Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
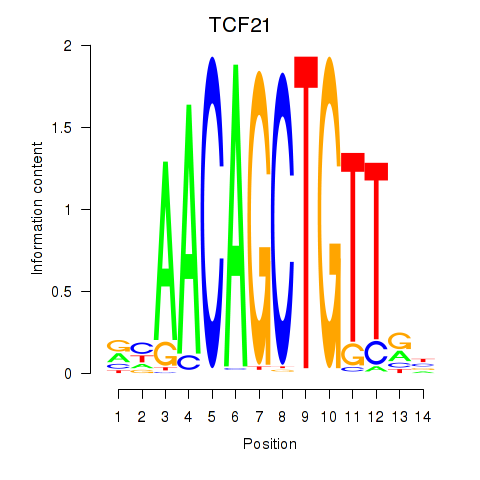
Results for TCF21
Z-value: 0.72
Transcription factors associated with TCF21
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TCF21
|
ENSG00000118526.6 | transcription factor 21 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TCF21 | hg19_v2_chr6_+_134210243_134210276 | -0.06 | 3.8e-01 | Click! |
Activity profile of TCF21 motif
Sorted Z-values of TCF21 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_79805146 | 19.62 |
ENST00000415593.1
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide |
| chrX_+_48432892 | 18.43 |
ENST00000376759.3
ENST00000430348.2 |
RBM3
|
RNA binding motif (RNP1, RRM) protein 3 |
| chr5_+_68462837 | 18.18 |
ENST00000256442.5
|
CCNB1
|
cyclin B1 |
| chr3_-_81792780 | 17.73 |
ENST00000489715.1
|
GBE1
|
glucan (1,4-alpha-), branching enzyme 1 |
| chrX_+_69509927 | 17.20 |
ENST00000374403.3
|
KIF4A
|
kinesin family member 4A |
| chr8_-_71519889 | 17.09 |
ENST00000521425.1
|
TRAM1
|
translocation associated membrane protein 1 |
| chr5_+_68462944 | 16.96 |
ENST00000506572.1
|
CCNB1
|
cyclin B1 |
| chr5_+_68463043 | 16.40 |
ENST00000508407.1
ENST00000505500.1 |
CCNB1
|
cyclin B1 |
| chr19_+_16186903 | 15.76 |
ENST00000588507.1
|
TPM4
|
tropomyosin 4 |
| chr5_-_95158644 | 15.17 |
ENST00000237858.6
|
GLRX
|
glutaredoxin (thioltransferase) |
| chr8_-_55014415 | 13.80 |
ENST00000522007.1
ENST00000521898.1 ENST00000518546.1 ENST00000316963.3 |
LYPLA1
|
lysophospholipase I |
| chr20_-_52210368 | 13.26 |
ENST00000371471.2
|
ZNF217
|
zinc finger protein 217 |
| chr5_+_135364584 | 12.94 |
ENST00000442011.2
ENST00000305126.8 |
TGFBI
|
transforming growth factor, beta-induced, 68kDa |
| chr1_-_94374946 | 12.54 |
ENST00000370238.3
|
GCLM
|
glutamate-cysteine ligase, modifier subunit |
| chr12_+_122326662 | 12.16 |
ENST00000261817.2
ENST00000538613.1 ENST00000542602.1 |
PSMD9
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 9 |
| chr22_+_40742512 | 12.07 |
ENST00000454266.2
ENST00000342312.6 |
ADSL
|
adenylosuccinate lyase |
| chr3_-_182833863 | 11.61 |
ENST00000492597.1
|
MCCC1
|
methylcrotonoyl-CoA carboxylase 1 (alpha) |
| chr1_-_150669500 | 11.55 |
ENST00000271732.3
|
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr12_-_50677255 | 11.00 |
ENST00000551691.1
ENST00000394943.3 ENST00000341247.4 |
LIMA1
|
LIM domain and actin binding 1 |
| chr22_+_38071615 | 10.95 |
ENST00000215909.5
|
LGALS1
|
lectin, galactoside-binding, soluble, 1 |
| chr7_-_99698338 | 10.83 |
ENST00000354230.3
ENST00000425308.1 |
MCM7
|
minichromosome maintenance complex component 7 |
| chr11_+_74660278 | 10.75 |
ENST00000263672.6
ENST00000530257.1 ENST00000526361.1 ENST00000532972.1 |
SPCS2
|
signal peptidase complex subunit 2 homolog (S. cerevisiae) |
| chr18_+_3247779 | 10.53 |
ENST00000578611.1
ENST00000583449.1 |
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr19_+_16187085 | 10.49 |
ENST00000300933.4
|
TPM4
|
tropomyosin 4 |
| chrX_-_16887963 | 10.37 |
ENST00000380084.4
|
RBBP7
|
retinoblastoma binding protein 7 |
| chr12_-_10875831 | 10.36 |
ENST00000279550.7
ENST00000228251.4 |
YBX3
|
Y box binding protein 3 |
| chr11_+_32112431 | 10.05 |
ENST00000054950.3
|
RCN1
|
reticulocalbin 1, EF-hand calcium binding domain |
| chr2_-_106054952 | 9.99 |
ENST00000336660.5
ENST00000393352.3 ENST00000607522.1 |
FHL2
|
four and a half LIM domains 2 |
| chr20_+_30327063 | 9.78 |
ENST00000300403.6
ENST00000340513.4 |
TPX2
|
TPX2, microtubule-associated |
| chr11_+_58910201 | 9.72 |
ENST00000528737.1
|
FAM111A
|
family with sequence similarity 111, member A |
| chr12_+_50144381 | 9.60 |
ENST00000552370.1
|
TMBIM6
|
transmembrane BAX inhibitor motif containing 6 |
| chr11_+_58910295 | 9.50 |
ENST00000420244.1
|
FAM111A
|
family with sequence similarity 111, member A |
| chr17_+_34848049 | 9.50 |
ENST00000588902.1
ENST00000591067.1 |
ZNHIT3
|
zinc finger, HIT-type containing 3 |
| chr16_-_9030515 | 9.39 |
ENST00000535863.1
ENST00000381886.4 |
USP7
|
ubiquitin specific peptidase 7 (herpes virus-associated) |
| chr15_+_65843130 | 9.23 |
ENST00000569894.1
|
PTPLAD1
|
protein tyrosine phosphatase-like A domain containing 1 |
| chr7_-_132766818 | 9.19 |
ENST00000262570.5
|
CHCHD3
|
coiled-coil-helix-coiled-coil-helix domain containing 3 |
| chr11_-_88070920 | 9.13 |
ENST00000524463.1
ENST00000227266.5 |
CTSC
|
cathepsin C |
| chr17_-_43025005 | 8.90 |
ENST00000587309.1
ENST00000593135.1 ENST00000339151.4 |
KIF18B
|
kinesin family member 18B |
| chr7_-_132766800 | 8.67 |
ENST00000542753.1
ENST00000448878.1 |
CHCHD3
|
coiled-coil-helix-coiled-coil-helix domain containing 3 |
| chr11_-_82997371 | 8.66 |
ENST00000525503.1
|
CCDC90B
|
coiled-coil domain containing 90B |
| chr11_+_125496619 | 8.57 |
ENST00000532669.1
ENST00000278916.3 |
CHEK1
|
checkpoint kinase 1 |
| chr1_-_9129895 | 8.50 |
ENST00000473209.1
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr6_-_31697255 | 8.33 |
ENST00000436437.1
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr15_+_63340734 | 8.30 |
ENST00000560959.1
|
TPM1
|
tropomyosin 1 (alpha) |
| chr5_-_98262240 | 8.30 |
ENST00000284049.3
|
CHD1
|
chromodomain helicase DNA binding protein 1 |
| chr2_-_17981462 | 8.26 |
ENST00000402989.1
ENST00000428868.1 |
SMC6
|
structural maintenance of chromosomes 6 |
| chr15_+_63340858 | 8.10 |
ENST00000560615.1
|
TPM1
|
tropomyosin 1 (alpha) |
| chr22_-_36924944 | 7.87 |
ENST00000405442.1
ENST00000402116.1 |
EIF3D
|
eukaryotic translation initiation factor 3, subunit D |
| chr11_-_67169253 | 7.80 |
ENST00000527663.1
ENST00000312989.7 |
PPP1CA
|
protein phosphatase 1, catalytic subunit, alpha isozyme |
| chr15_+_63796779 | 7.76 |
ENST00000561442.1
ENST00000560070.1 ENST00000540797.1 ENST00000380324.3 ENST00000268049.7 ENST00000536001.1 ENST00000539772.1 |
USP3
|
ubiquitin specific peptidase 3 |
| chr11_+_125496400 | 7.76 |
ENST00000524737.1
|
CHEK1
|
checkpoint kinase 1 |
| chr17_+_45286387 | 7.75 |
ENST00000572316.1
ENST00000354968.1 ENST00000576874.1 ENST00000536623.2 |
MYL4
|
myosin, light chain 4, alkali; atrial, embryonic |
| chr13_+_37574678 | 7.69 |
ENST00000389704.3
|
EXOSC8
|
exosome component 8 |
| chr4_-_140223670 | 7.56 |
ENST00000394228.1
ENST00000539387.1 |
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr16_-_31105870 | 7.46 |
ENST00000394971.3
|
VKORC1
|
vitamin K epoxide reductase complex, subunit 1 |
| chr17_+_6347729 | 7.45 |
ENST00000572447.1
|
FAM64A
|
family with sequence similarity 64, member A |
| chr12_-_6960407 | 7.42 |
ENST00000540683.1
ENST00000229265.6 ENST00000535406.1 ENST00000422785.3 |
CDCA3
|
cell division cycle associated 3 |
| chr15_+_63340647 | 7.38 |
ENST00000404484.4
|
TPM1
|
tropomyosin 1 (alpha) |
| chr22_+_44351301 | 7.26 |
ENST00000350028.4
|
SAMM50
|
SAMM50 sorting and assembly machinery component |
| chr16_-_46655538 | 7.23 |
ENST00000303383.3
|
SHCBP1
|
SHC SH2-domain binding protein 1 |
| chr2_-_234763147 | 7.21 |
ENST00000411486.2
ENST00000432087.1 ENST00000441687.1 ENST00000414924.1 |
HJURP
|
Holliday junction recognition protein |
| chr18_+_3448455 | 7.20 |
ENST00000549780.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr18_+_158513 | 7.16 |
ENST00000400266.3
ENST00000580410.1 ENST00000383589.2 ENST00000261601.7 |
USP14
|
ubiquitin specific peptidase 14 (tRNA-guanine transglycosylase) |
| chr17_+_6347761 | 7.13 |
ENST00000250056.8
ENST00000571373.1 ENST00000570337.2 ENST00000572595.2 ENST00000576056.1 |
FAM64A
|
family with sequence similarity 64, member A |
| chr3_+_23959185 | 7.02 |
ENST00000354811.5
|
RPL15
|
ribosomal protein L15 |
| chrX_-_107334750 | 7.01 |
ENST00000340200.5
ENST00000372296.1 ENST00000372295.1 ENST00000361815.5 |
PSMD10
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 10 |
| chr5_-_179050066 | 6.94 |
ENST00000329433.6
ENST00000510411.1 |
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr13_-_41635512 | 6.93 |
ENST00000405737.2
|
ELF1
|
E74-like factor 1 (ets domain transcription factor) |
| chrX_-_107334790 | 6.92 |
ENST00000217958.3
|
PSMD10
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 10 |
| chr7_-_6523755 | 6.80 |
ENST00000436575.1
ENST00000258739.4 |
DAGLB
KDELR2
|
diacylglycerol lipase, beta KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2 |
| chr15_+_89787180 | 6.68 |
ENST00000300027.8
ENST00000310775.7 ENST00000567891.1 ENST00000564920.1 ENST00000565255.1 ENST00000567996.1 ENST00000451393.2 ENST00000563250.1 |
FANCI
|
Fanconi anemia, complementation group I |
| chr3_+_197677379 | 6.59 |
ENST00000442341.1
|
RPL35A
|
ribosomal protein L35a |
| chr20_-_52790055 | 6.51 |
ENST00000395955.3
|
CYP24A1
|
cytochrome P450, family 24, subfamily A, polypeptide 1 |
| chr2_-_3595547 | 6.46 |
ENST00000438485.1
|
RP13-512J5.1
|
Uncharacterized protein |
| chr4_+_37892682 | 6.34 |
ENST00000508802.1
ENST00000261439.4 ENST00000402522.1 |
TBC1D1
|
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
| chr11_-_82997420 | 6.22 |
ENST00000455220.2
ENST00000529689.1 |
CCDC90B
|
coiled-coil domain containing 90B |
| chr15_+_63340553 | 6.14 |
ENST00000334895.5
|
TPM1
|
tropomyosin 1 (alpha) |
| chr19_-_4867665 | 6.13 |
ENST00000592528.1
ENST00000589494.1 ENST00000585479.1 ENST00000221957.4 |
PLIN3
|
perilipin 3 |
| chr15_-_72523924 | 6.07 |
ENST00000566809.1
ENST00000567087.1 ENST00000569050.1 ENST00000568883.1 |
PKM
|
pyruvate kinase, muscle |
| chr3_+_52245458 | 6.00 |
ENST00000459884.1
|
ALAS1
|
aminolevulinate, delta-, synthase 1 |
| chr6_-_31697563 | 5.97 |
ENST00000375789.2
ENST00000416410.1 |
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr11_-_82997013 | 5.83 |
ENST00000529073.1
ENST00000529611.1 |
CCDC90B
|
coiled-coil domain containing 90B |
| chr8_+_33342268 | 5.82 |
ENST00000360128.6
|
MAK16
|
MAK16 homolog (S. cerevisiae) |
| chr6_+_30585486 | 5.67 |
ENST00000259873.4
ENST00000506373.2 |
MRPS18B
|
mitochondrial ribosomal protein S18B |
| chr22_+_44351419 | 5.66 |
ENST00000396202.3
|
SAMM50
|
SAMM50 sorting and assembly machinery component |
| chr17_+_45286706 | 5.63 |
ENST00000393450.1
ENST00000572303.1 |
MYL4
|
myosin, light chain 4, alkali; atrial, embryonic |
| chr2_-_191115229 | 5.59 |
ENST00000409820.2
ENST00000410045.1 |
HIBCH
|
3-hydroxyisobutyryl-CoA hydrolase |
| chr11_-_104827425 | 5.54 |
ENST00000393150.3
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr11_+_125496124 | 5.54 |
ENST00000533778.2
ENST00000534070.1 |
CHEK1
|
checkpoint kinase 1 |
| chr15_+_63340775 | 5.53 |
ENST00000559281.1
ENST00000317516.7 |
TPM1
|
tropomyosin 1 (alpha) |
| chr2_+_231280908 | 5.53 |
ENST00000427101.2
ENST00000432979.1 |
SP100
|
SP100 nuclear antigen |
| chr17_-_7297833 | 5.48 |
ENST00000571802.1
ENST00000576201.1 ENST00000573213.1 ENST00000324822.11 |
TMEM256-PLSCR3
|
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr11_+_64948665 | 5.47 |
ENST00000533820.1
|
CAPN1
|
calpain 1, (mu/I) large subunit |
| chr20_-_54967187 | 5.38 |
ENST00000422322.1
ENST00000371356.2 ENST00000451915.1 ENST00000347343.2 ENST00000395911.1 ENST00000395907.1 ENST00000441357.1 ENST00000456249.1 ENST00000420474.1 ENST00000395909.4 ENST00000395914.1 ENST00000312783.6 ENST00000395915.3 ENST00000395913.3 |
AURKA
|
aurora kinase A |
| chr1_+_212738676 | 5.37 |
ENST00000366981.4
ENST00000366987.2 |
ATF3
|
activating transcription factor 3 |
| chr16_+_56970567 | 5.33 |
ENST00000563911.1
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr22_-_50964558 | 5.31 |
ENST00000535425.1
ENST00000439934.1 |
SCO2
|
SCO2 cytochrome c oxidase assembly protein |
| chr18_+_55888767 | 5.26 |
ENST00000431212.2
ENST00000586268.1 ENST00000587190.1 |
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr1_-_39339777 | 5.25 |
ENST00000397572.2
|
MYCBP
|
MYC binding protein |
| chr6_-_28891709 | 5.24 |
ENST00000377194.3
ENST00000377199.3 |
TRIM27
|
tripartite motif containing 27 |
| chr2_+_192110199 | 5.16 |
ENST00000304164.4
|
MYO1B
|
myosin IB |
| chr7_-_41742697 | 4.97 |
ENST00000242208.4
|
INHBA
|
inhibin, beta A |
| chr11_-_503521 | 4.93 |
ENST00000534797.1
|
RNH1
|
ribonuclease/angiogenin inhibitor 1 |
| chr1_+_79115503 | 4.87 |
ENST00000370747.4
ENST00000438486.1 ENST00000545124.1 |
IFI44
|
interferon-induced protein 44 |
| chr8_-_117886955 | 4.82 |
ENST00000297338.2
|
RAD21
|
RAD21 homolog (S. pombe) |
| chr1_+_155107820 | 4.77 |
ENST00000484157.1
|
SLC50A1
|
solute carrier family 50 (sugar efflux transporter), member 1 |
| chr20_-_43133491 | 4.74 |
ENST00000411544.1
|
SERINC3
|
serine incorporator 3 |
| chr1_-_202927490 | 4.74 |
ENST00000340990.5
|
ADIPOR1
|
adiponectin receptor 1 |
| chr3_+_141106643 | 4.70 |
ENST00000514251.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr3_+_49058444 | 4.69 |
ENST00000326925.6
ENST00000395458.2 |
NDUFAF3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr11_-_1643368 | 4.68 |
ENST00000399682.1
|
KRTAP5-4
|
keratin associated protein 5-4 |
| chr20_-_61847586 | 4.65 |
ENST00000370339.3
|
YTHDF1
|
YTH domain family, member 1 |
| chr14_-_50154921 | 4.58 |
ENST00000553805.2
ENST00000554396.1 ENST00000216367.5 ENST00000539565.2 |
POLE2
|
polymerase (DNA directed), epsilon 2, accessory subunit |
| chr16_-_31106048 | 4.48 |
ENST00000300851.6
|
VKORC1
|
vitamin K epoxide reductase complex, subunit 1 |
| chr19_-_43702231 | 4.44 |
ENST00000597374.1
ENST00000599371.1 |
PSG4
|
pregnancy specific beta-1-glycoprotein 4 |
| chr5_+_95066823 | 4.37 |
ENST00000506817.1
ENST00000379982.3 |
RHOBTB3
|
Rho-related BTB domain containing 3 |
| chrX_+_148622138 | 4.33 |
ENST00000450602.2
ENST00000441248.1 |
CXorf40A
|
chromosome X open reading frame 40A |
| chr21_-_33651324 | 4.31 |
ENST00000290130.3
|
MIS18A
|
MIS18 kinetochore protein A |
| chr17_-_77813186 | 4.21 |
ENST00000448310.1
ENST00000269397.4 |
CBX4
|
chromobox homolog 4 |
| chr11_-_87908600 | 4.09 |
ENST00000531138.1
ENST00000526372.1 ENST00000243662.6 |
RAB38
|
RAB38, member RAS oncogene family |
| chr16_-_31106211 | 4.08 |
ENST00000532364.1
ENST00000529564.1 ENST00000319788.7 ENST00000354895.4 ENST00000394975.2 |
RP11-196G11.1
VKORC1
|
Uncharacterized protein vitamin K epoxide reductase complex, subunit 1 |
| chr11_-_111957451 | 4.07 |
ENST00000504148.2
ENST00000541231.1 |
TIMM8B
|
translocase of inner mitochondrial membrane 8 homolog B (yeast) |
| chr11_-_62341445 | 4.02 |
ENST00000329251.4
|
EEF1G
|
eukaryotic translation elongation factor 1 gamma |
| chr1_+_148739443 | 3.98 |
ENST00000417839.1
|
NBPF16
|
neuroblastoma breakpoint family, member 16 |
| chr1_-_167522982 | 3.95 |
ENST00000370509.4
|
CREG1
|
cellular repressor of E1A-stimulated genes 1 |
| chr2_+_181845298 | 3.91 |
ENST00000410062.4
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr3_+_135969148 | 3.86 |
ENST00000251654.4
ENST00000490504.1 ENST00000483687.1 ENST00000468777.1 ENST00000462637.1 ENST00000466072.1 ENST00000482086.1 ENST00000471595.1 ENST00000469217.1 ENST00000465423.1 ENST00000478469.1 |
PCCB
|
propionyl CoA carboxylase, beta polypeptide |
| chr16_+_71929397 | 3.78 |
ENST00000537613.1
ENST00000424485.2 ENST00000606369.1 ENST00000329908.8 ENST00000538850.1 ENST00000541918.1 ENST00000534994.1 ENST00000378798.5 ENST00000539186.1 |
IST1
|
increased sodium tolerance 1 homolog (yeast) |
| chr1_-_206785898 | 3.77 |
ENST00000271764.2
|
EIF2D
|
eukaryotic translation initiation factor 2D |
| chr20_+_44044717 | 3.77 |
ENST00000279036.6
ENST00000279035.9 ENST00000372689.5 ENST00000545755.1 ENST00000341555.5 ENST00000535404.1 ENST00000543458.2 ENST00000432270.1 |
PIGT
|
phosphatidylinositol glycan anchor biosynthesis, class T |
| chr11_+_111957497 | 3.71 |
ENST00000375549.3
ENST00000528182.1 ENST00000528048.1 ENST00000528021.1 ENST00000526592.1 ENST00000525291.1 |
SDHD
|
succinate dehydrogenase complex, subunit D, integral membrane protein |
| chr1_-_206785789 | 3.68 |
ENST00000437518.1
ENST00000367114.3 |
EIF2D
|
eukaryotic translation initiation factor 2D |
| chr4_+_146403912 | 3.62 |
ENST00000507367.1
ENST00000394092.2 ENST00000515385.1 |
SMAD1
|
SMAD family member 1 |
| chr18_+_21529811 | 3.58 |
ENST00000588004.1
|
LAMA3
|
laminin, alpha 3 |
| chr2_-_158345462 | 3.57 |
ENST00000439355.1
ENST00000540637.1 |
CYTIP
|
cytohesin 1 interacting protein |
| chr1_-_9129631 | 3.57 |
ENST00000377414.3
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr22_+_23264766 | 3.54 |
ENST00000390331.2
|
IGLC7
|
immunoglobulin lambda constant 7 |
| chr10_-_51130715 | 3.48 |
ENST00000402038.3
|
PARG
|
poly (ADP-ribose) glycohydrolase |
| chr11_+_64002292 | 3.45 |
ENST00000426086.2
|
VEGFB
|
vascular endothelial growth factor B |
| chr1_+_144811943 | 3.38 |
ENST00000281815.8
|
NBPF9
|
neuroblastoma breakpoint family, member 9 |
| chr12_-_14133053 | 3.38 |
ENST00000609686.1
|
GRIN2B
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2B |
| chr3_+_49059038 | 3.36 |
ENST00000451378.2
|
NDUFAF3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chrX_+_148622513 | 3.33 |
ENST00000393985.3
ENST00000423421.1 ENST00000423540.2 ENST00000434353.2 ENST00000514208.1 |
CXorf40A
|
chromosome X open reading frame 40A |
| chr9_+_34646624 | 3.31 |
ENST00000450095.2
ENST00000556278.1 |
GALT
GALT
|
galactose-1-phosphate uridylyltransferase Uncharacterized protein |
| chr6_+_143772060 | 3.27 |
ENST00000367591.4
|
PEX3
|
peroxisomal biogenesis factor 3 |
| chr8_-_41522719 | 3.17 |
ENST00000335651.6
|
ANK1
|
ankyrin 1, erythrocytic |
| chr6_-_2842219 | 3.14 |
ENST00000380739.5
|
SERPINB1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr9_+_131133598 | 3.11 |
ENST00000372853.4
ENST00000452446.1 ENST00000372850.1 ENST00000372847.1 |
URM1
|
ubiquitin related modifier 1 |
| chr1_-_21113105 | 3.10 |
ENST00000375000.1
ENST00000419490.1 ENST00000414993.1 ENST00000443615.1 ENST00000312239.5 |
HP1BP3
|
heterochromatin protein 1, binding protein 3 |
| chr11_+_7597639 | 3.06 |
ENST00000533792.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chrX_-_102942961 | 3.04 |
ENST00000434230.1
ENST00000418819.1 ENST00000360458.1 |
MORF4L2
|
mortality factor 4 like 2 |
| chr8_-_82633476 | 2.94 |
ENST00000518419.1
ENST00000517588.1 ENST00000521895.1 ENST00000520076.1 ENST00000519523.1 ENST00000522520.1 ENST00000521287.1 ENST00000523096.1 ENST00000220669.5 ENST00000517450.1 |
ZFAND1
|
zinc finger, AN1-type domain 1 |
| chr7_-_76255444 | 2.92 |
ENST00000454397.1
|
POMZP3
|
POM121 and ZP3 fusion |
| chr11_-_615570 | 2.90 |
ENST00000525445.1
ENST00000348655.6 ENST00000397566.1 |
IRF7
|
interferon regulatory factor 7 |
| chr6_+_26458152 | 2.87 |
ENST00000312541.5
|
BTN2A1
|
butyrophilin, subfamily 2, member A1 |
| chr10_-_12084770 | 2.84 |
ENST00000357604.5
|
UPF2
|
UPF2 regulator of nonsense transcripts homolog (yeast) |
| chr4_+_47487285 | 2.82 |
ENST00000273859.3
ENST00000504445.1 |
ATP10D
|
ATPase, class V, type 10D |
| chr22_+_39052632 | 2.80 |
ENST00000411557.1
ENST00000396811.2 ENST00000216029.3 ENST00000416285.1 |
CBY1
|
chibby homolog 1 (Drosophila) |
| chr1_-_204135450 | 2.77 |
ENST00000272190.8
ENST00000367195.2 |
REN
|
renin |
| chr2_+_68384976 | 2.77 |
ENST00000263657.2
|
PNO1
|
partner of NOB1 homolog (S. cerevisiae) |
| chr2_-_152590946 | 2.76 |
ENST00000172853.10
|
NEB
|
nebulin |
| chr11_+_34127142 | 2.75 |
ENST00000257829.3
ENST00000531159.2 |
NAT10
|
N-acetyltransferase 10 (GCN5-related) |
| chr9_+_118950325 | 2.72 |
ENST00000534838.1
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1 |
| chr4_-_4543700 | 2.69 |
ENST00000505286.1
ENST00000306200.2 |
STX18
|
syntaxin 18 |
| chr17_-_26989136 | 2.69 |
ENST00000247020.4
|
SDF2
|
stromal cell-derived factor 2 |
| chr16_+_30383613 | 2.60 |
ENST00000568749.1
|
MYLPF
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr13_+_41635617 | 2.60 |
ENST00000542082.1
|
WBP4
|
WW domain binding protein 4 |
| chr17_-_7145106 | 2.57 |
ENST00000577035.1
|
GABARAP
|
GABA(A) receptor-associated protein |
| chr1_+_144151520 | 2.44 |
ENST00000369372.4
|
NBPF8
|
neuroblastoma breakpoint family, member 8 |
| chr15_-_42749711 | 2.41 |
ENST00000565611.1
ENST00000263805.4 ENST00000565948.1 |
ZNF106
|
zinc finger protein 106 |
| chr1_+_46049706 | 2.40 |
ENST00000527470.1
ENST00000525515.1 ENST00000537798.1 ENST00000402363.3 ENST00000528238.1 ENST00000350030.3 ENST00000470768.1 ENST00000372052.4 ENST00000351223.3 |
NASP
|
nuclear autoantigenic sperm protein (histone-binding) |
| chr4_+_37962018 | 2.28 |
ENST00000504686.1
|
PTTG2
|
pituitary tumor-transforming 2 |
| chr17_-_7145475 | 2.23 |
ENST00000571129.1
ENST00000571253.1 ENST00000573928.1 |
GABARAP
|
GABA(A) receptor-associated protein |
| chr11_+_71498552 | 2.19 |
ENST00000346333.6
ENST00000359244.4 ENST00000426628.2 |
FAM86C1
|
family with sequence similarity 86, member C1 |
| chr15_+_57998923 | 2.17 |
ENST00000380557.4
|
POLR2M
|
polymerase (RNA) II (DNA directed) polypeptide M |
| chr2_+_219264466 | 2.14 |
ENST00000273062.2
|
CTDSP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 |
| chr16_-_31439735 | 2.14 |
ENST00000287490.4
|
COX6A2
|
cytochrome c oxidase subunit VIa polypeptide 2 |
| chr17_+_4736627 | 2.14 |
ENST00000355280.6
ENST00000347992.7 |
MINK1
|
misshapen-like kinase 1 |
| chr5_-_140700322 | 2.13 |
ENST00000313368.5
|
TAF7
|
TAF7 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 55kDa |
| chr1_-_45476944 | 2.12 |
ENST00000372172.4
|
HECTD3
|
HECT domain containing E3 ubiquitin protein ligase 3 |
| chr2_+_232575128 | 2.07 |
ENST00000412128.1
|
PTMA
|
prothymosin, alpha |
| chr12_+_56473628 | 2.05 |
ENST00000549282.1
ENST00000549061.1 ENST00000267101.3 |
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr9_+_34646651 | 2.02 |
ENST00000378842.3
|
GALT
|
galactose-1-phosphate uridylyltransferase |
| chr19_-_41903161 | 1.97 |
ENST00000602129.1
ENST00000593771.1 ENST00000596905.1 ENST00000221233.4 |
EXOSC5
|
exosome component 5 |
| chr19_+_48216600 | 1.97 |
ENST00000263277.3
ENST00000538399.1 |
EHD2
|
EH-domain containing 2 |
| chr12_+_57828521 | 1.90 |
ENST00000309668.2
|
INHBC
|
inhibin, beta C |
| chr14_-_23288930 | 1.90 |
ENST00000554517.1
ENST00000285850.7 ENST00000397529.2 ENST00000555702.1 |
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr7_-_128045984 | 1.88 |
ENST00000470772.1
ENST00000480861.1 ENST00000496200.1 |
IMPDH1
|
IMP (inosine 5'-monophosphate) dehydrogenase 1 |
| chr7_-_25164868 | 1.86 |
ENST00000409409.1
ENST00000409764.1 ENST00000413447.1 |
CYCS
|
cytochrome c, somatic |
| chr2_+_61244697 | 1.86 |
ENST00000401576.1
ENST00000295030.5 ENST00000414712.2 |
PEX13
|
peroxisomal biogenesis factor 13 |
| chr6_+_150070857 | 1.85 |
ENST00000544496.1
|
PCMT1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr9_+_125137565 | 1.82 |
ENST00000373698.5
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr4_-_89152474 | 1.78 |
ENST00000515655.1
|
ABCG2
|
ATP-binding cassette, sub-family G (WHITE), member 2 |
| chr7_+_99102573 | 1.76 |
ENST00000394170.2
|
ZKSCAN5
|
zinc finger with KRAB and SCAN domains 5 |
| chrX_+_9431324 | 1.74 |
ENST00000407597.2
ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X
|
transducin (beta)-like 1X-linked |
| chr15_+_57998821 | 1.73 |
ENST00000299638.3
|
POLR2M
|
polymerase (RNA) II (DNA directed) polypeptide M |
| chr1_+_25870070 | 1.71 |
ENST00000374338.4
|
LDLRAP1
|
low density lipoprotein receptor adaptor protein 1 |
| chr18_-_77748527 | 1.70 |
ENST00000591711.1
ENST00000588162.1 ENST00000269601.5 |
TXNL4A
|
thioredoxin-like 4A |
| chr6_+_46761118 | 1.67 |
ENST00000230588.4
|
MEP1A
|
meprin A, alpha (PABA peptide hydrolase) |
| chr19_+_11546093 | 1.66 |
ENST00000591462.1
|
PRKCSH
|
protein kinase C substrate 80K-H |
Network of associatons between targets according to the STRING database.
First level regulatory network of TCF21
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 17.2 | 51.5 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) response to DDT(GO:0046680) histone H3-S10 phosphorylation involved in chromosome condensation(GO:2000775) |
| 5.2 | 26.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 4.5 | 35.9 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 4.0 | 12.1 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 3.6 | 21.9 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 3.1 | 12.5 | GO:1904386 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 3.1 | 30.6 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 2.8 | 13.8 | GO:0002084 | protein depalmitoylation(GO:0002084) negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 2.6 | 7.9 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 2.4 | 7.2 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 2.4 | 4.8 | GO:1901656 | glycoside transport(GO:1901656) |
| 2.3 | 9.4 | GO:0035616 | maintenance of DNA methylation(GO:0010216) histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 2.2 | 6.5 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
| 2.1 | 10.4 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 2.0 | 10.0 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 2.0 | 7.8 | GO:0036496 | regulation of translational initiation by eIF2 alpha dephosphorylation(GO:0036496) |
| 1.9 | 7.7 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 1.8 | 5.4 | GO:1900195 | spindle assembly involved in female meiosis I(GO:0007057) positive regulation of oocyte maturation(GO:1900195) |
| 1.8 | 5.4 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 1.8 | 5.3 | GO:0006258 | UDP-glucose catabolic process(GO:0006258) galactose catabolic process via UDP-galactose(GO:0033499) |
| 1.7 | 12.2 | GO:0098704 | fructose transport(GO:0015755) fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 1.7 | 6.8 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 1.7 | 5.0 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 1.6 | 16.0 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 1.6 | 9.6 | GO:1904721 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) negative regulation of immunoglobulin secretion(GO:0051025) negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 1.3 | 3.9 | GO:0051685 | maintenance of ER location(GO:0051685) |
| 1.3 | 10.4 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 1.3 | 11.6 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 1.3 | 3.9 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 1.2 | 3.7 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 1.2 | 12.9 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 1.1 | 17.2 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 1.1 | 5.6 | GO:0006574 | valine catabolic process(GO:0006574) |
| 1.1 | 10.8 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 1.0 | 5.2 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 1.0 | 3.1 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 1.0 | 6.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 1.0 | 4.8 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.9 | 7.4 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.9 | 8.3 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.9 | 2.7 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.9 | 14.3 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.8 | 15.2 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.7 | 18.3 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.7 | 2.8 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.7 | 3.4 | GO:2000110 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.7 | 4.7 | GO:0009597 | detection of virus(GO:0009597) |
| 0.7 | 2.0 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.7 | 2.0 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.6 | 1.9 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.6 | 3.8 | GO:0009838 | abscission(GO:0009838) |
| 0.6 | 3.8 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.6 | 1.9 | GO:0060152 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.6 | 6.8 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.6 | 5.5 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.6 | 4.7 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.6 | 4.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.6 | 40.7 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.5 | 3.3 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.5 | 5.3 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.5 | 10.8 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.5 | 2.9 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.5 | 2.4 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.5 | 1.4 | GO:0032289 | central nervous system myelin formation(GO:0032289) detection of bacterial lipoprotein(GO:0042494) detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.5 | 2.8 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.5 | 6.0 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.5 | 5.5 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.5 | 3.6 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.4 | 2.7 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.4 | 1.7 | GO:0090118 | receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) |
| 0.4 | 11.5 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.4 | 3.5 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.4 | 1.5 | GO:1901804 | beta-glucoside metabolic process(GO:1901804) beta-glucoside catabolic process(GO:1901805) positive regulation of neuronal action potential(GO:1904457) |
| 0.3 | 2.4 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.3 | 7.8 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.3 | 1.9 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.3 | 1.5 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.3 | 6.9 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.3 | 5.3 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.3 | 2.6 | GO:0030091 | protein repair(GO:0030091) |
| 0.3 | 4.8 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.3 | 0.6 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.3 | 6.7 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.3 | 4.6 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.2 | 4.4 | GO:0007565 | female pregnancy(GO:0007565) |
| 0.2 | 2.7 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.2 | 3.4 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.2 | 9.8 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.2 | 4.5 | GO:0045199 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.2 | 0.7 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.2 | 0.5 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 0.2 | 4.9 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.2 | 15.8 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.2 | 17.9 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.2 | 2.1 | GO:0015846 | polyamine transport(GO:0015846) |
| 0.2 | 1.0 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.2 | 11.0 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.2 | 15.6 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.2 | 2.4 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.2 | 30.7 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.2 | 4.0 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 4.1 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.2 | 0.4 | GO:2000321 | positive regulation of T-helper 17 cell differentiation(GO:2000321) |
| 0.2 | 3.1 | GO:0070828 | heterochromatin organization(GO:0070828) |
| 0.2 | 1.4 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.2 | 1.4 | GO:0002326 | B cell lineage commitment(GO:0002326) immunoglobulin V(D)J recombination(GO:0033152) |
| 0.2 | 1.2 | GO:0034127 | regulation of MyD88-independent toll-like receptor signaling pathway(GO:0034127) negative regulation of MyD88-independent toll-like receptor signaling pathway(GO:0034128) positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.2 | 8.6 | GO:0001913 | T cell mediated cytotoxicity(GO:0001913) |
| 0.2 | 0.8 | GO:0090650 | renal water absorption(GO:0070295) response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.1 | 4.6 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.1 | 1.6 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 2.7 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) |
| 0.1 | 2.4 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.1 | 1.3 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.1 | 1.6 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.1 | 3.6 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 10.9 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.1 | 1.4 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 2.6 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 0.7 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.1 | 3.0 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 1.3 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.1 | 15.4 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.1 | 1.8 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 5.0 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 0.5 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 2.1 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 2.0 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 3.9 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 0.9 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 0.9 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.1 | 0.2 | GO:0010041 | response to iron(III) ion(GO:0010041) |
| 0.1 | 0.8 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 0.2 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.1 | 9.0 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.1 | 3.4 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 5.7 | GO:0070125 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 0.1 | 0.3 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 0.6 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.1 | 1.3 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.1 | 2.2 | GO:0032259 | methylation(GO:0032259) |
| 0.1 | 2.3 | GO:2000816 | negative regulation of mitotic sister chromatid separation(GO:2000816) |
| 0.1 | 0.9 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 1.7 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 2.6 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 4.4 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.1 | 2.8 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 1.0 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.1 | 2.0 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.3 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 6.5 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 4.0 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.3 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.0 | 0.4 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 2.1 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 1.2 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.6 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 1.6 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 3.6 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 2.8 | GO:0006402 | mRNA catabolic process(GO:0006402) |
| 0.0 | 0.3 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 0.0 | 0.7 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.0 | 1.4 | GO:0033146 | regulation of intracellular estrogen receptor signaling pathway(GO:0033146) |
| 0.0 | 1.7 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 1.1 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.1 | GO:0046886 | positive regulation of hormone biosynthetic process(GO:0046886) |
| 0.0 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 3.4 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.0 | 0.2 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.0 | 0.6 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 1.7 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.4 | GO:0001569 | patterning of blood vessels(GO:0001569) |
| 0.0 | 0.4 | GO:0060259 | regulation of feeding behavior(GO:0060259) |
| 0.0 | 0.8 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 0.8 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 1.3 | GO:0055007 | cardiac muscle cell differentiation(GO:0055007) |
| 0.0 | 3.7 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.8 | GO:0001525 | angiogenesis(GO:0001525) |
| 0.0 | 7.7 | GO:0043312 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.0 | 0.1 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.4 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.0 | 0.6 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.3 | 51.5 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 6.5 | 19.6 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 3.3 | 62.2 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 2.6 | 17.9 | GO:0061617 | MICOS complex(GO:0061617) |
| 2.3 | 11.6 | GO:1905202 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 2.1 | 8.3 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 1.8 | 5.4 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 1.7 | 5.0 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 1.5 | 10.8 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 1.4 | 26.1 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 1.3 | 3.8 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 1.2 | 13.3 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 1.2 | 4.8 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 1.1 | 8.9 | GO:0005818 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 1.0 | 6.8 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.9 | 3.7 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.9 | 5.5 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.9 | 4.6 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.9 | 5.3 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.9 | 9.7 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.9 | 7.9 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.8 | 10.8 | GO:0042555 | MCM complex(GO:0042555) |
| 0.8 | 4.9 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.8 | 4.1 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.8 | 7.8 | GO:0042587 | glycogen granule(GO:0042587) PTW/PP1 phosphatase complex(GO:0072357) |
| 0.8 | 7.8 | GO:0070938 | contractile ring(GO:0070938) |
| 0.7 | 5.4 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.5 | 4.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.5 | 1.4 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.4 | 3.6 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.4 | 10.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.3 | 19.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.3 | 30.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.3 | 4.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.3 | 3.8 | GO:0090543 | Flemming body(GO:0090543) |
| 0.3 | 1.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.3 | 21.9 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.3 | 4.8 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.2 | 1.7 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.2 | 2.6 | GO:0043203 | axon hillock(GO:0043203) |
| 0.2 | 5.7 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.2 | 3.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 13.2 | GO:0005844 | polysome(GO:0005844) |
| 0.2 | 2.1 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.2 | 4.5 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.2 | 32.0 | GO:0015934 | large ribosomal subunit(GO:0015934) |
| 0.2 | 5.1 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.2 | 30.6 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.2 | 15.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.2 | 11.0 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.2 | 3.6 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.2 | 2.1 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 15.8 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.1 | 7.4 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.6 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 7.6 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 5.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 3.0 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 0.7 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 12.2 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.1 | 7.2 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.1 | 2.7 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 5.3 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 8.8 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 1.7 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 0.4 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.1 | 1.6 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 4.3 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 3.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 8.0 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 0.3 | GO:0070081 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.1 | 11.5 | GO:1904813 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.1 | 1.2 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.1 | 1.6 | GO:0030990 | intraciliary transport particle(GO:0030990) |
| 0.1 | 19.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 1.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.4 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 2.7 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 3.4 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 5.2 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.9 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 3.5 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 1.0 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 22.8 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.0 | 3.4 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 1.0 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.4 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 3.9 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.0 | 2.0 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 2.6 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.5 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 7.2 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 18.1 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 16.3 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 4.5 | GO:0005925 | focal adhesion(GO:0005925) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.4 | 51.5 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 4.0 | 16.0 | GO:0016900 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 3.9 | 19.6 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 3.6 | 10.9 | GO:0048030 | disaccharide binding(GO:0048030) |
| 3.6 | 21.9 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 3.3 | 13.4 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 2.9 | 14.3 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 2.4 | 7.2 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 2.3 | 11.6 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 2.0 | 6.0 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 2.0 | 9.8 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 1.7 | 12.2 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 1.7 | 13.8 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 1.7 | 6.8 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 1.7 | 5.0 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 1.6 | 9.6 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 1.5 | 6.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 1.5 | 15.2 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 1.5 | 12.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 1.3 | 6.7 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 1.3 | 3.8 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 1.2 | 3.7 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 1.2 | 4.9 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 1.0 | 4.1 | GO:0035651 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.9 | 2.6 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.8 | 6.8 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.7 | 67.6 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.7 | 5.3 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.6 | 3.9 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.6 | 1.9 | GO:0045155 | electron transporter, transferring electrons from CoQH2-cytochrome c reductase complex and cytochrome c oxidase complex activity(GO:0045155) |
| 0.6 | 5.4 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.6 | 3.5 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.6 | 12.8 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.6 | 17.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.5 | 2.1 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.5 | 5.3 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.5 | 5.3 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.5 | 1.9 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.5 | 1.4 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.5 | 1.8 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.4 | 2.7 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.4 | 9.1 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.4 | 19.1 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.4 | 2.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.4 | 5.5 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.4 | 3.4 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.4 | 10.8 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.4 | 1.5 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.4 | 7.9 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.4 | 4.6 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.4 | 8.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.3 | 2.9 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.3 | 1.3 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.3 | 5.5 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.3 | 7.8 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.3 | 11.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.3 | 9.4 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.3 | 1.0 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.3 | 2.0 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.2 | 4.7 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.2 | 4.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 9.7 | GO:0004532 | exoribonuclease activity(GO:0004532) |
| 0.2 | 4.2 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.2 | 4.8 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.2 | 1.8 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.2 | 21.3 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.2 | 1.6 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.2 | 2.4 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.2 | 1.6 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.2 | 10.4 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.2 | 0.8 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.2 | 1.4 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.2 | 13.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.2 | 4.7 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 3.1 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.2 | 15.1 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.2 | 1.9 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.2 | 0.9 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 0.7 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.2 | 3.9 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 0.9 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.2 | 5.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 1.8 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.2 | 7.5 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.2 | 4.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.2 | 8.2 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 13.8 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.1 | 7.6 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 7.6 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.5 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.1 | 4.8 | GO:0051119 | sugar transmembrane transporter activity(GO:0051119) |
| 0.1 | 2.8 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 19.2 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 1.2 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 2.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 2.4 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 0.6 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.1 | 4.9 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.1 | 0.6 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 4.4 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 10.4 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.1 | 4.2 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 0.3 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.1 | 0.4 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.1 | 2.8 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 1.0 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.1 | 1.5 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 4.0 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 3.6 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 0.9 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 3.9 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.1 | 2.7 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 3.8 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 0.6 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.1 | 3.6 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 2.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 12.7 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 3.9 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 1.7 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.3 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 3.4 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 1.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.2 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 1.6 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.4 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.4 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 2.1 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.8 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 2.4 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.8 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.6 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 1.0 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 8.5 | GO:0008134 | transcription factor binding(GO:0008134) |
| 0.0 | 0.4 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.4 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.5 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.8 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 2.2 | GO:0008168 | methyltransferase activity(GO:0008168) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 21.9 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 1.1 | 66.7 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.4 | 16.4 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.3 | 10.0 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.2 | 12.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 4.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.2 | 6.7 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.2 | 28.3 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.2 | 3.5 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.2 | 3.6 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 6.8 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.2 | 6.9 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 18.5 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 9.4 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 5.5 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 2.7 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 4.2 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 2.1 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 10.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 3.4 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 6.4 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 5.3 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 2.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 5.9 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 7.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 4.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 2.0 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 5.8 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 1.9 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.1 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 1.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.4 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.9 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 2.1 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.7 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.7 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.7 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 2.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.8 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 73.4 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 1.1 | 78.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 1.1 | 9.7 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 1.1 | 16.0 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 1.0 | 21.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.8 | 14.0 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.7 | 10.8 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.7 | 17.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.6 | 17.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.6 | 10.8 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.6 | 12.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.5 | 6.9 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.4 | 15.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.4 | 10.7 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.4 | 13.8 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.4 | 12.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.4 | 26.1 | REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | Genes involved in p53-Independent G1/S DNA damage checkpoint |
| 0.4 | 21.9 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.3 | 7.8 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.3 | 4.6 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.3 | 17.0 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.3 | 4.6 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.3 | 3.4 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.2 | 12.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.2 | 6.0 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 3.9 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.2 | 3.7 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.2 | 3.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.2 | 1.8 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.2 | 6.2 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.2 | 43.3 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.2 | 5.7 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.2 | 3.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.2 | 7.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.2 | 16.3 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.1 | 4.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.4 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.1 | 2.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 9.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 3.8 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.1 | 5.5 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.1 | 1.7 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 10.8 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 12.2 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 2.8 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.1 | 4.8 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 2.0 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 6.8 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 2.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 1.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 8.3 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.4 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.0 | 1.6 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 1.1 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 1.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.8 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 3.6 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 1.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.9 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 1.2 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 1.9 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.1 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.8 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.9 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |