Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for TBP
Z-value: 0.55
Transcription factors associated with TBP
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBP
|
ENSG00000112592.8 | TATA-box binding protein |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TBP | hg19_v2_chr6_+_170863421_170863484 | -0.35 | 1.2e-07 | Click! |
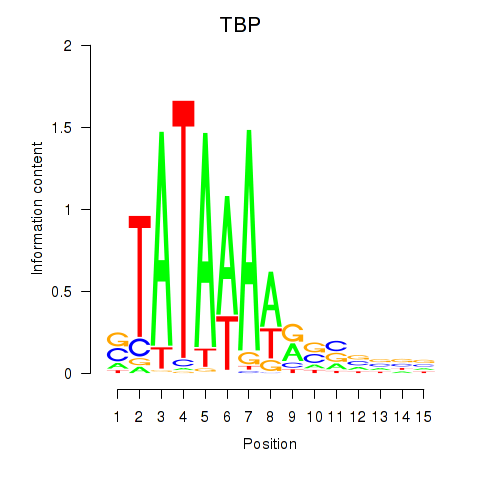
Activity profile of TBP motif
Sorted Z-values of TBP motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TBP
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 16.3 | 48.9 | GO:0001798 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 14.2 | 56.9 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 12.5 | 49.8 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 11.5 | 57.6 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 9.0 | 26.9 | GO:0048611 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 8.9 | 8.9 | GO:0003129 | heart induction(GO:0003129) |
| 8.5 | 59.2 | GO:0032119 | sequestering of zinc ion(GO:0032119) |
| 7.7 | 107.5 | GO:0015671 | oxygen transport(GO:0015671) |
| 7.5 | 45.3 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 7.4 | 14.7 | GO:0045168 | cell-cell signaling involved in cell fate commitment(GO:0045168) |
| 7.2 | 28.7 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 6.9 | 27.5 | GO:1900154 | regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 6.6 | 52.6 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) |
| 6.5 | 25.9 | GO:0070426 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070426) positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070434) |
| 5.8 | 163.3 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 5.7 | 17.0 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) |
| 5.6 | 16.7 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 5.4 | 32.2 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 5.3 | 15.8 | GO:0070295 | renal water absorption(GO:0070295) |
| 5.2 | 15.7 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 5.0 | 20.2 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) |
| 4.9 | 19.7 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 4.9 | 24.4 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 4.9 | 43.9 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 4.8 | 4.8 | GO:0086073 | cardiac muscle cell-cardiac muscle cell adhesion(GO:0086042) bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 4.4 | 52.4 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 4.2 | 12.5 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 4.1 | 24.8 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) |
| 4.1 | 20.6 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 4.1 | 16.4 | GO:0044010 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 4.1 | 12.2 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 4.0 | 12.0 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 3.8 | 11.3 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 3.7 | 18.5 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) cellular response to vitamin E(GO:0071306) response to fluoride(GO:1902617) |
| 3.6 | 18.2 | GO:0003431 | growth plate cartilage chondrocyte growth(GO:0003430) growth plate cartilage chondrocyte development(GO:0003431) Harderian gland development(GO:0070384) |
| 3.6 | 10.7 | GO:0021571 | rhombomere 5 development(GO:0021571) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 3.5 | 13.9 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 3.4 | 10.3 | GO:0018153 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 3.3 | 9.8 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 3.3 | 16.4 | GO:1903906 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 3.3 | 16.3 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 3.2 | 9.7 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 3.0 | 9.1 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 3.0 | 9.1 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 2.9 | 17.7 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 2.7 | 8.2 | GO:1990869 | nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) negative regulation by host of viral process(GO:0044793) negative regulation by host of viral genome replication(GO:0044828) response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) |
| 2.7 | 8.1 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 2.7 | 8.1 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 2.6 | 7.9 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 2.6 | 15.6 | GO:0034371 | chylomicron remodeling(GO:0034371) |
| 2.5 | 7.6 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 2.5 | 7.5 | GO:2000547 | dendritic cell dendrite assembly(GO:0097026) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 2.5 | 15.0 | GO:0042713 | sperm ejaculation(GO:0042713) |
| 2.4 | 65.9 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 2.3 | 11.7 | GO:0060356 | leucine import(GO:0060356) |
| 2.3 | 18.5 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 2.2 | 15.6 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 2.2 | 17.5 | GO:1904469 | positive regulation of tumor necrosis factor secretion(GO:1904469) |
| 2.2 | 6.5 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 2.1 | 6.4 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 2.0 | 8.1 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 2.0 | 13.9 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 2.0 | 29.5 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 1.9 | 19.3 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 1.9 | 20.7 | GO:0000050 | urea cycle(GO:0000050) |
| 1.9 | 11.3 | GO:0050666 | regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 1.9 | 5.6 | GO:0043163 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 1.9 | 7.5 | GO:0002434 | immune complex clearance(GO:0002434) immune complex clearance by monocytes and macrophages(GO:0002436) response to high density lipoprotein particle(GO:0055099) T cell extravasation(GO:0072683) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) |
| 1.9 | 3.7 | GO:0051795 | positive regulation of catagen(GO:0051795) |
| 1.9 | 61.2 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 1.8 | 12.8 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 1.8 | 14.3 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 1.8 | 5.3 | GO:1904437 | antigen processing and presentation of peptide antigen via MHC class Ib(GO:0002428) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) |
| 1.8 | 5.3 | GO:0051039 | histone displacement(GO:0001207) regulation of transcription involved in meiotic cell cycle(GO:0051037) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 1.7 | 7.0 | GO:0002725 | negative regulation of T cell cytokine production(GO:0002725) |
| 1.7 | 5.2 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 1.7 | 5.1 | GO:0001694 | histamine biosynthetic process(GO:0001694) |
| 1.7 | 3.3 | GO:0035106 | operant conditioning(GO:0035106) |
| 1.6 | 6.6 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 1.6 | 6.5 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 1.6 | 9.7 | GO:1903301 | fructose 2,6-bisphosphate metabolic process(GO:0006003) positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 1.6 | 9.6 | GO:0060613 | fat pad development(GO:0060613) |
| 1.6 | 74.1 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 1.5 | 10.6 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 1.5 | 4.6 | GO:0034308 | primary alcohol metabolic process(GO:0034308) |
| 1.5 | 4.6 | GO:2000510 | positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 1.5 | 9.1 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 1.5 | 5.9 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 1.4 | 5.7 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 1.4 | 5.7 | GO:0035962 | response to interleukin-13(GO:0035962) |
| 1.4 | 16.9 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 1.4 | 34.9 | GO:0001502 | cartilage condensation(GO:0001502) |
| 1.4 | 2.8 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 1.4 | 31.4 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) positive regulation of neutrophil migration(GO:1902624) |
| 1.3 | 6.6 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 1.3 | 11.8 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 1.3 | 10.5 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 1.3 | 5.2 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 1.3 | 8.9 | GO:0019532 | oxalate transport(GO:0019532) |
| 1.3 | 22.8 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 1.3 | 3.8 | GO:0035993 | subthalamic nucleus development(GO:0021763) deltoid tuberosity development(GO:0035993) prolactin secreting cell differentiation(GO:0060127) left lung morphogenesis(GO:0060460) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) cell proliferation involved in outflow tract morphogenesis(GO:0061325) |
| 1.2 | 18.7 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 1.2 | 12.4 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 1.2 | 4.9 | GO:0002215 | defense response to nematode(GO:0002215) |
| 1.2 | 4.8 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 1.2 | 7.2 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 1.2 | 4.7 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 1.2 | 10.5 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 1.2 | 9.2 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 1.1 | 4.6 | GO:0035425 | autocrine signaling(GO:0035425) |
| 1.1 | 4.4 | GO:0019627 | urea metabolic process(GO:0019627) |
| 1.0 | 4.1 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 1.0 | 21.0 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 1.0 | 32.9 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 1.0 | 1.9 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.9 | 20.9 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.9 | 4.7 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.9 | 5.6 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.9 | 7.5 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.9 | 12.2 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.9 | 20.2 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.9 | 9.1 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.9 | 76.8 | GO:0070268 | cornification(GO:0070268) |
| 0.9 | 8.7 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.8 | 8.4 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.8 | 4.8 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.8 | 23.0 | GO:0050716 | positive regulation of interleukin-1 secretion(GO:0050716) |
| 0.7 | 8.2 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.7 | 5.2 | GO:0032097 | positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) |
| 0.7 | 10.4 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.7 | 2.8 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.7 | 5.3 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.6 | 4.5 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.6 | 2.6 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.6 | 5.0 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.6 | 17.9 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.6 | 2.4 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.6 | 5.8 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.6 | 12.3 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.6 | 1.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.5 | 10.4 | GO:0031424 | keratinization(GO:0031424) |
| 0.5 | 5.8 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.5 | 6.6 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.5 | 19.0 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.5 | 2.5 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.5 | 5.3 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.5 | 6.1 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.5 | 5.4 | GO:0030728 | ovulation(GO:0030728) |
| 0.4 | 3.4 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) |
| 0.4 | 2.5 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.4 | 33.9 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.4 | 2.6 | GO:0035934 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 0.4 | 4.4 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.4 | 2.9 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.4 | 1.8 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.4 | 12.9 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.3 | 6.8 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.3 | 1.0 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.3 | 19.5 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.3 | 2.0 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.3 | 0.7 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.3 | 8.4 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.3 | 8.2 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.3 | 1.8 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.3 | 1.4 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.3 | 6.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.3 | 8.7 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.3 | 7.1 | GO:0034694 | response to prostaglandin(GO:0034694) |
| 0.3 | 1.0 | GO:0035356 | cellular triglyceride homeostasis(GO:0035356) negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.3 | 7.8 | GO:1903963 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.3 | 2.0 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.3 | 4.6 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.2 | 28.1 | GO:0007586 | digestion(GO:0007586) |
| 0.2 | 10.2 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.2 | 11.7 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.2 | 10.4 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.2 | 4.0 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.2 | 6.0 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.2 | 0.6 | GO:0042421 | norepinephrine biosynthetic process(GO:0042421) |
| 0.2 | 4.8 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.2 | 1.9 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.2 | 11.7 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.2 | 2.2 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.2 | 5.3 | GO:0006875 | cellular metal ion homeostasis(GO:0006875) |
| 0.2 | 2.9 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.2 | 4.2 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) |
| 0.2 | 1.0 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.2 | 4.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 5.1 | GO:0042269 | regulation of natural killer cell mediated cytotoxicity(GO:0042269) |
| 0.2 | 1.4 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.2 | 3.9 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.2 | 2.3 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.1 | 1.2 | GO:0061000 | negative regulation of dendritic spine development(GO:0061000) |
| 0.1 | 3.6 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.1 | 0.9 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.1 | 4.2 | GO:0019730 | antimicrobial humoral response(GO:0019730) |
| 0.1 | 3.7 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 0.7 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.1 | 16.4 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.1 | 3.3 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.1 | 4.1 | GO:0007140 | male meiosis(GO:0007140) |
| 0.1 | 5.0 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.1 | 4.6 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 1.0 | GO:0072678 | T cell migration(GO:0072678) |
| 0.1 | 3.4 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.1 | 3.0 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 13.5 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 8.3 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.1 | 3.4 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) |
| 0.1 | 2.4 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.1 | 8.5 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 0.9 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.1 | 1.2 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 3.2 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 6.6 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 1.3 | GO:0036258 | multivesicular body assembly(GO:0036258) |
| 0.0 | 4.6 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 1.5 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.3 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 0.0 | 2.2 | GO:0034620 | cellular response to unfolded protein(GO:0034620) |
| 0.0 | 1.7 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 3.9 | GO:0001837 | epithelial to mesenchymal transition(GO:0001837) |
| 0.0 | 1.3 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 2.1 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.0 | 0.6 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 2.7 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 3.5 | GO:0043542 | endothelial cell migration(GO:0043542) |
| 0.0 | 6.1 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 1.1 | GO:0019233 | sensory perception of pain(GO:0019233) |
| 0.0 | 2.6 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.0 | 0.6 | GO:0015813 | acidic amino acid transport(GO:0015800) L-glutamate transport(GO:0015813) |
| 0.0 | 0.1 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.0 | 2.0 | GO:0034332 | adherens junction organization(GO:0034332) |
| 0.0 | 1.7 | GO:0050906 | detection of stimulus involved in sensory perception(GO:0050906) |
| 0.0 | 0.7 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.0 | 0.4 | GO:0032410 | negative regulation of transporter activity(GO:0032410) |
| 0.0 | 0.1 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 1.5 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 3.0 | GO:0051480 | regulation of cytosolic calcium ion concentration(GO:0051480) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.7 | 38.1 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 12.5 | 49.8 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 9.0 | 107.5 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 6.7 | 26.9 | GO:1990745 | EARP complex(GO:1990745) |
| 6.1 | 24.4 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 3.7 | 52.4 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 3.3 | 9.8 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 2.7 | 52.2 | GO:0097386 | glial cell projection(GO:0097386) |
| 2.6 | 15.6 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 2.5 | 7.6 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 2.1 | 22.8 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 2.1 | 18.5 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 2.0 | 12.2 | GO:1990037 | Lewy body core(GO:1990037) |
| 2.0 | 112.3 | GO:0000786 | nucleosome(GO:0000786) |
| 2.0 | 17.6 | GO:0005579 | membrane attack complex(GO:0005579) |
| 1.8 | 82.7 | GO:0045095 | keratin filament(GO:0045095) |
| 1.8 | 14.3 | GO:0035976 | AP1 complex(GO:0035976) |
| 1.6 | 151.7 | GO:0035580 | specific granule lumen(GO:0035580) |
| 1.6 | 27.5 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 1.5 | 10.7 | GO:0072687 | meiotic spindle(GO:0072687) |
| 1.5 | 76.2 | GO:0001533 | cornified envelope(GO:0001533) |
| 1.4 | 16.4 | GO:0036038 | MKS complex(GO:0036038) |
| 1.3 | 22.3 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 1.3 | 57.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 1.2 | 23.3 | GO:0097225 | sperm midpiece(GO:0097225) |
| 1.2 | 12.0 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 1.2 | 10.4 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 1.1 | 16.4 | GO:0030478 | actin cap(GO:0030478) |
| 0.9 | 6.5 | GO:0001939 | female pronucleus(GO:0001939) |
| 0.9 | 7.0 | GO:0045179 | apical cortex(GO:0045179) |
| 0.9 | 5.2 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.8 | 4.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.8 | 8.9 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.8 | 10.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.7 | 2.2 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.7 | 54.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.7 | 29.9 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.7 | 47.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.7 | 15.7 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.6 | 15.1 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.6 | 16.7 | GO:0032982 | myosin filament(GO:0032982) |
| 0.6 | 8.5 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.6 | 115.4 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.6 | 10.6 | GO:0042599 | lamellar body(GO:0042599) |
| 0.6 | 4.4 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.5 | 29.1 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.5 | 19.2 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.5 | 16.8 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.5 | 4.8 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.5 | 5.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.5 | 12.0 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.5 | 1.4 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.4 | 1.3 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.4 | 5.6 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.4 | 12.5 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.4 | 2.9 | GO:0030057 | desmosome(GO:0030057) |
| 0.4 | 15.8 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.4 | 25.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.4 | 5.9 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.3 | 130.0 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.3 | 3.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.3 | 351.6 | GO:0005615 | extracellular space(GO:0005615) |
| 0.3 | 9.7 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.2 | 2.0 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 2.6 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.2 | 12.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.2 | 5.7 | GO:0031430 | M band(GO:0031430) |
| 0.2 | 10.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.2 | 9.5 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.2 | 4.3 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.2 | 112.0 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.1 | 1.0 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.1 | 3.0 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 4.0 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 5.8 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 2.1 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 7.9 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 3.9 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 6.6 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 3.4 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 1.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.6 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 1.5 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 5.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.9 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 18.1 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 7.4 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 1.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.7 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.9 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.2 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 19.7 | 59.2 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 14.2 | 56.9 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 12.7 | 38.1 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 12.1 | 157.4 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 9.6 | 28.7 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 5.6 | 16.7 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 5.0 | 20.2 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 4.0 | 32.2 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 4.0 | 7.9 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) indanol dehydrogenase activity(GO:0047718) |
| 4.0 | 15.8 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 3.9 | 15.6 | GO:0035473 | lipase binding(GO:0035473) |
| 3.8 | 11.3 | GO:0047150 | betaine-homocysteine S-methyltransferase activity(GO:0047150) |
| 3.4 | 16.9 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 3.1 | 24.6 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 2.7 | 8.1 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 2.6 | 10.4 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 2.5 | 7.5 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 2.5 | 7.5 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 2.5 | 12.4 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 2.4 | 25.9 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 2.2 | 6.5 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 2.0 | 31.4 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 2.0 | 5.9 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 1.6 | 9.7 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 1.6 | 8.0 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 1.5 | 12.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 1.5 | 29.0 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 1.5 | 9.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 1.5 | 8.9 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 1.5 | 5.9 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 1.4 | 18.5 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 1.4 | 5.7 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 1.4 | 8.3 | GO:0045569 | TRAIL binding(GO:0045569) |
| 1.4 | 16.4 | GO:0045159 | myosin II binding(GO:0045159) |
| 1.3 | 22.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 1.3 | 6.3 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 1.2 | 158.5 | GO:0005179 | hormone activity(GO:0005179) |
| 1.2 | 4.9 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 1.2 | 8.5 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 1.2 | 86.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 1.2 | 11.7 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 1.2 | 5.8 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 1.1 | 8.9 | GO:0039706 | co-receptor binding(GO:0039706) |
| 1.1 | 12.0 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 1.1 | 9.7 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 1.1 | 19.3 | GO:0019864 | IgG binding(GO:0019864) |
| 1.1 | 13.8 | GO:0015250 | water channel activity(GO:0015250) |
| 1.0 | 28.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 1.0 | 10.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.9 | 6.6 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.9 | 17.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.9 | 32.9 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.9 | 20.3 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.9 | 30.0 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.9 | 7.1 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.8 | 17.0 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.8 | 5.6 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.8 | 4.6 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.7 | 44.0 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.7 | 8.2 | GO:0008061 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 0.7 | 19.3 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.7 | 10.0 | GO:0031432 | titin binding(GO:0031432) |
| 0.7 | 12.8 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.7 | 5.6 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.7 | 58.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.6 | 10.3 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.6 | 7.0 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.6 | 10.5 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.6 | 1.8 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.6 | 96.4 | GO:0002020 | protease binding(GO:0002020) |
| 0.6 | 47.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.6 | 8.7 | GO:0016918 | retinal binding(GO:0016918) |
| 0.6 | 3.4 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.6 | 15.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.5 | 4.8 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.5 | 11.8 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.5 | 38.7 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.5 | 3.4 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.5 | 6.3 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.5 | 7.7 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.5 | 8.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.5 | 2.9 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.5 | 10.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.4 | 4.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.4 | 12.7 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.4 | 13.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.4 | 5.2 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.4 | 3.3 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.4 | 3.5 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.4 | 2.3 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.4 | 4.2 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.4 | 7.4 | GO:0016208 | AMP binding(GO:0016208) |
| 0.4 | 6.6 | GO:0010857 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) calcium-dependent protein kinase activity(GO:0010857) |
| 0.4 | 1.8 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.4 | 3.9 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.3 | 8.7 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.3 | 6.8 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.3 | 2.0 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.3 | 19.3 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.3 | 2.5 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.3 | 1.2 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.3 | 2.4 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.3 | 0.6 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.3 | 31.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.3 | 3.7 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.3 | 1.1 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.3 | 1.4 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.3 | 2.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.3 | 5.8 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.3 | 26.9 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.3 | 1.6 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.3 | 2.6 | GO:0015386 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.2 | 4.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.2 | 4.7 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.2 | 7.1 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.2 | 4.5 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.2 | 4.3 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.2 | 5.5 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.2 | 8.5 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.2 | 7.4 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.2 | 23.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.2 | 2.4 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.2 | 0.7 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.2 | 3.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.2 | 10.2 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.2 | 2.6 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 22.8 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 17.9 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 2.0 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 0.9 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.1 | 1.0 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 1.8 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 3.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 30.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 19.9 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 5.7 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 9.6 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 87.3 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.1 | 0.9 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.1 | 77.1 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.1 | 3.0 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.4 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.1 | 7.3 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 7.3 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.1 | 6.8 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 2.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 1.1 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 2.3 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.9 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 4.7 | GO:0016782 | transferase activity, transferring sulfur-containing groups(GO:0016782) |
| 0.1 | 0.9 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.6 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.1 | 2.5 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 18.9 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 1.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 2.0 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 1.3 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 5.9 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 20.2 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.7 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 1.1 | GO:0004385 | guanylate kinase activity(GO:0004385) ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 10.9 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 11.4 | GO:0008289 | lipid binding(GO:0008289) |
| 0.0 | 5.3 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 1.8 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 9.2 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 4.6 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 2.9 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 1.2 | GO:0008083 | growth factor activity(GO:0008083) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 82.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 2.2 | 50.9 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 1.8 | 28.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 1.6 | 114.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 1.4 | 12.5 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 1.2 | 30.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.8 | 14.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.7 | 20.6 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.7 | 50.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.7 | 27.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.7 | 29.4 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.7 | 19.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.6 | 32.4 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.6 | 49.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.6 | 18.5 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.6 | 19.1 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.6 | 16.4 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.6 | 11.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.6 | 30.2 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.5 | 38.2 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.5 | 7.0 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.5 | 15.0 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.5 | 127.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.5 | 28.3 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.5 | 16.9 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.4 | 16.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.4 | 18.5 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.4 | 123.6 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.3 | 9.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.3 | 4.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.3 | 5.3 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.2 | 6.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.2 | 11.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.2 | 8.3 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.2 | 8.1 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.2 | 3.8 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.2 | 7.4 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.2 | 5.3 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.2 | 10.1 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.2 | 6.3 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.2 | 7.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 5.1 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.2 | 26.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.2 | 3.8 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.1 | 4.5 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 4.6 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 9.1 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 1.3 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.1 | 4.2 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 3.8 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.1 | 3.8 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 17.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 4.1 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 4.3 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 3.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.9 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 46.5 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 2.3 | 48.3 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 2.1 | 29.6 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 1.9 | 52.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 1.9 | 48.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 1.8 | 167.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 1.7 | 16.7 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 1.6 | 46.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 1.5 | 22.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 1.3 | 26.0 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 1.2 | 16.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 1.1 | 18.5 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.9 | 32.9 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.9 | 20.9 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.8 | 16.8 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.8 | 24.5 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.7 | 2.2 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.7 | 113.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.7 | 14.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.7 | 54.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.7 | 19.5 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.7 | 10.1 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.6 | 7.9 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.5 | 4.2 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.5 | 33.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.5 | 9.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.5 | 17.5 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.5 | 29.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.5 | 6.8 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.4 | 8.9 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.4 | 21.3 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.4 | 51.7 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.4 | 5.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.4 | 12.9 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.4 | 7.0 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.4 | 32.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.3 | 29.3 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.3 | 3.6 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.3 | 2.6 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.3 | 11.7 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.3 | 13.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 2.1 | REACTOME FORMATION OF RNA POL II ELONGATION COMPLEX | Genes involved in Formation of RNA Pol II elongation complex |
| 0.2 | 4.8 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 0.2 | 2.5 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.2 | 2.8 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.2 | 3.0 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.2 | 7.5 | REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | Genes involved in Lipid digestion, mobilization, and transport |
| 0.2 | 7.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.2 | 2.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.2 | 5.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 7.6 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.1 | 3.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 12.0 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 1.4 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.1 | 3.0 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 2.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 8.1 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.1 | 2.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 7.6 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 1.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 2.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 4.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 2.8 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 2.6 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.1 | 4.4 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 1.5 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.6 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.9 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |