Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
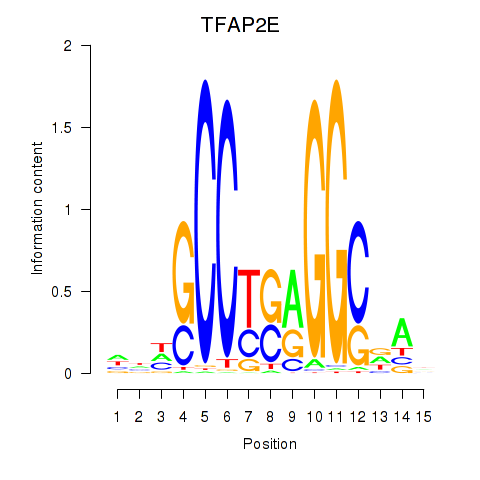
Results for TFAP2E
Z-value: 1.06
Transcription factors associated with TFAP2E
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TFAP2E
|
ENSG00000116819.6 | transcription factor AP-2 epsilon |
Activity profile of TFAP2E motif
Sorted Z-values of TFAP2E motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_56623433 | 23.44 |
ENST00000570176.1
|
MT3
|
metallothionein 3 |
| chr17_-_42992856 | 23.01 |
ENST00000588316.1
ENST00000435360.2 ENST00000586793.1 ENST00000588735.1 ENST00000588037.1 ENST00000592320.1 ENST00000253408.5 |
GFAP
|
glial fibrillary acidic protein |
| chr11_+_82612740 | 22.18 |
ENST00000524921.1
ENST00000528759.1 ENST00000525361.1 ENST00000430323.2 ENST00000533655.1 ENST00000532764.1 ENST00000532589.1 ENST00000525388.1 |
C11orf82
|
chromosome 11 open reading frame 82 |
| chr12_+_53773944 | 20.48 |
ENST00000551969.1
ENST00000327443.4 |
SP1
|
Sp1 transcription factor |
| chr1_-_11866034 | 16.08 |
ENST00000376590.3
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr1_-_11865982 | 15.86 |
ENST00000418034.1
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr5_-_115152651 | 15.38 |
ENST00000250535.4
|
CDO1
|
cysteine dioxygenase type 1 |
| chr2_-_177502254 | 13.20 |
ENST00000339037.3
|
AC017048.3
|
long intergenic non-protein coding RNA 1116 |
| chr15_+_23810853 | 13.12 |
ENST00000568252.1
|
MKRN3
|
makorin ring finger protein 3 |
| chr14_+_24837226 | 12.20 |
ENST00000554050.1
ENST00000554903.1 ENST00000554779.1 ENST00000250373.4 ENST00000553708.1 |
NFATC4
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 |
| chr1_+_9599540 | 11.91 |
ENST00000302692.6
|
SLC25A33
|
solute carrier family 25 (pyrimidine nucleotide carrier), member 33 |
| chr6_+_7108210 | 10.85 |
ENST00000467782.1
ENST00000334984.6 ENST00000349384.6 |
RREB1
|
ras responsive element binding protein 1 |
| chr3_+_49591881 | 10.14 |
ENST00000296452.4
|
BSN
|
bassoon presynaptic cytomatrix protein |
| chr1_+_9005917 | 9.94 |
ENST00000549778.1
ENST00000480186.3 ENST00000377443.2 ENST00000377436.3 ENST00000377442.2 |
CA6
|
carbonic anhydrase VI |
| chrX_+_152990302 | 9.42 |
ENST00000218104.3
|
ABCD1
|
ATP-binding cassette, sub-family D (ALD), member 1 |
| chr4_-_82393009 | 9.10 |
ENST00000436139.2
|
RASGEF1B
|
RasGEF domain family, member 1B |
| chr17_+_3627185 | 8.84 |
ENST00000325418.4
|
GSG2
|
germ cell associated 2 (haspin) |
| chr15_+_27112058 | 8.81 |
ENST00000355395.5
|
GABRA5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| chr3_+_4535155 | 8.76 |
ENST00000544951.1
|
ITPR1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr11_-_47870091 | 8.65 |
ENST00000526870.1
|
NUP160
|
nucleoporin 160kDa |
| chr12_+_53443963 | 8.59 |
ENST00000546602.1
ENST00000552570.1 ENST00000549700.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr15_+_27112251 | 8.22 |
ENST00000400081.3
|
GABRA5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| chr7_-_140624499 | 7.92 |
ENST00000288602.6
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B |
| chr15_+_27112380 | 7.91 |
ENST00000554596.1
|
GABRA5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| chr6_+_7107830 | 7.43 |
ENST00000379933.3
|
RREB1
|
ras responsive element binding protein 1 |
| chr15_-_65503801 | 7.36 |
ENST00000261883.4
|
CILP
|
cartilage intermediate layer protein, nucleotide pyrophosphohydrolase |
| chr15_+_27112194 | 7.36 |
ENST00000555182.1
|
GABRA5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| chr15_+_27112296 | 6.99 |
ENST00000554038.1
|
GABRA5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| chr10_+_99894399 | 6.95 |
ENST00000298999.3
ENST00000314594.5 |
R3HCC1L
|
R3H domain and coiled-coil containing 1-like |
| chr19_-_11039188 | 6.93 |
ENST00000588347.1
|
YIPF2
|
Yip1 domain family, member 2 |
| chr2_+_239335449 | 6.74 |
ENST00000264607.4
|
ASB1
|
ankyrin repeat and SOCS box containing 1 |
| chr10_+_99894380 | 6.73 |
ENST00000370584.3
|
R3HCC1L
|
R3H domain and coiled-coil containing 1-like |
| chr12_+_75784850 | 6.57 |
ENST00000550916.1
ENST00000435775.1 ENST00000378689.2 ENST00000378692.3 ENST00000320460.4 ENST00000547164.1 |
GLIPR1L2
|
GLI pathogenesis-related 1 like 2 |
| chr17_-_47755338 | 6.51 |
ENST00000508805.1
ENST00000515508.2 ENST00000451526.2 ENST00000507970.1 |
SPOP
|
speckle-type POZ protein |
| chr19_-_46389359 | 6.45 |
ENST00000302165.3
|
IRF2BP1
|
interferon regulatory factor 2 binding protein 1 |
| chr17_+_29158962 | 6.43 |
ENST00000321990.4
|
ATAD5
|
ATPase family, AAA domain containing 5 |
| chr20_+_58508817 | 6.23 |
ENST00000358293.3
|
FAM217B
|
family with sequence similarity 217, member B |
| chr10_-_112678976 | 5.93 |
ENST00000448814.1
|
BBIP1
|
BBSome interacting protein 1 |
| chr1_-_11865351 | 5.63 |
ENST00000413656.1
ENST00000376585.1 ENST00000423400.1 ENST00000431243.1 |
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr19_+_4969116 | 5.19 |
ENST00000588337.1
ENST00000159111.4 ENST00000381759.4 |
KDM4B
|
lysine (K)-specific demethylase 4B |
| chr13_-_27334879 | 5.17 |
ENST00000405846.3
|
GPR12
|
G protein-coupled receptor 12 |
| chr19_+_36195429 | 5.16 |
ENST00000392197.2
|
ZBTB32
|
zinc finger and BTB domain containing 32 |
| chr11_-_47870019 | 5.09 |
ENST00000378460.2
|
NUP160
|
nucleoporin 160kDa |
| chr3_+_4535025 | 5.02 |
ENST00000302640.8
ENST00000354582.6 ENST00000423119.2 ENST00000357086.4 ENST00000456211.2 |
ITPR1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr4_-_85419603 | 4.84 |
ENST00000295886.4
|
NKX6-1
|
NK6 homeobox 1 |
| chr11_+_116700614 | 4.73 |
ENST00000375345.1
|
APOC3
|
apolipoprotein C-III |
| chr19_+_36195467 | 4.71 |
ENST00000426659.2
|
ZBTB32
|
zinc finger and BTB domain containing 32 |
| chr1_+_35225339 | 4.65 |
ENST00000339480.1
|
GJB4
|
gap junction protein, beta 4, 30.3kDa |
| chr11_+_116700600 | 4.52 |
ENST00000227667.3
|
APOC3
|
apolipoprotein C-III |
| chr3_+_155860751 | 4.35 |
ENST00000471742.1
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr1_+_17634689 | 4.22 |
ENST00000375453.1
ENST00000375448.4 |
PADI4
|
peptidyl arginine deiminase, type IV |
| chr10_-_98273668 | 4.09 |
ENST00000357947.3
|
TLL2
|
tolloid-like 2 |
| chr9_-_21077939 | 4.07 |
ENST00000380232.2
|
IFNB1
|
interferon, beta 1, fibroblast |
| chrX_+_128872998 | 3.99 |
ENST00000371106.3
|
XPNPEP2
|
X-prolyl aminopeptidase (aminopeptidase P) 2, membrane-bound |
| chr3_+_51428704 | 3.85 |
ENST00000323686.4
|
RBM15B
|
RNA binding motif protein 15B |
| chr20_-_13619550 | 3.80 |
ENST00000455532.1
ENST00000544472.1 ENST00000539805.1 ENST00000337743.4 |
TASP1
|
taspase, threonine aspartase, 1 |
| chr2_-_45236540 | 3.79 |
ENST00000303077.6
|
SIX2
|
SIX homeobox 2 |
| chr19_-_43969796 | 3.65 |
ENST00000244333.3
|
LYPD3
|
LY6/PLAUR domain containing 3 |
| chr9_-_34662651 | 3.63 |
ENST00000259631.4
|
CCL27
|
chemokine (C-C motif) ligand 27 |
| chr11_-_113345995 | 3.62 |
ENST00000355319.2
ENST00000542616.1 |
DRD2
|
dopamine receptor D2 |
| chr20_+_55205825 | 3.54 |
ENST00000544508.1
|
TFAP2C
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr5_+_133984462 | 3.54 |
ENST00000398844.2
ENST00000322887.4 |
SEC24A
|
SEC24 family member A |
| chr12_+_57984965 | 3.51 |
ENST00000540759.2
ENST00000551772.1 ENST00000550465.1 ENST00000354947.5 |
PIP4K2C
|
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma |
| chr8_+_17780483 | 3.49 |
ENST00000517730.1
ENST00000518537.1 ENST00000523055.1 ENST00000519253.1 |
PCM1
|
pericentriolar material 1 |
| chr17_-_39769005 | 3.32 |
ENST00000301653.4
ENST00000593067.1 |
KRT16
|
keratin 16 |
| chr17_+_72428218 | 3.30 |
ENST00000392628.2
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr11_-_35440796 | 3.20 |
ENST00000278379.3
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr6_+_37787262 | 3.01 |
ENST00000287218.4
|
ZFAND3
|
zinc finger, AN1-type domain 3 |
| chr19_-_39264072 | 3.01 |
ENST00000599035.1
ENST00000378626.4 |
LGALS7
|
lectin, galactoside-binding, soluble, 7 |
| chr3_+_52813932 | 3.00 |
ENST00000537050.1
|
ITIH1
|
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr19_-_11039261 | 2.97 |
ENST00000590329.1
ENST00000587943.1 ENST00000585858.1 ENST00000586748.1 ENST00000586575.1 ENST00000253031.2 |
YIPF2
|
Yip1 domain family, member 2 |
| chr1_+_17248418 | 2.85 |
ENST00000375541.5
|
CROCC
|
ciliary rootlet coiled-coil, rootletin |
| chr1_-_201346761 | 2.76 |
ENST00000455702.1
ENST00000422165.1 ENST00000367318.5 ENST00000367320.2 ENST00000438742.1 ENST00000412633.1 ENST00000458432.2 ENST00000421663.2 ENST00000367322.1 ENST00000509001.1 |
TNNT2
|
troponin T type 2 (cardiac) |
| chr19_+_45504688 | 2.73 |
ENST00000221452.8
ENST00000540120.1 ENST00000505236.1 |
RELB
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr11_-_118305921 | 2.65 |
ENST00000532619.1
|
RP11-770J1.4
|
RP11-770J1.4 |
| chr11_-_66056478 | 2.62 |
ENST00000431556.2
ENST00000528575.1 |
YIF1A
|
Yip1 interacting factor homolog A (S. cerevisiae) |
| chr14_+_21214039 | 2.57 |
ENST00000326842.2
|
EDDM3A
|
epididymal protein 3A |
| chr12_+_72666407 | 2.52 |
ENST00000261180.4
|
TRHDE
|
thyrotropin-releasing hormone degrading enzyme |
| chr17_-_31620006 | 2.43 |
ENST00000225823.2
|
ASIC2
|
acid-sensing (proton-gated) ion channel 2 |
| chr3_-_48130314 | 2.35 |
ENST00000439356.1
ENST00000395734.3 ENST00000426837.2 |
MAP4
|
microtubule-associated protein 4 |
| chr11_-_35441524 | 2.19 |
ENST00000395750.1
ENST00000449068.1 |
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr11_-_47869865 | 2.16 |
ENST00000530326.1
ENST00000532747.1 |
NUP160
|
nucleoporin 160kDa |
| chr2_+_38893047 | 2.16 |
ENST00000272252.5
|
GALM
|
galactose mutarotase (aldose 1-epimerase) |
| chr22_+_29601840 | 2.07 |
ENST00000334018.6
ENST00000429226.1 ENST00000404755.3 ENST00000404820.3 ENST00000430127.1 |
EMID1
|
EMI domain containing 1 |
| chr3_-_48632593 | 1.98 |
ENST00000454817.1
ENST00000328333.8 |
COL7A1
|
collagen, type VII, alpha 1 |
| chr9_+_19230433 | 1.93 |
ENST00000434457.2
ENST00000602925.1 |
DENND4C
|
DENN/MADD domain containing 4C |
| chr11_+_118478313 | 1.89 |
ENST00000356063.5
|
PHLDB1
|
pleckstrin homology-like domain, family B, member 1 |
| chr5_-_151304337 | 1.80 |
ENST00000455880.2
ENST00000545569.1 ENST00000274576.4 |
GLRA1
|
glycine receptor, alpha 1 |
| chr9_+_35605274 | 1.79 |
ENST00000336395.5
|
TESK1
|
testis-specific kinase 1 |
| chrX_+_152783131 | 1.75 |
ENST00000349466.2
ENST00000370186.1 |
ATP2B3
|
ATPase, Ca++ transporting, plasma membrane 3 |
| chr2_+_241375069 | 1.75 |
ENST00000264039.2
|
GPC1
|
glypican 1 |
| chr2_+_233243233 | 1.70 |
ENST00000392027.2
|
ALPP
|
alkaline phosphatase, placental |
| chr2_+_74741569 | 1.69 |
ENST00000233638.7
|
TLX2
|
T-cell leukemia homeobox 2 |
| chr16_+_69600058 | 1.69 |
ENST00000393742.2
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive |
| chr6_+_99282570 | 1.46 |
ENST00000328345.5
|
POU3F2
|
POU class 3 homeobox 2 |
| chr2_-_241831424 | 1.44 |
ENST00000402775.2
ENST00000307486.8 |
C2orf54
|
chromosome 2 open reading frame 54 |
| chr4_-_101439242 | 1.41 |
ENST00000296420.4
|
EMCN
|
endomucin |
| chr16_+_69599899 | 1.39 |
ENST00000567239.1
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive |
| chr3_+_110790590 | 1.38 |
ENST00000485303.1
|
PVRL3
|
poliovirus receptor-related 3 |
| chr3_-_136471204 | 1.17 |
ENST00000480733.1
ENST00000383202.2 ENST00000236698.5 ENST00000434713.2 |
STAG1
|
stromal antigen 1 |
| chr6_+_13272904 | 1.12 |
ENST00000379335.3
ENST00000379329.1 |
PHACTR1
|
phosphatase and actin regulator 1 |
| chr14_-_25519317 | 0.94 |
ENST00000323944.5
|
STXBP6
|
syntaxin binding protein 6 (amisyn) |
| chr6_+_292051 | 0.89 |
ENST00000344450.5
|
DUSP22
|
dual specificity phosphatase 22 |
| chrX_+_115567767 | 0.86 |
ENST00000371900.4
|
SLC6A14
|
solute carrier family 6 (amino acid transporter), member 14 |
| chr11_-_82612727 | 0.82 |
ENST00000531128.1
ENST00000535099.1 ENST00000527444.1 |
PRCP
|
prolylcarboxypeptidase (angiotensinase C) |
| chr19_+_50936142 | 0.76 |
ENST00000357701.5
|
MYBPC2
|
myosin binding protein C, fast type |
| chr12_-_48213568 | 0.73 |
ENST00000080059.7
ENST00000354334.3 ENST00000430670.1 ENST00000552960.1 ENST00000440293.1 |
HDAC7
|
histone deacetylase 7 |
| chr22_+_31608219 | 0.43 |
ENST00000406516.1
ENST00000444929.2 ENST00000331728.4 |
LIMK2
|
LIM domain kinase 2 |
| chr17_+_37824411 | 0.43 |
ENST00000269582.2
|
PNMT
|
phenylethanolamine N-methyltransferase |
| chr8_+_17780346 | 0.34 |
ENST00000325083.8
|
PCM1
|
pericentriolar material 1 |
| chr17_+_37824700 | 0.32 |
ENST00000581428.1
|
PNMT
|
phenylethanolamine N-methyltransferase |
| chr17_+_37824217 | 0.24 |
ENST00000394246.1
|
PNMT
|
phenylethanolamine N-methyltransferase |
| chr1_-_154531095 | 0.14 |
ENST00000292211.4
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1 |
| chr9_+_136501478 | 0.11 |
ENST00000393056.2
ENST00000263611.2 |
DBH
|
dopamine beta-hydroxylase (dopamine beta-monooxygenase) |
| chr19_+_49458107 | 0.02 |
ENST00000539787.1
ENST00000345358.7 ENST00000391871.3 ENST00000415969.2 ENST00000354470.3 ENST00000506183.1 ENST00000293288.8 |
BAX
|
BCL2-associated X protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of TFAP2E
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.8 | 23.4 | GO:0097212 | cadmium ion homeostasis(GO:0055073) lysosomal membrane organization(GO:0097212) negative regulation of hydrogen peroxide catabolic process(GO:2000296) regulation of oxygen metabolic process(GO:2000374) |
| 7.5 | 37.6 | GO:0033274 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 4.1 | 16.4 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 4.0 | 11.9 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 3.8 | 15.4 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 3.4 | 13.8 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 3.4 | 20.5 | GO:1900239 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 3.1 | 9.3 | GO:0060620 | regulation of cholesterol import(GO:0060620) negative regulation of cholesterol import(GO:0060621) regulation of sterol import(GO:2000909) negative regulation of sterol import(GO:2000910) |
| 2.4 | 12.2 | GO:1904637 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 2.4 | 9.4 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 1.6 | 4.8 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) type B pancreatic cell maturation(GO:0072560) |
| 1.6 | 7.9 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 1.3 | 3.8 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 1.2 | 23.0 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 1.0 | 39.3 | GO:0060384 | innervation(GO:0060384) |
| 1.0 | 4.1 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 1.0 | 3.8 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.9 | 3.6 | GO:0044467 | glial cell-derived neurotrophic factor secretion(GO:0044467) regulation of glial cell-derived neurotrophic factor secretion(GO:1900166) positive regulation of glial cell-derived neurotrophic factor secretion(GO:1900168) |
| 0.8 | 4.2 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.8 | 8.8 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.7 | 2.2 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.7 | 2.9 | GO:0032053 | ciliary basal body organization(GO:0032053) positive regulation of protein localization to cilium(GO:1903566) |
| 0.7 | 2.8 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.7 | 4.1 | GO:0002725 | negative regulation of T cell cytokine production(GO:0002725) |
| 0.7 | 5.4 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.7 | 7.4 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.6 | 5.2 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.6 | 3.9 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.6 | 4.6 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.4 | 3.5 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.4 | 6.4 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.4 | 2.3 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.4 | 9.9 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.3 | 1.7 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.3 | 1.7 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.3 | 2.4 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.3 | 15.9 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.3 | 6.7 | GO:0030539 | male genitalia development(GO:0030539) |
| 0.3 | 1.8 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.3 | 3.5 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.3 | 4.4 | GO:1902260 | diaphragm development(GO:0060539) negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.3 | 2.5 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.2 | 1.0 | GO:0042418 | epinephrine biosynthetic process(GO:0042418) |
| 0.2 | 1.5 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.2 | 3.3 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.2 | 2.6 | GO:0007320 | insemination(GO:0007320) |
| 0.2 | 0.8 | GO:0002155 | regulation of thyroid hormone mediated signaling pathway(GO:0002155) kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 0.2 | 9.9 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.2 | 1.9 | GO:2001197 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.2 | 2.7 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.1 | 1.7 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.1 | 0.9 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 8.3 | GO:0035418 | protein localization to synapse(GO:0035418) |
| 0.1 | 7.6 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 0.9 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.1 | 0.7 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 3.9 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 2.7 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.1 | 3.0 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 0.8 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 2.5 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 2.0 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 0.1 | GO:0042421 | norepinephrine biosynthetic process(GO:0042421) |
| 0.0 | 3.6 | GO:0008584 | male gonad development(GO:0008584) development of primary male sexual characteristics(GO:0046546) |
| 0.0 | 0.9 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.1 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.0 | 3.6 | GO:0060326 | cell chemotaxis(GO:0060326) |
| 0.0 | 1.2 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 9.5 | GO:0016567 | protein ubiquitination(GO:0016567) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 23.4 | GO:0097449 | astrocyte projection(GO:0097449) |
| 1.5 | 13.8 | GO:0005955 | calcineurin complex(GO:0005955) |
| 1.4 | 39.3 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 1.2 | 9.3 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 1.0 | 15.9 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.6 | 4.4 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.5 | 13.7 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.5 | 2.8 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.4 | 9.4 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.3 | 2.9 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.3 | 2.5 | GO:0034464 | BBSome(GO:0034464) |
| 0.3 | 12.0 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.3 | 4.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 1.9 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.2 | 2.7 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.2 | 26.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.2 | 3.8 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.2 | 5.4 | GO:0030673 | axolemma(GO:0030673) |
| 0.2 | 3.5 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.2 | 1.8 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 0.9 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 3.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 16.4 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 20.5 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.1 | 3.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 7.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 2.0 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.0 | 3.6 | GO:0016328 | lateral plasma membrane(GO:0016328) ciliary membrane(GO:0060170) |
| 0.0 | 9.1 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 3.5 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 2.3 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 8.8 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 0.1 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 2.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.8 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 11.9 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 21.5 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 0.8 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 31.7 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.9 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 3.0 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 7.8 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.2 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.5 | 37.6 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 4.0 | 11.9 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 3.4 | 13.8 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 2.3 | 9.3 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 1.9 | 39.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 1.8 | 23.4 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 1.4 | 4.2 | GO:0034618 | arginine binding(GO:0034618) |
| 1.3 | 9.4 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 1.3 | 9.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 1.2 | 3.5 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.9 | 3.6 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.9 | 12.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.9 | 20.5 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.8 | 8.8 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.8 | 5.4 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.6 | 5.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.6 | 15.4 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.6 | 9.9 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.5 | 3.6 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.5 | 2.4 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.4 | 15.9 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.4 | 4.1 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.4 | 1.8 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.3 | 2.8 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.3 | 1.0 | GO:0004603 | phenylethanolamine N-methyltransferase activity(GO:0004603) |
| 0.3 | 6.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.3 | 7.9 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.2 | 5.2 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.2 | 4.4 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.2 | 2.2 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 5.4 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 26.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 3.8 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 1.7 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.9 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 2.9 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 8.4 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 0.7 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.8 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 8.4 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 5.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 12.0 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.9 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 4.3 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 1.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 1.5 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 4.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.8 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 3.9 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 3.0 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 5.6 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 1.8 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 7.2 | GO:0005088 | Ras guanyl-nucleotide exchange factor activity(GO:0005088) |
| 0.0 | 0.9 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 16.5 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.4 | 20.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.4 | 7.9 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.3 | 4.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 11.0 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 3.6 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 1.7 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 3.9 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 3.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 2.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.7 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 1.4 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 2.7 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 7.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 4.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 4.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.7 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 39.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.8 | 9.9 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.7 | 9.9 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.5 | 37.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.5 | 9.4 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.4 | 9.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.4 | 20.5 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.4 | 15.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.4 | 7.9 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.3 | 23.0 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.3 | 4.6 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.2 | 4.1 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.2 | 1.8 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.2 | 4.8 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 6.2 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 6.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 4.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 3.6 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 1.7 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 1.7 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 1.8 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 3.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 3.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.1 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 1.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 3.8 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.8 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 6.6 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.2 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |