase 1 |
| chr11_+_125496400 | 10.77 |
ENST00000524737.1
|
CHEK1
|
checkpoint kinase 1 |
| chr8_+_104310661 | 10.76 |
ENST00000522566.1
|
FZD6
|
frizzled family receptor 6 |
| chr2_+_201994042 | 10.72 |
ENST00000417748.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr6_-_150067696 | 10.62 |
ENST00000340413.2
ENST00000367403.3 |
NUP43
|
nucleoporin 43kDa |
| chr11_-_33744487 | 10.52 |
ENST00000426650.2
|
CD59
|
CD59 molecule, complement regulatory protein |
| chr6_+_31371337 | 10.44 |
ENST00000449934.2
ENST00000421350.1 |
MICA
|
MHC class I polypeptide-related sequence A |
| chr10_-_91174215 | 10.43 |
ENST00000371837.1
|
LIPA
|
lipase A, lysosomal acid, cholesterol esterase |
| chr5_+_32585605 | 10.17 |
ENST00000265073.4
ENST00000515355.1 ENST00000502897.1 ENST00000510442.1 |
SUB1
|
SUB1 homolog (S. cerevisiae) |
| chrX_+_118708517 | 10.05 |
ENST00000346330.3
|
UBE2A
|
ubiquitin-conjugating enzyme E2A |
| chr5_-_68665296 | 10.02 |
ENST00000512152.1
ENST00000503245.1 ENST00000512561.1 ENST00000380822.4 |
TAF9
|
TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa |
| chr5_+_151151504 | 10.00 |
ENST00000356245.3
ENST00000507878.2 |
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr19_-_55919087 | 9.99 |
ENST00000587845.1
ENST00000589978.1 ENST00000264552.9 |
UBE2S
|
ubiquitin-conjugating enzyme E2S |
| chr15_-_66797172 | 9.99 |
ENST00000569438.1
ENST00000569696.1 ENST00000307961.6 |
RPL4
|
ribosomal protein L4 |
| chr1_-_94344754 | 9.92 |
ENST00000436063.2
|
DNTTIP2
|
deoxynucleotidyltransferase, terminal, interacting protein 2 |
| chrX_-_10588459 | 9.88 |
ENST00000380782.2
|
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr11_-_33744256 | 9.83 |
ENST00000415002.2
ENST00000437761.2 ENST00000445143.2 |
CD59
|
CD59 molecule, complement regulatory protein |
| chr5_+_151151471 | 9.82 |
ENST00000394123.3
ENST00000543466.1 |
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr2_+_192109911 | 9.74 |
ENST00000418908.1
ENST00000339514.4 ENST00000392318.3 |
MYO1B
|
myosin IB |
| chrX_-_153707246 | 9.69 |
ENST00000407062.1
|
LAGE3
|
L antigen family, member 3 |
| chr17_+_57784826 | 9.66 |
ENST00000262291.4
|
VMP1
|
vacuole membrane protein 1 |
| chr14_-_23504087 | 9.57 |
ENST00000493471.2
ENST00000460922.2 |
PSMB5
|
proteasome (prosome, macropain) subunit, beta type, 5 |
| chr7_-_91764108 | 9.55 |
ENST00000450723.1
|
CYP51A1
|
cytochrome P450, family 51, subfamily A, polypeptide 1 |
| chr11_-_65626753 | 9.50 |
ENST00000526975.1
ENST00000531413.1 |
CFL1
|
cofilin 1 (non-muscle) |
| chr20_+_47835884 | 9.45 |
ENST00000371764.4
|
DDX27
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 27 |
| chr10_+_45495898 | 9.43 |
ENST00000298299.3
|
ZNF22
|
zinc finger protein 22 |
| chr12_+_95867727 | 9.36 |
ENST00000323666.5
ENST00000546753.1 |
METAP2
|
methionyl aminopeptidase 2 |
| chr17_-_685559 | 9.36 |
ENST00000301329.6
|
GLOD4
|
glyoxalase domain containing 4 |
| chr15_+_59397275 | 9.31 |
ENST00000288207.2
|
CCNB2
|
cyclin B2 |
| chr5_+_172386419 | 9.30 |
ENST00000265100.2
ENST00000519239.1 |
RPL26L1
|
ribosomal protein L26-like 1 |
| chr6_+_44214824 | 9.27 |
ENST00000371646.5
ENST00000353801.3 |
HSP90AB1
|
heat shock protein 90kDa alpha (cytosolic), class B member 1 |
| chr9_-_139343294 | 9.22 |
ENST00000313084.5
|
SEC16A
|
SEC16 homolog A (S. cerevisiae) |
| chr20_-_31989307 | 9.17 |
ENST00000473997.1
ENST00000544843.1 ENST00000346416.2 ENST00000357886.4 ENST00000339269.5 |
CDK5RAP1
|
CDK5 regulatory subunit associated protein 1 |
| chr14_-_54908043 | 9.15 |
ENST00000556113.1
ENST00000553660.1 ENST00000395573.4 ENST00000557690.1 ENST00000216416.4 |
CNIH1
|
cornichon family AMPA receptor auxiliary protein 1 |
| chr11_+_108535752 | 9.14 |
ENST00000322536.3
|
DDX10
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 10 |
| chr4_+_41540160 | 9.09 |
ENST00000503057.1
ENST00000511496.1 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr5_-_68665084 | 9.08 |
ENST00000509462.1
|
TAF9
|
TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa |
| chr2_+_200820269 | 8.96 |
ENST00000392290.1
|
C2orf47
|
chromosome 2 open reading frame 47 |
| chr21_-_30445886 | 8.87 |
ENST00000431234.1
ENST00000540844.1 ENST00000286788.4 |
CCT8
|
chaperonin containing TCP1, subunit 8 (theta) |
| chr9_+_33264861 | 8.79 |
ENST00000223500.8
|
CHMP5
|
charged multivesicular body protein 5 |
| chrX_-_15511438 | 8.73 |
ENST00000380420.5
|
PIR
|
pirin (iron-binding nuclear protein) |
| chr14_-_75530693 | 8.56 |
ENST00000555135.1
ENST00000357971.3 ENST00000553302.1 ENST00000555694.1 ENST00000238618.3 |
ACYP1
|
acylphosphatase 1, erythrocyte (common) type |
| chr13_-_36944307 | 8.55 |
ENST00000355182.4
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome) |
| chr2_+_192110199 | 8.53 |
ENST00000304164.4
|
MYO1B
|
myosin IB |
| chrX_-_10588595 | 8.45 |
ENST00000423614.1
ENST00000317552.4 |
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chrX_+_11129388 | 8.40 |
ENST00000321143.4
ENST00000380763.3 ENST00000380762.4 |
HCCS
|
holocytochrome c synthase |
| chr1_-_6420737 | 8.39 |
ENST00000541130.1
ENST00000377845.3 |
ACOT7
|
acyl-CoA thioesterase 7 |
| chr3_-_64009658 | 8.38 |
ENST00000394431.2
|
PSMD6
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 |
| chr21_+_34602680 | 8.35 |
ENST00000447980.1
|
IFNAR2
|
interferon (alpha, beta and omega) receptor 2 |
| chr1_+_161284047 | 8.34 |
ENST00000367975.2
ENST00000342751.4 ENST00000432287.2 ENST00000392169.2 ENST00000513009.1 |
SDHC
|
succinate dehydrogenase complex, subunit C, integral membrane protein, 15kDa |
| chr6_+_10747986 | 8.26 |
ENST00000379542.5
|
TMEM14B
|
transmembrane protein 14B |
| chr16_-_30441293 | 8.25 |
ENST00000565758.1
ENST00000567983.1 ENST00000319285.4 |
DCTPP1
|
dCTP pyrophosphatase 1 |
| chr20_-_49575058 | 8.07 |
ENST00000371584.4
ENST00000371583.5 ENST00000413082.1 |
DPM1
|
dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit |
| chr12_+_4758264 | 8.05 |
ENST00000266544.5
|
NDUFA9
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9, 39kDa |
| chr1_-_95392635 | 7.88 |
ENST00000538964.1
ENST00000394202.4 ENST00000370206.4 |
CNN3
|
calponin 3, acidic |
| chr15_+_59397298 | 7.83 |
ENST00000559622.1
|
CCNB2
|
cyclin B2 |
| chr5_+_169010638 | 7.82 |
ENST00000265295.4
ENST00000506574.1 ENST00000515224.1 ENST00000508247.1 ENST00000513941.1 |
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr19_+_33072373 | 7.80 |
ENST00000586035.1
|
PDCD5
|
programmed cell death 5 |
| chr21_+_40759684 | 7.79 |
ENST00000380708.1
|
WRB
|
tryptophan rich basic protein |
| chr14_-_23504337 | 7.74 |
ENST00000361611.6
|
PSMB5
|
proteasome (prosome, macropain) subunit, beta type, 5 |
| chr3_-_176914238 | 7.73 |
ENST00000430069.1
ENST00000428970.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr8_+_110552337 | 7.72 |
ENST00000337573.5
|
EBAG9
|
estrogen receptor binding site associated, antigen, 9 |
| chr10_-_74927810 | 7.55 |
ENST00000372979.4
ENST00000430082.2 ENST00000454759.2 ENST00000413026.1 ENST00000453402.1 |
ECD
|
ecdysoneless homolog (Drosophila) |
| chr1_+_153606532 | 7.51 |
ENST00000403433.1
|
CHTOP
|
chromatin target of PRMT1 |
| chr2_-_55496174 | 7.48 |
ENST00000417363.1
ENST00000412530.1 ENST00000394600.3 ENST00000366137.2 ENST00000420637.1 |
MTIF2
|
mitochondrial translational initiation factor 2 |
| chrX_-_20159934 | 7.47 |
ENST00000379593.1
ENST00000379607.5 |
EIF1AX
|
eukaryotic translation initiation factor 1A, X-linked |
| chr12_+_56435637 | 7.44 |
ENST00000356464.5
ENST00000552361.1 |
RPS26
|
ribosomal protein S26 |
| chr9_-_95087604 | 7.42 |
ENST00000542613.1
ENST00000542053.1 ENST00000358855.4 ENST00000545558.1 ENST00000432670.2 ENST00000433029.2 ENST00000411621.2 |
NOL8
|
nucleolar protein 8 |
| chr4_+_41937131 | 7.40 |
ENST00000504986.1
ENST00000508448.1 ENST00000513702.1 ENST00000325094.5 |
TMEM33
|
transmembrane protein 33 |
| chrX_-_77395186 | 7.40 |
ENST00000341864.5
|
TAF9B
|
TAF9B RNA polymerase II, TATA box binding protein (TBP)-associated factor, 31kDa |
| chr17_+_65713925 | 7.35 |
ENST00000253247.4
|
NOL11
|
nucleolar protein 11 |
| chr1_-_193074504 | 7.32 |
ENST00000367439.3
|
GLRX2
|
glutaredoxin 2 |
| chr19_-_46195029 | 7.30 |
ENST00000588599.1
ENST00000585392.1 ENST00000590212.1 ENST00000587367.1 ENST00000391932.3 |
SNRPD2
|
small nuclear ribonucleoprotein D2 polypeptide 16.5kDa |
| chr14_+_51706886 | 7.28 |
ENST00000457354.2
|
TMX1
|
thioredoxin-related transmembrane protein 1 |
| chrX_+_48755183 | 7.28 |
ENST00000376563.1
ENST00000376566.4 |
PQBP1
|
polyglutamine binding protein 1 |
| chr2_+_177134134 | 7.27 |
ENST00000249442.6
ENST00000392529.2 ENST00000443241.1 |
MTX2
|
metaxin 2 |
| chr13_-_31736027 | 7.24 |
ENST00000380406.5
ENST00000320027.5 ENST00000380405.4 |
HSPH1
|
heat shock 105kDa/110kDa protein 1 |
| chr22_+_24309089 | 7.23 |
ENST00000215770.5
|
DDTL
|
D-dopachrome tautomerase-like |
| chr11_-_47447970 | 7.21 |
ENST00000298852.3
ENST00000530912.1 |
PSMC3
|
proteasome (prosome, macropain) 26S subunit, ATPase, 3 |
| chr21_-_43346790 | 7.19 |
ENST00000329623.7
|
C2CD2
|
C2 calcium-dependent domain containing 2 |
| chr8_+_101162812 | 7.17 |
ENST00000353107.3
ENST00000522439.1 |
POLR2K
|
polymerase (RNA) II (DNA directed) polypeptide K, 7.0kDa |
| chr11_-_62439012 | 7.16 |
ENST00000532208.1
ENST00000377954.2 ENST00000415855.2 ENST00000431002.2 ENST00000354588.3 |
C11orf48
|
chromosome 11 open reading frame 48 |
| chrX_+_77154935 | 7.09 |
ENST00000481445.1
|
COX7B
|
cytochrome c oxidase subunit VIIb |
| chr14_-_23504432 | 7.08 |
ENST00000425762.2
|
PSMB5
|
proteasome (prosome, macropain) subunit, beta type, 5 |
| chr8_-_74205851 | 7.04 |
ENST00000396467.1
|
RPL7
|
ribosomal protein L7 |
| chr1_-_165738072 | 7.02 |
ENST00000481278.1
|
TMCO1
|
transmembrane and coiled-coil domains 1 |
| chr2_+_122513109 | 6.99 |
ENST00000389682.3
ENST00000536142.1 |
TSN
|
translin |
| chr19_+_21688366 | 6.95 |
ENST00000358491.4
ENST00000597078.1 |
ZNF429
|
zinc finger protein 429 |
| chr10_+_51187938 | 6.90 |
ENST00000311663.5
|
FAM21D
|
family with sequence similarity 21, member D |
| chr17_-_685493 | 6.77 |
ENST00000536578.1
ENST00000301328.5 ENST00000576419.1 |
GLOD4
|
glyoxalase domain containing 4 |
| chr1_-_236767779 | 6.64 |
ENST00000366579.1
ENST00000366582.3 ENST00000366581.2 |
HEATR1
|
HEAT repeat containing 1 |
| chr20_-_36156293 | 6.63 |
ENST00000373537.2
ENST00000414542.2 |
BLCAP
|
bladder cancer associated protein |
| chrX_-_23926004 | 6.61 |
ENST00000379226.4
ENST00000379220.3 |
APOO
|
apolipoprotein O |
| chr5_+_172386517 | 6.58 |
ENST00000519522.1
|
RPL26L1
|
ribosomal protein L26-like 1 |
| chrX_+_48755202 | 6.55 |
ENST00000447146.2
ENST00000376548.5 ENST00000247140.4 |
PQBP1
|
polyglutamine binding protein 1 |
| chr21_+_40752170 | 6.53 |
ENST00000333781.5
ENST00000541890.1 |
WRB
|
tryptophan rich basic protein |
| chr17_-_30228678 | 6.51 |
ENST00000261708.4
|
UTP6
|
UTP6, small subunit (SSU) processome component, homolog (yeast) |
| chr8_+_110552831 | 6.41 |
ENST00000530629.1
|
EBAG9
|
estrogen receptor binding site associated, antigen, 9 |
| chr13_+_48611665 | 6.40 |
ENST00000258662.2
|
NUDT15
|
nudix (nucleoside diphosphate linked moiety X)-type motif 15 |
| chr2_+_85132749 | 6.25 |
ENST00000233143.4
|
TMSB10
|
thymosin beta 10 |
| chr2_+_74056147 | 6.22 |
ENST00000394070.2
ENST00000536064.1 |
STAMBP
|
STAM binding protein |
| chr1_-_235324772 | 6.21 |
ENST00000408888.3
|
RBM34
|
RNA binding motif protein 34 |
| chr3_-_57583185 | 6.17 |
ENST00000463880.1
|
ARF4
|
ADP-ribosylation factor 4 |
| chr5_+_6714718 | 6.16 |
ENST00000230859.6
ENST00000515721.1 |
PAPD7
|
PAP associated domain containing 7 |
| chr1_-_3566627 | 6.07 |
ENST00000419924.2
ENST00000270708.7 |
WRAP73
|
WD repeat containing, antisense to TP73 |
| chr2_-_70520832 | 6.06 |
ENST00000454893.1
ENST00000272348.2 |
SNRPG
|
small nuclear ribonucleoprotein polypeptide G |
| chr12_-_32908809 | 5.96 |
ENST00000324868.8
|
YARS2
|
tyrosyl-tRNA synthetase 2, mitochondrial |
| chr11_-_77790865 | 5.96 |
ENST00000534029.1
ENST00000525085.1 ENST00000527806.1 ENST00000528164.1 ENST00000528251.1 ENST00000530054.1 |
NDUFC2
NDUFC2-KCTD14
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2, 14.5kDa NDUFC2-KCTD14 readthrough |
| chrX_-_153707545 | 5.90 |
ENST00000357360.4
|
LAGE3
|
L antigen family, member 3 |
| chr14_+_55518349 | 5.89 |
ENST00000395468.4
|
MAPK1IP1L
|
mitogen-activated protein kinase 1 interacting protein 1-like |
| chr15_-_55541227 | 5.88 |
ENST00000566877.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr10_-_96122682 | 5.80 |
ENST00000371361.3
|
NOC3L
|
nucleolar complex associated 3 homolog (S. cerevisiae) |
| chr20_-_36156125 | 5.79 |
ENST00000397135.1
ENST00000397137.1 |
BLCAP
|
bladder cancer associated protein |
| chr11_+_125496124 | 5.78 |
ENST00000533778.2
ENST00000534070.1 |
CHEK1
|
checkpoint kinase 1 |
| chr2_-_47403642 | 5.71 |
ENST00000456319.1
ENST00000409563.1 ENST00000272298.7 |
CALM2
|
calmodulin 2 (phosphorylase kinase, delta) |
| chr18_+_3247779 | 5.63 |
ENST00000578611.1
ENST00000583449.1 |
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr6_-_127664475 | 5.61 |
ENST00000474289.2
ENST00000534442.1 ENST00000368289.2 ENST00000525745.1 ENST00000430841.2 |
ECHDC1
|
enoyl CoA hydratase domain containing 1 |
| chr2_+_201994208 | 5.49 |
ENST00000440180.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr5_+_95998746 | 5.48 |
ENST00000508608.1
|
CAST
|
calpastatin |
| chr3_-_79816965 | 5.45 |
ENST00000464233.1
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr13_+_50656307 | 5.39 |
ENST00000378180.4
|
DLEU1
|
deleted in lymphocytic leukemia 1 (non-protein coding) |
| chr1_-_220220000 | 5.37 |
ENST00000366923.3
|
EPRS
|
glutamyl-prolyl-tRNA synthetase |
| chr15_-_101835414 | 5.32 |
ENST00000254193.6
|
SNRPA1
|
small nuclear ribonucleoprotein polypeptide A' |
| chr15_-_90777277 | 5.28 |
ENST00000328649.6
|
CIB1
|
calcium and integrin binding 1 (calmyrin) |
| chr16_-_18812746 | 5.27 |
ENST00000546206.2
ENST00000562819.1 ENST00000562234.2 ENST00000304414.7 ENST00000567078.2 |
ARL6IP1
RP11-1035H13.3
|
ADP-ribosylation factor-like 6 interacting protein 1 Uncharacterized protein |
| chr22_-_32058166 | 5.25 |
ENST00000435900.1
ENST00000336566.4 |
PISD
|
phosphatidylserine decarboxylase |
| chr2_-_37068530 | 5.23 |
ENST00000593798.1
|
AC007382.1
|
Uncharacterized protein |
| chr10_+_112327425 | 5.21 |
ENST00000361804.4
|
SMC3
|
structural maintenance of chromosomes 3 |
| chr10_-_35379524 | 5.17 |
ENST00000374751.3
ENST00000374742.1 ENST00000602371.1 |
CUL2
|
cullin 2 |
| chr7_-_124569991 | 5.06 |
ENST00000446993.1
ENST00000357628.3 ENST00000393329.1 |
POT1
|
protection of telomeres 1 |
| chr17_-_15587602 | 5.05 |
ENST00000416464.2
ENST00000578237.1 ENST00000581200.1 |
TRIM16
|
tripartite motif containing 16 |
| chr3_-_11685345 | 5.02 |
ENST00000430365.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr10_+_115439282 | 5.01 |
ENST00000369321.2
ENST00000345633.4 |
CASP7
|
caspase 7, apoptosis-related cysteine peptidase |
| chr8_+_59465728 | 4.96 |
ENST00000260130.4
ENST00000422546.2 ENST00000447182.2 ENST00000413219.2 ENST00000424270.2 ENST00000523483.1 ENST00000520168.1 |
SDCBP
|
syndecan binding protein (syntenin) |
| chr8_-_117768023 | 4.92 |
ENST00000518949.1
ENST00000522453.1 ENST00000521861.1 ENST00000518995.1 |
EIF3H
|
eukaryotic translation initiation factor 3, subunit H |
| chr11_-_62607036 | 4.90 |
ENST00000311713.7
ENST00000278856.4 |
WDR74
|
WD repeat domain 74 |
| chr20_-_34252759 | 4.89 |
ENST00000414711.1
ENST00000416778.1 ENST00000397442.1 ENST00000440240.1 ENST00000412056.1 ENST00000352393.4 ENST00000458038.1 ENST00000420363.1 ENST00000434795.1 ENST00000437100.1 ENST00000414664.1 ENST00000359646.1 ENST00000424458.1 ENST00000374104.3 ENST00000374114.3 |
CPNE1
RBM12
|
copine I RNA binding motif protein 12 |
| chr11_-_111741994 | 4.88 |
ENST00000398006.2
|
ALG9
|
ALG9, alpha-1,2-mannosyltransferase |
| chr17_-_62502022 | 4.86 |
ENST00000578804.1
|
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr8_-_59572301 | 4.86 |
ENST00000038176.3
|
NSMAF
|
neutral sphingomyelinase (N-SMase) activation associated factor |
| chr3_+_142720366 | 4.85 |
ENST00000493782.1
ENST00000397933.2 ENST00000473835.2 ENST00000493598.2 |
U2SURP
|
U2 snRNP-associated SURP domain containing |
| chr20_-_49575081 | 4.80 |
ENST00000371588.5
ENST00000371582.4 |
DPM1
|
dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit |
| chrX_-_99891796 | 4.78 |
ENST00000373020.4
|
TSPAN6
|
tetraspanin 6 |
| chr11_+_10772534 | 4.62 |
ENST00000361367.2
|
CTR9
|
CTR9, Paf1/RNA polymerase II complex component |
| chr22_-_29107919 | 4.62 |
ENST00000434810.1
ENST00000456369.1 |
CHEK2
|
checkpoint kinase 2 |
| chr8_-_95487331 | 4.61 |
ENST00000336148.5
|
RAD54B
|
RAD54 homolog B (S. cerevisiae) |
| chr11_+_125495862 | 4.60 |
ENST00000428830.2
ENST00000544373.1 ENST00000527013.1 ENST00000526937.1 ENST00000534685.1 |
CHEK1
|
checkpoint kinase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SP100
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.5 | 37.4 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 6.4 | 25.5 | GO:0043335 | protein unfolding(GO:0043335) |
| 5.3 | 32.0 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 5.1 | 20.3 | GO:0001971 | negative regulation of activation of membrane attack complex(GO:0001971) |
| 4.0 | 71.5 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 3.3 | 10.0 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 2.8 | 11.4 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 2.8 | 8.3 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 2.7 | 10.8 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 2.6 | 10.4 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 2.6 | 54.5 | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 2.6 | 23.3 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 2.6 | 12.9 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 2.5 | 20.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 2.5 | 7.4 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 2.4 | 14.3 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 2.3 | 13.8 | GO:1902957 | negative regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902957) |
| 2.2 | 6.6 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 2.2 | 22.0 | GO:0042262 | DNA protection(GO:0042262) |
| 2.1 | 8.6 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 2.1 | 8.4 | GO:0036116 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 2.0 | 12.3 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 2.0 | 10.0 | GO:0051510 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) regulation of barbed-end actin filament capping(GO:2000812) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 2.0 | 8.0 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 2.0 | 17.9 | GO:0030091 | protein repair(GO:0030091) |
| 1.9 | 7.8 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 1.8 | 9.2 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 1.8 | 5.5 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 1.8 | 26.6 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 1.8 | 5.3 | GO:0038163 | endomitotic cell cycle(GO:0007113) thrombopoietin-mediated signaling pathway(GO:0038163) |
| 1.6 | 13.0 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 1.6 | 14.1 | GO:0051503 | ATP transport(GO:0015867) adenine nucleotide transport(GO:0051503) |
| 1.5 | 4.6 | GO:2001160 | regulation of genetic imprinting(GO:2000653) regulation of histone H3-K79 methylation(GO:2001160) positive regulation of histone H3-K79 methylation(GO:2001162) |
| 1.5 | 4.6 | GO:1903925 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 1.5 | 4.6 | GO:1901420 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) negative regulation of response to alcohol(GO:1901420) |
| 1.5 | 11.9 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 1.5 | 8.9 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 1.5 | 14.6 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 1.4 | 4.3 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 1.4 | 4.3 | GO:0036509 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 1.4 | 18.3 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 1.4 | 4.2 | GO:0006059 | hexitol metabolic process(GO:0006059) response to methylglyoxal(GO:0051595) |
| 1.4 | 16.7 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 1.3 | 12.8 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 1.3 | 12.6 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 1.3 | 16.3 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 1.2 | 3.7 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 1.2 | 5.0 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 1.2 | 12.3 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 1.2 | 3.6 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 1.2 | 3.6 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 1.2 | 18.6 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 1.1 | 4.6 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 1.1 | 3.4 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 1.1 | 3.4 | GO:0034471 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 1.1 | 8.9 | GO:1903750 | positive regulation of MHC class I biosynthetic process(GO:0045345) negative regulation of establishment of protein localization to mitochondrion(GO:1903748) regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 1.1 | 4.4 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 1.1 | 4.2 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 1.0 | 3.1 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 1.0 | 5.2 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 1.0 | 8.1 | GO:0006983 | ER overload response(GO:0006983) |
| 1.0 | 4.0 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 1.0 | 6.0 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 1.0 | 7.8 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.9 | 7.4 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.9 | 11.8 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.9 | 25.1 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.9 | 42.0 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.9 | 4.3 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.9 | 35.9 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.8 | 2.5 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.8 | 3.3 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.8 | 80.5 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.8 | 9.4 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.8 | 20.6 | GO:2000637 | positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.7 | 4.3 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.7 | 2.2 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.7 | 2.9 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.7 | 5.0 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.7 | 4.3 | GO:0032596 | protein transport within lipid bilayer(GO:0032594) protein transport into membrane raft(GO:0032596) |
| 0.7 | 5.5 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.7 | 3.4 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.7 | 8.9 | GO:1904869 | regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.7 | 2.7 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.7 | 2.0 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.7 | 3.9 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.6 | 6.2 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.6 | 21.2 | GO:0033522 | histone H2A ubiquitination(GO:0033522) |
| 0.6 | 6.2 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.6 | 4.3 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.6 | 7.3 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.6 | 4.3 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.6 | 71.9 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.6 | 11.9 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.6 | 7.0 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.6 | 19.8 | GO:0033048 | negative regulation of sister chromatid segregation(GO:0033046) negative regulation of mitotic sister chromatid segregation(GO:0033048) negative regulation of mitotic sister chromatid separation(GO:2000816) |
| 0.6 | 2.3 | GO:0051030 | snRNA export from nucleus(GO:0006408) snRNA transport(GO:0051030) |
| 0.6 | 5.0 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.6 | 6.2 | GO:0031584 | activation of phospholipase D activity(GO:0031584) apical protein localization(GO:0045176) |
| 0.5 | 10.1 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.5 | 17.5 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.5 | 7.7 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.5 | 6.1 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.5 | 4.5 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.5 | 29.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.5 | 3.9 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.5 | 5.7 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.5 | 21.4 | GO:0007077 | mitotic nuclear envelope disassembly(GO:0007077) |
| 0.5 | 36.1 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.5 | 1.8 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.4 | 1.8 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.4 | 11.6 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.4 | 5.5 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.4 | 2.1 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.4 | 4.9 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.4 | 4.0 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.4 | 7.9 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.4 | 2.4 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.4 | 1.2 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.4 | 23.0 | GO:0032508 | DNA duplex unwinding(GO:0032508) |
| 0.4 | 6.4 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.3 | 4.9 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.3 | 8.3 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.3 | 4.0 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.3 | 15.2 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.3 | 3.3 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) |
| 0.3 | 6.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.3 | 12.4 | GO:0030262 | apoptotic nuclear changes(GO:0030262) |
| 0.3 | 2.1 | GO:1990564 | protein ufmylation(GO:0071569) protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.3 | 1.1 | GO:0061643 | chemorepulsion of dopaminergic neuron axon(GO:0036518) chemorepulsion of axon(GO:0061643) |
| 0.3 | 10.4 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.3 | 4.6 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.2 | 2.5 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.2 | 3.7 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.2 | 8.8 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.2 | 4.1 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.2 | 1.7 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.2 | 1.3 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.2 | 1.2 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.2 | 7.4 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.2 | 7.2 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.2 | 1.0 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.2 | 1.9 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.2 | 0.8 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.2 | 0.4 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.2 | 3.1 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.2 | 11.3 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.2 | 2.8 | GO:0018126 | protein hydroxylation(GO:0018126) |
| 0.2 | 15.2 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.2 | 3.7 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.2 | 4.2 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.2 | 6.4 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.2 | 1.2 | GO:0015816 | glycine transport(GO:0015816) |
| 0.2 | 1.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 1.2 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.1 | 5.5 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 7.0 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.1 | 0.8 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.1 | 9.6 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 1.1 | GO:0070374 | positive regulation of ERK1 and ERK2 cascade(GO:0070374) |
| 0.1 | 8.2 | GO:0071349 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.1 | 3.8 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.1 | 6.0 | GO:0043038 | amino acid activation(GO:0043038) tRNA aminoacylation(GO:0043039) |
| 0.1 | 1.8 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.1 | 2.4 | GO:0044764 | multi-organism cellular process(GO:0044764) |
| 0.1 | 1.0 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.1 | 3.3 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.1 | 3.4 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.1 | 3.7 | GO:0045661 | regulation of myoblast differentiation(GO:0045661) |
| 0.1 | 7.8 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.1 | 0.3 | GO:0038169 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.1 | 4.2 | GO:0090114 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) COPII-coated vesicle budding(GO:0090114) |
| 0.1 | 12.8 | GO:0061572 | actin filament bundle assembly(GO:0051017) actin filament bundle organization(GO:0061572) |
| 0.1 | 6.5 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.1 | 0.4 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.5 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.0 | 1.2 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 1.0 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 6.5 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.0 | 4.9 | GO:2000045 | regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
| 0.0 | 1.2 | GO:0000060 | protein import into nucleus, translocation(GO:0000060) |
| 0.0 | 1.2 | GO:0033146 | regulation of intracellular estrogen receptor signaling pathway(GO:0033146) |
| 0.0 | 1.2 | GO:0043086 | negative regulation of catalytic activity(GO:0043086) |
| 0.0 | 2.4 | GO:1990823 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 2.1 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 2.2 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 1.8 | GO:0019827 | stem cell population maintenance(GO:0019827) maintenance of cell number(GO:0098727) |
| 0.0 | 2.3 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.5 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 1.4 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.1 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.9 | GO:0018108 | peptidyl-tyrosine phosphorylation(GO:0018108) |
| 0.0 | 0.3 | GO:0051591 | response to cAMP(GO:0051591) |
| 0.0 | 0.1 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 1.3 | GO:0006626 | protein targeting to mitochondrion(GO:0006626) |
| 0.0 | 0.6 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 1.2 | GO:0050871 | positive regulation of B cell activation(GO:0050871) |
| 0.0 | 0.2 | GO:0052695 | cellular glucuronidation(GO:0052695) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.9 | 43.6 | GO:1990423 | RZZ complex(GO:1990423) |
| 8.3 | 33.2 | GO:0031417 | NatC complex(GO:0031417) |
| 4.7 | 14.0 | GO:0034455 | t-UTP complex(GO:0034455) |
| 4.3 | 12.9 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 4.2 | 21.2 | GO:0033503 | HULC complex(GO:0033503) |
| 3.5 | 13.8 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 3.1 | 15.6 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 3.1 | 24.5 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 2.5 | 17.5 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 2.4 | 19.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 2.4 | 28.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 2.3 | 9.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 2.1 | 8.3 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 1.8 | 17.6 | GO:0034719 | SMN-Sm protein complex(GO:0034719) |
| 1.7 | 5.2 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 1.7 | 27.2 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 1.3 | 6.5 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 1.3 | 13.0 | GO:0001739 | sex chromatin(GO:0001739) |
| 1.3 | 20.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 1.1 | 34.5 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 1.1 | 62.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 1.1 | 4.2 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 1.0 | 5.2 | GO:0030891 | VCB complex(GO:0030891) |
| 0.9 | 7.5 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.9 | 6.6 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.9 | 2.8 | GO:1903349 | omegasome membrane(GO:1903349) |
| 0.9 | 5.5 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.9 | 5.3 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.8 | 15.8 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.8 | 14.6 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.8 | 2.4 | GO:0031523 | Myb complex(GO:0031523) |
| 0.8 | 4.7 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.8 | 5.4 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.8 | 20.7 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.7 | 4.5 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.7 | 8.9 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.7 | 4.3 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.7 | 4.3 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.7 | 10.6 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.7 | 8.9 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.7 | 7.4 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.7 | 4.6 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.6 | 4.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.6 | 4.3 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.6 | 4.3 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.6 | 6.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.5 | 4.9 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.5 | 3.8 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.5 | 3.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.5 | 6.1 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.5 | 5.1 | GO:0070187 | telosome(GO:0070187) |
| 0.5 | 3.5 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.5 | 7.1 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.5 | 13.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.5 | 31.0 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.5 | 4.6 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.5 | 4.1 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.4 | 11.4 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.4 | 9.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.4 | 9.4 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.4 | 3.3 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.4 | 7.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.4 | 31.7 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.4 | 4.0 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.4 | 5.3 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.4 | 37.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.3 | 22.9 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.3 | 5.0 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.3 | 5.3 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.3 | 18.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.3 | 10.8 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.3 | 10.0 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.3 | 18.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.3 | 9.2 | GO:0005844 | polysome(GO:0005844) |
| 0.3 | 7.7 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.2 | 7.8 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.2 | 36.7 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.2 | 14.0 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.2 | 4.9 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.2 | 2.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 4.4 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.2 | 1.2 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.2 | 17.8 | GO:0030175 | filopodium(GO:0030175) |
| 0.2 | 1.7 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.2 | 1.8 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.2 | 2.9 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.2 | 2.0 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 7.7 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.1 | 21.4 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 7.1 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 20.9 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.1 | 6.2 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 1.6 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 1.0 | GO:0016011 | dystroglycan complex(GO:0016011) sarcoglycan complex(GO:0016012) |
| 0.1 | 2.3 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 2.2 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 1.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 1.1 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 9.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 4.9 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 9.7 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 7.2 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 3.0 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 4.3 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 25.9 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 4.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.6 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 1.0 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 2.5 | GO:0043657 | host(GO:0018995) host cell(GO:0043657) |
| 0.1 | 41.4 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 3.0 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 2.4 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 3.5 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 2.2 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 12.8 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 1.2 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 9.3 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 1.2 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 1.7 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 1.5 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 17.3 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 24.4 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.4 | GO:0031526 | brush border membrane(GO:0031526) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.5 | 37.4 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 9.0 | 27.1 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 7.8 | 23.3 | GO:0098626 | methylselenol reductase activity(GO:0098625) methylseleninic acid reductase activity(GO:0098626) |
| 5.3 | 32.0 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 4.3 | 12.9 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 4.0 | 11.9 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 3.6 | 17.9 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 3.5 | 34.7 | GO:0030911 | TPR domain binding(GO:0030911) |
| 3.3 | 13.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 3.2 | 12.8 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 2.8 | 11.4 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 2.6 | 7.8 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 2.4 | 11.9 | GO:0070404 | NADH binding(GO:0070404) |
| 2.2 | 11.2 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 2.1 | 6.4 | GO:0008413 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) NADH pyrophosphatase activity(GO:0035529) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 2.1 | 10.4 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 2.1 | 8.3 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 2.1 | 18.7 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 1.9 | 5.6 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 1.7 | 5.2 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 1.7 | 5.0 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 1.7 | 8.3 | GO:0004905 | type I interferon receptor activity(GO:0004905) |
| 1.7 | 5.0 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 1.6 | 4.9 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 1.6 | 9.4 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 1.5 | 12.3 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 1.5 | 4.6 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 1.5 | 12.1 | GO:0015217 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine ribonucleotide transmembrane transporter activity(GO:0005346) ATP transmembrane transporter activity(GO:0005347) purine nucleotide transmembrane transporter activity(GO:0015216) ADP transmembrane transporter activity(GO:0015217) |
| 1.5 | 7.3 | GO:0030613 | oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) |
| 1.4 | 13.0 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 1.4 | 20.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 1.4 | 8.6 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 1.4 | 4.3 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 1.3 | 5.2 | GO:0036033 | mediator complex binding(GO:0036033) |
| 1.3 | 14.0 | GO:0046790 | virion binding(GO:0046790) |
| 1.2 | 12.3 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 1.2 | 3.6 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 1.2 | 8.3 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 1.0 | 6.2 | GO:0046979 | TAP2 binding(GO:0046979) |
| 1.0 | 14.3 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 1.0 | 9.5 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.9 | 8.5 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.9 | 10.1 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.9 | 5.5 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.9 | 5.5 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.9 | 27.2 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.9 | 5.3 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.8 | 8.4 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.8 | 3.3 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.8 | 18.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.8 | 6.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.8 | 8.4 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.8 | 17.4 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.7 | 31.2 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.7 | 21.4 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.6 | 14.1 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.6 | 24.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.6 | 6.9 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.6 | 2.4 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.6 | 7.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.6 | 2.4 | GO:0016972 | flavin-linked sulfhydryl oxidase activity(GO:0016971) thiol oxidase activity(GO:0016972) |
| 0.6 | 47.0 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.6 | 18.4 | GO:0001848 | complement binding(GO:0001848) |
| 0.6 | 29.7 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.5 | 22.3 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.5 | 4.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.5 | 3.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.5 | 15.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.5 | 11.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.5 | 3.9 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.5 | 4.2 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.4 | 7.3 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.4 | 7.2 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.4 | 4.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.4 | 7.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.4 | 4.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.4 | 1.8 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.3 | 1.0 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.3 | 8.9 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.3 | 2.0 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.3 | 72.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.3 | 23.3 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.3 | 3.3 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.3 | 10.3 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.3 | 7.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.3 | 7.9 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.3 | 4.1 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.3 | 4.6 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.3 | 17.1 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.3 | 8.5 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.3 | 11.0 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.3 | 8.9 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.2 | 24.6 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.2 | 11.3 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.2 | 6.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 0.7 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.2 | 16.2 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.2 | 2.5 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.2 | 0.6 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.2 | 1.2 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.2 | 1.8 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.2 | 18.2 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.2 | 4.9 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.2 | 7.8 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.2 | 2.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.2 | 1.2 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.2 | 7.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 2.8 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.2 | 18.3 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.2 | 0.5 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.2 | 9.2 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.2 | 3.7 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.2 | 3.9 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.2 | 3.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.2 | 4.3 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.2 | 5.7 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.1 | 0.7 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.1 | 3.9 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 2.1 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 5.4 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 1.2 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.1 | 1.3 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 10.9 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 6.1 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.1 | 1.7 | GO:0005536 | glucose binding(GO:0005536) |
| 0.1 | 0.4 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 1.3 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 2.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 7.5 | GO:1902936 | phosphatidylinositol bisphosphate binding(GO:1902936) |
| 0.1 | 6.2 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.1 | 1.6 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.1 | 0.8 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 0.7 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 0.8 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 1.2 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 1.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.6 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.6 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.5 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 1.2 | GO:0019783 | ubiquitin-like protein-specific protease activity(GO:0019783) |
| 0.0 | 1.5 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 3.7 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 3.4 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 4.4 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 4.6 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.1 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 4.3 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 39.4 | GO:0003723 | RNA binding(GO:0003723) |
| 0.0 | 6.3 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 2.5 | GO:0005496 | steroid binding(GO:0005496) |
| 0.0 | 6.3 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 1.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.1 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.9 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 2.9 | GO:0016887 | ATPase activity(GO:0016887) |
| 0.0 | 1.3 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 1.0 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.8 | GO:0047485 | protein N-terminus binding(GO:0047485) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 32.0 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 1.0 | 63.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.8 | 24.6 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.7 | 25.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.5 | 17.5 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.5 | 24.9 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.5 | 23.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.4 | 22.3 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.4 | 5.0 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.4 | 11.9 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.3 | 13.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.3 | 5.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.2 | 16.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.2 | 5.0 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.2 | 5.2 | PID ATM PATHWAY | ATM pathway |
| 0.2 | 26.3 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.2 | 15.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.2 | 10.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.2 | 4.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.2 | 5.0 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 12.0 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 5.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 9.3 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 1.7 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 7.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 1.8 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 11.6 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 5.0 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 3.0 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 4.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 2.7 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 2.0 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 6.2 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 4.0 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 2.4 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 2.6 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.0 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.1 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 59.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 1.9 | 36.6 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 1.4 | 44.8 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 1.3 | 24.1 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 1.2 | 41.1 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 1.2 | 42.7 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 1.1 | 15.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 1.0 | 80.5 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 1.0 | 24.2 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 1.0 | 17.1 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.9 | 17.5 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.9 | 59.2 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.8 | 69.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.8 | 12.8 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.7 | 16.2 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF MITOTIC PROTEINS | Genes involved in APC/C:Cdc20 mediated degradation of mitotic proteins |
| 0.6 | 6.2 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.6 | 27.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.6 | 47.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.5 | 9.7 | REACTOME PROCESSIVE SYNTHESIS ON THE LAGGING STRAND | Genes involved in Processive synthesis on the lagging strand |
| 0.5 | 14.6 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.5 | 37.0 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.5 | 4.9 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.4 | 46.6 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.4 | 11.8 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.4 | 9.5 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.3 | 11.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.3 | 20.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.3 | 6.0 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.3 | 3.8 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.3 | 7.2 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.3 | 7.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.3 | 5.5 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.3 | 4.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.3 | 7.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.3 | 5.7 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.2 | 4.0 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.2 | 8.1 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.2 | 1.8 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.2 | 6.9 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.2 | 4.1 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.2 | 36.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.2 | 4.2 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.2 | 8.7 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.2 | 1.8 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.2 | 5.0 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 3.1 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 3.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.7 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 2.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 2.0 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 2.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 3.3 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 9.4 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 4.3 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.1 | 9.6 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 2.1 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 1.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 1.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 2.1 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 5.5 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 3.4 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 3.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |