Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
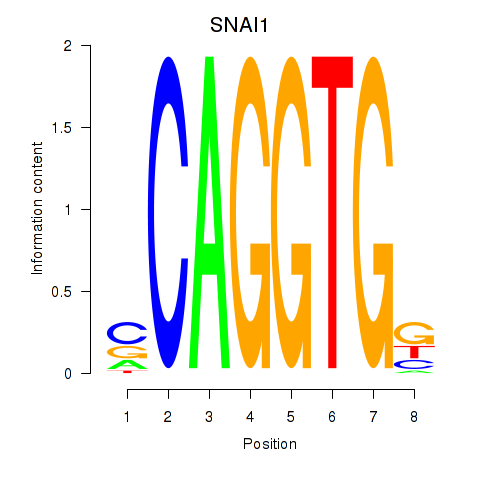
Results for TWIST1_SNAI1
Z-value: 0.12
Transcription factors associated with TWIST1_SNAI1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TWIST1
|
ENSG00000122691.8 | twist family bHLH transcription factor 1 |
|
SNAI1
|
ENSG00000124216.3 | snail family transcriptional repressor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TWIST1 | hg19_v2_chr7_-_19157248_19157295 | -0.02 | 7.9e-01 | Click! |
| SNAI1 | hg19_v2_chr20_+_48599506_48599536 | 0.02 | 8.3e-01 | Click! |
Activity profile of TWIST1_SNAI1 motif
Sorted Z-values of TWIST1_SNAI1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_175870085 | 13.93 |
ENST00000409156.3
|
CHN1
|
chimerin 1 |
| chr19_+_36359341 | 12.88 |
ENST00000221891.4
|
APLP1
|
amyloid beta (A4) precursor-like protein 1 |
| chr9_+_17579084 | 12.43 |
ENST00000380607.4
|
SH3GL2
|
SH3-domain GRB2-like 2 |
| chr3_-_149688896 | 12.33 |
ENST00000239940.7
|
PFN2
|
profilin 2 |
| chr12_+_52626898 | 11.15 |
ENST00000331817.5
|
KRT7
|
keratin 7 |
| chr17_-_57184260 | 10.43 |
ENST00000376149.3
ENST00000393066.3 |
TRIM37
|
tripartite motif containing 37 |
| chr12_+_53342625 | 9.94 |
ENST00000388837.2
ENST00000550600.1 ENST00000388835.3 |
KRT18
|
keratin 18 |
| chr6_-_3227877 | 8.77 |
ENST00000259818.7
|
TUBB2B
|
tubulin, beta 2B class IIb |
| chr17_+_52978156 | 8.29 |
ENST00000348161.4
|
TOM1L1
|
target of myb1 (chicken)-like 1 |
| chr18_+_29077990 | 8.24 |
ENST00000261590.8
|
DSG2
|
desmoglein 2 |
| chr18_-_71959159 | 7.65 |
ENST00000494131.2
ENST00000397914.4 ENST00000340533.4 |
CYB5A
|
cytochrome b5 type A (microsomal) |
| chr20_-_60942361 | 7.59 |
ENST00000252999.3
|
LAMA5
|
laminin, alpha 5 |
| chr5_-_132112921 | 7.47 |
ENST00000378721.4
ENST00000378701.1 |
SEPT8
|
septin 8 |
| chr2_+_47596287 | 7.47 |
ENST00000263735.4
|
EPCAM
|
epithelial cell adhesion molecule |
| chr12_+_6309517 | 6.76 |
ENST00000382519.4
ENST00000009180.4 |
CD9
|
CD9 molecule |
| chr16_-_74808710 | 6.66 |
ENST00000219368.3
ENST00000544337.1 |
FA2H
|
fatty acid 2-hydroxylase |
| chr15_+_63340647 | 6.42 |
ENST00000404484.4
|
TPM1
|
tropomyosin 1 (alpha) |
| chr4_+_166300084 | 6.36 |
ENST00000402744.4
|
CPE
|
carboxypeptidase E |
| chr3_-_149688502 | 6.12 |
ENST00000481767.1
ENST00000475518.1 |
PFN2
|
profilin 2 |
| chr19_+_54926601 | 6.01 |
ENST00000301194.4
|
TTYH1
|
tweety family member 1 |
| chr20_+_17550691 | 6.00 |
ENST00000474024.1
|
DSTN
|
destrin (actin depolymerizing factor) |
| chr14_+_104029278 | 5.90 |
ENST00000472726.2
ENST00000409074.2 ENST00000440963.1 ENST00000556253.2 ENST00000247618.4 |
RP11-73M18.2
APOPT1
|
Kinesin light chain 1 apoptogenic 1, mitochondrial |
| chr11_+_86511569 | 5.89 |
ENST00000441050.1
|
PRSS23
|
protease, serine, 23 |
| chr3_-_149688655 | 5.84 |
ENST00000461930.1
ENST00000423691.2 ENST00000490975.1 ENST00000461868.1 ENST00000452853.2 |
PFN2
|
profilin 2 |
| chr17_-_39684550 | 5.75 |
ENST00000455635.1
ENST00000361566.3 |
KRT19
|
keratin 19 |
| chr19_+_54926621 | 5.69 |
ENST00000376530.3
ENST00000445095.1 ENST00000391739.3 ENST00000376531.3 |
TTYH1
|
tweety family member 1 |
| chr6_+_7541808 | 5.69 |
ENST00000379802.3
|
DSP
|
desmoplakin |
| chr5_-_132113083 | 5.55 |
ENST00000296873.7
|
SEPT8
|
septin 8 |
| chr19_+_35783047 | 5.49 |
ENST00000595791.1
ENST00000597035.1 ENST00000537831.2 |
MAG
|
myelin associated glycoprotein |
| chr6_+_7541845 | 5.49 |
ENST00000418664.2
|
DSP
|
desmoplakin |
| chr19_+_35630926 | 5.48 |
ENST00000588081.1
ENST00000589121.1 |
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr17_-_42992856 | 5.40 |
ENST00000588316.1
ENST00000435360.2 ENST00000586793.1 ENST00000588735.1 ENST00000588037.1 ENST00000592320.1 ENST00000253408.5 |
GFAP
|
glial fibrillary acidic protein |
| chr17_-_57184064 | 5.21 |
ENST00000262294.7
|
TRIM37
|
tripartite motif containing 37 |
| chr15_+_63334831 | 5.16 |
ENST00000288398.6
ENST00000358278.3 ENST00000560445.1 ENST00000403994.3 ENST00000357980.4 ENST00000559556.1 ENST00000559397.1 ENST00000267996.7 ENST00000560970.1 |
TPM1
|
tropomyosin 1 (alpha) |
| chr19_+_35630628 | 5.14 |
ENST00000588715.1
ENST00000588607.1 |
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr5_-_176056974 | 5.02 |
ENST00000510387.1
ENST00000506696.1 |
SNCB
|
synuclein, beta |
| chr15_+_63340858 | 4.95 |
ENST00000560615.1
|
TPM1
|
tropomyosin 1 (alpha) |
| chr19_+_19322758 | 4.95 |
ENST00000252575.6
|
NCAN
|
neurocan |
| chr2_-_106054952 | 4.94 |
ENST00000336660.5
ENST00000393352.3 ENST00000607522.1 |
FHL2
|
four and a half LIM domains 2 |
| chr1_-_156675368 | 4.83 |
ENST00000368222.3
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr19_+_35783037 | 4.79 |
ENST00000361922.4
|
MAG
|
myelin associated glycoprotein |
| chr7_+_116166331 | 4.77 |
ENST00000393468.1
ENST00000393467.1 |
CAV1
|
caveolin 1, caveolae protein, 22kDa |
| chr2_-_175869936 | 4.75 |
ENST00000409900.3
|
CHN1
|
chimerin 1 |
| chrX_+_114795489 | 4.74 |
ENST00000355899.3
ENST00000537301.1 ENST00000289290.3 |
PLS3
|
plastin 3 |
| chr15_+_63340734 | 4.65 |
ENST00000560959.1
|
TPM1
|
tropomyosin 1 (alpha) |
| chr3_-_134093275 | 4.65 |
ENST00000513145.1
ENST00000422605.2 |
AMOTL2
|
angiomotin like 2 |
| chr9_+_133320301 | 4.65 |
ENST00000352480.5
|
ASS1
|
argininosuccinate synthase 1 |
| chr9_+_133320339 | 4.62 |
ENST00000372394.1
ENST00000372393.3 ENST00000422569.1 |
ASS1
|
argininosuccinate synthase 1 |
| chr7_+_73242069 | 4.59 |
ENST00000435050.1
|
CLDN4
|
claudin 4 |
| chr2_+_17721920 | 4.57 |
ENST00000295156.4
|
VSNL1
|
visinin-like 1 |
| chr3_+_167453493 | 4.37 |
ENST00000295777.5
ENST00000472747.2 |
SERPINI1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr1_-_156675535 | 4.36 |
ENST00000368221.1
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr8_+_22429205 | 4.32 |
ENST00000520207.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr19_+_10217270 | 4.17 |
ENST00000446223.1
|
PPAN
|
peter pan homolog (Drosophila) |
| chr2_+_201170596 | 4.12 |
ENST00000439084.1
ENST00000409718.1 |
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr11_-_119293872 | 4.09 |
ENST00000524970.1
|
THY1
|
Thy-1 cell surface antigen |
| chr15_+_75287861 | 4.07 |
ENST00000425597.3
ENST00000562327.1 ENST00000568018.1 ENST00000562212.1 ENST00000567920.1 ENST00000566872.1 ENST00000361900.6 ENST00000545456.1 |
SCAMP5
|
secretory carrier membrane protein 5 |
| chr17_+_52978107 | 4.07 |
ENST00000445275.2
|
TOM1L1
|
target of myb1 (chicken)-like 1 |
| chr7_+_73242490 | 4.05 |
ENST00000431918.1
|
CLDN4
|
claudin 4 |
| chr7_+_116165754 | 4.04 |
ENST00000405348.1
|
CAV1
|
caveolin 1, caveolae protein, 22kDa |
| chr17_-_7165662 | 4.02 |
ENST00000571881.2
ENST00000360325.7 |
CLDN7
|
claudin 7 |
| chr19_-_51471362 | 3.94 |
ENST00000376853.4
ENST00000424910.2 |
KLK6
|
kallikrein-related peptidase 6 |
| chr19_-_51471381 | 3.94 |
ENST00000594641.1
|
KLK6
|
kallikrein-related peptidase 6 |
| chr22_-_36220420 | 3.89 |
ENST00000473487.2
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr3_+_111717600 | 3.88 |
ENST00000273368.4
|
TAGLN3
|
transgelin 3 |
| chr19_-_13068012 | 3.85 |
ENST00000316939.1
|
GADD45GIP1
|
growth arrest and DNA-damage-inducible, gamma interacting protein 1 |
| chr15_+_63340553 | 3.85 |
ENST00000334895.5
|
TPM1
|
tropomyosin 1 (alpha) |
| chr16_+_330448 | 3.83 |
ENST00000447871.1
|
ARHGDIG
|
Rho GDP dissociation inhibitor (GDI) gamma |
| chr1_-_113249948 | 3.82 |
ENST00000339083.7
ENST00000369642.3 |
RHOC
|
ras homolog family member C |
| chr8_-_81083890 | 3.82 |
ENST00000518937.1
|
TPD52
|
tumor protein D52 |
| chr11_-_6502534 | 3.81 |
ENST00000254584.2
ENST00000525235.1 ENST00000445086.2 |
ARFIP2
|
ADP-ribosylation factor interacting protein 2 |
| chr2_+_201170770 | 3.79 |
ENST00000409988.3
ENST00000409385.1 |
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr20_+_17550489 | 3.79 |
ENST00000246069.7
|
DSTN
|
destrin (actin depolymerizing factor) |
| chr16_+_330581 | 3.78 |
ENST00000219409.3
|
ARHGDIG
|
Rho GDP dissociation inhibitor (GDI) gamma |
| chr2_-_71454185 | 3.75 |
ENST00000244221.8
|
PAIP2B
|
poly(A) binding protein interacting protein 2B |
| chr22_+_31489344 | 3.74 |
ENST00000404574.1
|
SMTN
|
smoothelin |
| chr3_+_111718036 | 3.71 |
ENST00000455401.2
|
TAGLN3
|
transgelin 3 |
| chr7_+_75677465 | 3.67 |
ENST00000432020.2
|
MDH2
|
malate dehydrogenase 2, NAD (mitochondrial) |
| chr19_+_10217364 | 3.67 |
ENST00000430370.1
|
PPAN
|
peter pan homolog (Drosophila) |
| chr15_+_41136586 | 3.66 |
ENST00000431806.1
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr5_+_76506706 | 3.65 |
ENST00000340978.3
ENST00000346042.3 ENST00000264917.5 ENST00000342343.4 ENST00000333194.4 |
PDE8B
|
phosphodiesterase 8B |
| chr12_+_125549925 | 3.65 |
ENST00000316519.6
|
AACS
|
acetoacetyl-CoA synthetase |
| chr1_-_113249678 | 3.63 |
ENST00000369633.2
ENST00000425265.2 ENST00000369632.2 ENST00000436685.2 |
RHOC
|
ras homolog family member C |
| chr11_-_61646054 | 3.58 |
ENST00000527379.1
|
FADS3
|
fatty acid desaturase 3 |
| chr11_+_66824276 | 3.57 |
ENST00000308831.2
|
RHOD
|
ras homolog family member D |
| chr19_+_35532612 | 3.56 |
ENST00000600390.1
ENST00000597419.1 |
HPN
|
hepsin |
| chr3_-_111314230 | 3.55 |
ENST00000317012.4
|
ZBED2
|
zinc finger, BED-type containing 2 |
| chr11_+_114168085 | 3.52 |
ENST00000541754.1
|
NNMT
|
nicotinamide N-methyltransferase |
| chr7_+_75677354 | 3.49 |
ENST00000461263.2
ENST00000315758.5 ENST00000443006.1 |
MDH2
|
malate dehydrogenase 2, NAD (mitochondrial) |
| chr7_+_16793160 | 3.48 |
ENST00000262067.4
|
TSPAN13
|
tetraspanin 13 |
| chr14_+_105331596 | 3.45 |
ENST00000556508.1
ENST00000414716.3 ENST00000453495.1 ENST00000418279.1 |
CEP170B
|
centrosomal protein 170B |
| chr19_+_38755042 | 3.40 |
ENST00000301244.7
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2 |
| chr7_+_130020180 | 3.34 |
ENST00000481342.1
ENST00000011292.3 ENST00000604896.1 |
CPA1
|
carboxypeptidase A1 (pancreatic) |
| chr17_-_78450398 | 3.32 |
ENST00000306773.4
|
NPTX1
|
neuronal pentraxin I |
| chr17_-_73844722 | 3.30 |
ENST00000586257.1
|
WBP2
|
WW domain binding protein 2 |
| chr16_+_318638 | 3.27 |
ENST00000412541.1
ENST00000435035.1 |
ARHGDIG
|
Rho GDP dissociation inhibitor (GDI) gamma |
| chr2_+_173600514 | 3.27 |
ENST00000264111.6
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr12_+_57984965 | 3.25 |
ENST00000540759.2
ENST00000551772.1 ENST00000550465.1 ENST00000354947.5 |
PIP4K2C
|
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma |
| chr3_-_124606074 | 3.20 |
ENST00000296181.4
|
ITGB5
|
integrin, beta 5 |
| chr12_+_6309963 | 3.16 |
ENST00000382515.2
|
CD9
|
CD9 molecule |
| chr5_-_42811986 | 3.14 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr12_+_125549973 | 3.12 |
ENST00000536752.1
ENST00000261686.6 |
AACS
|
acetoacetyl-CoA synthetase |
| chr1_+_99127265 | 3.07 |
ENST00000306121.3
|
SNX7
|
sorting nexin 7 |
| chr19_-_7939319 | 3.06 |
ENST00000539422.1
|
CTD-3193O13.9
|
Protein FLJ22184 |
| chr8_+_22436248 | 3.03 |
ENST00000308354.7
|
PDLIM2
|
PDZ and LIM domain 2 (mystique) |
| chr12_+_79258547 | 3.02 |
ENST00000457153.2
|
SYT1
|
synaptotagmin I |
| chrX_+_105969893 | 3.02 |
ENST00000255499.2
|
RNF128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr13_+_110959598 | 3.02 |
ENST00000360467.5
|
COL4A2
|
collagen, type IV, alpha 2 |
| chr1_-_24126023 | 3.02 |
ENST00000429356.1
|
GALE
|
UDP-galactose-4-epimerase |
| chr3_+_50712672 | 3.00 |
ENST00000266037.9
|
DOCK3
|
dedicator of cytokinesis 3 |
| chr2_-_190044480 | 2.99 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr1_+_19923454 | 2.94 |
ENST00000602662.1
ENST00000602293.1 ENST00000322753.6 |
MINOS1-NBL1
MINOS1
|
MINOS1-NBL1 readthrough mitochondrial inner membrane organizing system 1 |
| chr15_+_63340775 | 2.92 |
ENST00000559281.1
ENST00000317516.7 |
TPM1
|
tropomyosin 1 (alpha) |
| chr8_+_22438009 | 2.91 |
ENST00000409417.1
|
PDLIM2
|
PDZ and LIM domain 2 (mystique) |
| chr8_-_110656995 | 2.90 |
ENST00000276646.9
ENST00000533065.1 |
SYBU
|
syntabulin (syntaxin-interacting) |
| chr11_-_65667997 | 2.90 |
ENST00000312562.2
ENST00000534222.1 |
FOSL1
|
FOS-like antigen 1 |
| chr1_+_15256230 | 2.89 |
ENST00000376028.4
ENST00000400798.2 |
KAZN
|
kazrin, periplakin interacting protein |
| chr16_+_14980632 | 2.85 |
ENST00000565655.1
|
NOMO1
|
NODAL modulator 1 |
| chr10_-_118764862 | 2.84 |
ENST00000260777.10
|
KIAA1598
|
KIAA1598 |
| chr15_-_71055878 | 2.84 |
ENST00000322954.6
|
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr1_+_87794150 | 2.84 |
ENST00000370544.5
|
LMO4
|
LIM domain only 4 |
| chrX_+_134166333 | 2.77 |
ENST00000257013.7
|
FAM127A
|
family with sequence similarity 127, member A |
| chr4_+_4387983 | 2.77 |
ENST00000397958.1
|
NSG1
|
Homo sapiens neuron specific gene family member 1 (NSG1), transcript variant 3, mRNA. |
| chr1_-_95392635 | 2.74 |
ENST00000538964.1
ENST00000394202.4 ENST00000370206.4 |
CNN3
|
calponin 3, acidic |
| chr7_-_73184588 | 2.74 |
ENST00000395145.2
|
CLDN3
|
claudin 3 |
| chr1_+_236558694 | 2.72 |
ENST00000359362.5
|
EDARADD
|
EDAR-associated death domain |
| chr5_-_132112907 | 2.71 |
ENST00000458488.2
|
SEPT8
|
septin 8 |
| chr3_-_120170052 | 2.71 |
ENST00000295633.3
|
FSTL1
|
follistatin-like 1 |
| chr3_-_62861012 | 2.70 |
ENST00000357948.3
ENST00000383710.4 |
CADPS
|
Ca++-dependent secretion activator |
| chr1_-_95007193 | 2.68 |
ENST00000370207.4
ENST00000334047.7 |
F3
|
coagulation factor III (thromboplastin, tissue factor) |
| chr3_-_149095652 | 2.63 |
ENST00000305366.3
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr6_-_84419101 | 2.62 |
ENST00000520302.1
ENST00000520213.1 ENST00000439399.2 ENST00000428679.2 ENST00000437520.1 |
SNAP91
|
synaptosomal-associated protein, 91kDa |
| chr1_-_40782938 | 2.61 |
ENST00000372736.3
ENST00000372748.3 |
COL9A2
|
collagen, type IX, alpha 2 |
| chr11_-_66115032 | 2.60 |
ENST00000311181.4
|
B3GNT1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1 |
| chr9_+_80912059 | 2.59 |
ENST00000347159.2
ENST00000376588.3 |
PSAT1
|
phosphoserine aminotransferase 1 |
| chr19_+_35783028 | 2.58 |
ENST00000600291.1
ENST00000392213.3 |
MAG
|
myelin associated glycoprotein |
| chr1_-_201368707 | 2.57 |
ENST00000391967.2
|
LAD1
|
ladinin 1 |
| chr12_-_63328817 | 2.56 |
ENST00000228705.6
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H |
| chr15_+_51973680 | 2.56 |
ENST00000542355.2
|
SCG3
|
secretogranin III |
| chr16_-_19897455 | 2.56 |
ENST00000568214.1
ENST00000569479.1 |
GPRC5B
|
G protein-coupled receptor, family C, group 5, member B |
| chr1_-_183387723 | 2.54 |
ENST00000287713.6
|
NMNAT2
|
nicotinamide nucleotide adenylyltransferase 2 |
| chr7_+_76054224 | 2.52 |
ENST00000394857.3
|
ZP3
|
zona pellucida glycoprotein 3 (sperm receptor) |
| chr19_+_35609380 | 2.52 |
ENST00000604621.1
|
FXYD3
|
FXYD domain containing ion transport regulator 3 |
| chr13_+_113633620 | 2.52 |
ENST00000421756.1
ENST00000375601.3 |
MCF2L
|
MCF.2 cell line derived transforming sequence-like |
| chr8_-_26371608 | 2.51 |
ENST00000522362.2
|
PNMA2
|
paraneoplastic Ma antigen 2 |
| chr15_+_51973550 | 2.51 |
ENST00000220478.3
|
SCG3
|
secretogranin III |
| chr10_+_18429671 | 2.49 |
ENST00000282343.8
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr16_+_3068393 | 2.48 |
ENST00000573001.1
|
TNFRSF12A
|
tumor necrosis factor receptor superfamily, member 12A |
| chr18_-_812517 | 2.47 |
ENST00000584307.1
|
YES1
|
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 |
| chr13_+_113622810 | 2.47 |
ENST00000397030.1
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like |
| chr4_+_113739244 | 2.47 |
ENST00000503271.1
ENST00000503423.1 ENST00000506722.1 |
ANK2
|
ankyrin 2, neuronal |
| chr1_+_1981890 | 2.46 |
ENST00000378567.3
ENST00000468310.1 |
PRKCZ
|
protein kinase C, zeta |
| chr16_-_67493110 | 2.46 |
ENST00000602876.1
|
ATP6V0D1
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d1 |
| chr3_+_172468472 | 2.44 |
ENST00000232458.5
ENST00000392692.3 |
ECT2
|
epithelial cell transforming sequence 2 oncogene |
| chr20_+_19867150 | 2.43 |
ENST00000255006.6
|
RIN2
|
Ras and Rab interactor 2 |
| chr7_+_872107 | 2.43 |
ENST00000405266.1
ENST00000401592.1 ENST00000403868.1 ENST00000425407.2 |
SUN1
|
Sad1 and UNC84 domain containing 1 |
| chrX_-_13956497 | 2.42 |
ENST00000398361.3
|
GPM6B
|
glycoprotein M6B |
| chr3_-_127541679 | 2.40 |
ENST00000265052.5
|
MGLL
|
monoglyceride lipase |
| chr14_+_90863327 | 2.39 |
ENST00000356978.4
|
CALM1
|
calmodulin 1 (phosphorylase kinase, delta) |
| chr11_-_65667884 | 2.37 |
ENST00000448083.2
ENST00000531493.1 ENST00000532401.1 |
FOSL1
|
FOS-like antigen 1 |
| chr2_-_9770706 | 2.35 |
ENST00000381844.4
|
YWHAQ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta |
| chr10_+_75910960 | 2.34 |
ENST00000539909.1
ENST00000286621.2 |
ADK
|
adenosine kinase |
| chr8_+_104152922 | 2.33 |
ENST00000309982.5
ENST00000438105.2 ENST00000297574.6 |
BAALC
|
brain and acute leukemia, cytoplasmic |
| chr1_-_150669500 | 2.31 |
ENST00000271732.3
|
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr12_-_50677255 | 2.30 |
ENST00000551691.1
ENST00000394943.3 ENST00000341247.4 |
LIMA1
|
LIM domain and actin binding 1 |
| chr17_+_39411636 | 2.30 |
ENST00000394008.1
|
KRTAP9-9
|
keratin associated protein 9-9 |
| chr3_-_48481434 | 2.30 |
ENST00000395694.2
ENST00000447018.1 ENST00000442740.1 |
CCDC51
|
coiled-coil domain containing 51 |
| chr16_-_66785497 | 2.29 |
ENST00000440564.2
ENST00000379482.2 ENST00000443351.2 ENST00000566150.1 |
DYNC1LI2
|
dynein, cytoplasmic 1, light intermediate chain 2 |
| chr18_-_812231 | 2.28 |
ENST00000314574.4
|
YES1
|
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 |
| chrX_+_47082408 | 2.28 |
ENST00000518022.1
ENST00000276052.6 |
CDK16
|
cyclin-dependent kinase 16 |
| chr1_+_35247859 | 2.27 |
ENST00000373362.3
|
GJB3
|
gap junction protein, beta 3, 31kDa |
| chr4_+_166128735 | 2.26 |
ENST00000226725.6
|
KLHL2
|
kelch-like family member 2 |
| chr5_+_94890778 | 2.25 |
ENST00000380009.4
|
ARSK
|
arylsulfatase family, member K |
| chrX_-_13956737 | 2.24 |
ENST00000454189.2
|
GPM6B
|
glycoprotein M6B |
| chr1_+_156030937 | 2.24 |
ENST00000361084.5
|
RAB25
|
RAB25, member RAS oncogene family |
| chr16_+_68679193 | 2.22 |
ENST00000581171.1
|
CDH3
|
cadherin 3, type 1, P-cadherin (placental) |
| chr7_+_73245193 | 2.21 |
ENST00000340958.2
|
CLDN4
|
claudin 4 |
| chr1_+_202995611 | 2.20 |
ENST00000367240.2
|
PPFIA4
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 4 |
| chr1_+_33352036 | 2.20 |
ENST00000373467.3
|
HPCA
|
hippocalcin |
| chr1_+_233749739 | 2.20 |
ENST00000366621.3
|
KCNK1
|
potassium channel, subfamily K, member 1 |
| chr8_+_26371763 | 2.18 |
ENST00000521913.1
|
DPYSL2
|
dihydropyrimidinase-like 2 |
| chr7_-_122526499 | 2.17 |
ENST00000412584.2
|
CADPS2
|
Ca++-dependent secretion activator 2 |
| chr15_+_68924327 | 2.16 |
ENST00000543950.1
|
CORO2B
|
coronin, actin binding protein, 2B |
| chr2_+_173600565 | 2.15 |
ENST00000397081.3
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr8_-_22089533 | 2.15 |
ENST00000321613.3
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein |
| chr19_+_38755203 | 2.14 |
ENST00000587090.1
ENST00000454580.3 |
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2 |
| chr1_-_201368653 | 2.13 |
ENST00000367313.3
|
LAD1
|
ladinin 1 |
| chr7_-_75677251 | 2.12 |
ENST00000431581.1
ENST00000359697.3 ENST00000451157.1 ENST00000340062.5 ENST00000360591.3 ENST00000248600.1 |
STYXL1
|
serine/threonine/tyrosine interacting-like 1 |
| chr5_-_16936340 | 2.12 |
ENST00000507288.1
ENST00000513610.1 |
MYO10
|
myosin X |
| chr4_+_169418195 | 2.12 |
ENST00000261509.6
ENST00000335742.7 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr17_+_49230897 | 2.11 |
ENST00000393196.3
ENST00000336097.3 ENST00000480143.1 ENST00000511355.1 ENST00000013034.3 ENST00000393198.3 ENST00000608447.1 ENST00000393193.2 ENST00000376392.6 ENST00000555572.1 |
NME1
NME1-NME2
NME2
|
NME/NM23 nucleoside diphosphate kinase 1 NME1-NME2 readthrough NME/NM23 nucleoside diphosphate kinase 2 |
| chr16_-_4987065 | 2.10 |
ENST00000590782.2
ENST00000345988.2 |
PPL
|
periplakin |
| chr16_-_21289627 | 2.09 |
ENST00000396023.2
ENST00000415987.2 |
CRYM
|
crystallin, mu |
| chr10_-_126849588 | 2.06 |
ENST00000411419.2
|
CTBP2
|
C-terminal binding protein 2 |
| chr15_-_52587945 | 2.03 |
ENST00000443683.2
ENST00000558479.1 ENST00000261839.7 |
MYO5C
|
myosin VC |
| chr19_-_10697895 | 2.02 |
ENST00000591240.1
ENST00000589684.1 ENST00000591676.1 ENST00000250244.6 ENST00000590923.1 |
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr17_-_39677971 | 2.02 |
ENST00000393976.2
|
KRT15
|
keratin 15 |
| chr11_+_27062502 | 2.01 |
ENST00000263182.3
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr6_-_30043539 | 2.01 |
ENST00000376751.3
ENST00000244360.6 |
RNF39
|
ring finger protein 39 |
| chr3_-_48481518 | 1.99 |
ENST00000412398.2
ENST00000395696.1 |
CCDC51
|
coiled-coil domain containing 51 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TWIST1_SNAI1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 15.6 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 3.5 | 28.0 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 2.9 | 8.8 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 2.6 | 12.9 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 2.5 | 7.5 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 2.2 | 13.2 | GO:0010734 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 2.2 | 19.6 | GO:0086042 | cardiac muscle cell-cardiac muscle cell adhesion(GO:0086042) bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 2.0 | 24.3 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 1.7 | 3.5 | GO:1903169 | regulation of calcium ion transmembrane transport(GO:1903169) |
| 1.6 | 9.4 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 1.5 | 4.6 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) response to DDT(GO:0046680) histone H3-S10 phosphorylation involved in chromosome condensation(GO:2000775) |
| 1.4 | 5.5 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 1.4 | 12.4 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 1.4 | 4.1 | GO:0051796 | regulation of monophenol monooxygenase activity(GO:0032771) positive regulation of monophenol monooxygenase activity(GO:0032773) negative regulation of catagen(GO:0051796) regulation of hair cycle by canonical Wnt signaling pathway(GO:0060901) positive regulation of melanosome transport(GO:1902910) |
| 1.4 | 4.1 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 1.3 | 11.8 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 1.3 | 6.5 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 1.3 | 6.4 | GO:0030070 | insulin processing(GO:0030070) |
| 1.1 | 3.4 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 1.1 | 6.8 | GO:0034201 | response to oleic acid(GO:0034201) |
| 1.0 | 3.1 | GO:0021592 | fourth ventricle development(GO:0021592) third ventricle development(GO:0021678) |
| 1.0 | 3.0 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 1.0 | 12.9 | GO:0048711 | positive regulation of astrocyte differentiation(GO:0048711) |
| 0.9 | 10.4 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.9 | 1.9 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.9 | 3.7 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.9 | 9.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.9 | 9.9 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.9 | 2.7 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.9 | 5.3 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.9 | 1.7 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.8 | 2.5 | GO:2000360 | positive regulation of female gonad development(GO:2000196) negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.8 | 6.7 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.8 | 4.9 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.8 | 9.7 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.8 | 3.1 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.8 | 2.3 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.8 | 3.9 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.7 | 3.0 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 0.7 | 10.2 | GO:1902669 | positive regulation of axon guidance(GO:1902669) |
| 0.7 | 12.4 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.7 | 3.3 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.7 | 2.0 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.7 | 3.3 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.6 | 3.9 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.6 | 18.7 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.6 | 1.3 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.6 | 3.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.6 | 2.4 | GO:0044821 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.6 | 3.6 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) regulation of thyroid hormone generation(GO:2000609) positive regulation of thyroid hormone generation(GO:2000611) |
| 0.6 | 1.8 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.6 | 1.8 | GO:1904351 | negative regulation of protein catabolic process in the vacuole(GO:1904351) negative regulation of lysosomal protein catabolic process(GO:1905166) |
| 0.6 | 16.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.6 | 3.4 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) |
| 0.6 | 0.6 | GO:2000359 | regulation of binding of sperm to zona pellucida(GO:2000359) |
| 0.5 | 3.2 | GO:0015853 | adenine transport(GO:0015853) |
| 0.5 | 5.4 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.5 | 2.6 | GO:0099540 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.5 | 8.9 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.5 | 1.6 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.5 | 2.6 | GO:0042816 | vitamin B6 metabolic process(GO:0042816) |
| 0.5 | 1.5 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 0.5 | 7.6 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.5 | 2.0 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.5 | 4.5 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.5 | 1.5 | GO:1901624 | negative regulation of lymphocyte chemotaxis(GO:1901624) |
| 0.5 | 0.5 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.5 | 4.7 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.4 | 5.4 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.4 | 1.8 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.4 | 1.3 | GO:0035606 | induction of programmed cell death(GO:0012502) peptidyl-cysteine S-trans-nitrosylation(GO:0035606) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) |
| 0.4 | 1.7 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.4 | 0.8 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.4 | 1.3 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.4 | 1.2 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.4 | 1.6 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.4 | 11.8 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.4 | 1.2 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.4 | 1.5 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.4 | 0.8 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.4 | 1.9 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.4 | 5.9 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.4 | 1.8 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.4 | 1.8 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.4 | 2.9 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) |
| 0.4 | 2.8 | GO:0042659 | regulation of cell fate specification(GO:0042659) |
| 0.4 | 0.4 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.4 | 1.1 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.4 | 1.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.3 | 2.4 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.3 | 0.7 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.3 | 1.3 | GO:0019087 | transformation of host cell by virus(GO:0019087) |
| 0.3 | 1.0 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.3 | 1.9 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.3 | 9.0 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.3 | 1.3 | GO:1902361 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.3 | 1.3 | GO:0034343 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.3 | 2.2 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.3 | 2.5 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.3 | 0.9 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.3 | 33.2 | GO:0070268 | cornification(GO:0070268) |
| 0.3 | 0.9 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.3 | 3.6 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.3 | 1.8 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.3 | 1.1 | GO:0030836 | positive regulation of actin filament depolymerization(GO:0030836) |
| 0.3 | 0.8 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.3 | 4.5 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.3 | 2.2 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.3 | 2.4 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.3 | 3.2 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.3 | 0.8 | GO:2000854 | positive regulation of corticosterone secretion(GO:2000854) |
| 0.3 | 1.6 | GO:0030421 | defecation(GO:0030421) |
| 0.3 | 1.3 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.3 | 2.8 | GO:0099639 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.3 | 2.8 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.2 | 1.7 | GO:0060897 | neural plate anterior/posterior regionalization(GO:0021999) neural plate regionalization(GO:0060897) |
| 0.2 | 4.5 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.2 | 1.0 | GO:0001189 | RNA polymerase I transcriptional preinitiation complex assembly(GO:0001188) RNA polymerase I transcriptional preinitiation complex assembly at the promoter for the nuclear large rRNA transcript(GO:0001189) |
| 0.2 | 0.7 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.2 | 1.2 | GO:0072248 | metanephric glomerular epithelium development(GO:0072244) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.2 | 1.5 | GO:0071680 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.2 | 0.7 | GO:0070781 | response to biotin(GO:0070781) |
| 0.2 | 3.3 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.2 | 2.1 | GO:0006554 | lysine catabolic process(GO:0006554) |
| 0.2 | 1.6 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.2 | 2.7 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.2 | 2.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.2 | 0.9 | GO:0042421 | norepinephrine biosynthetic process(GO:0042421) |
| 0.2 | 0.7 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.2 | 1.6 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.2 | 1.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.2 | 2.2 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.2 | 1.7 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.2 | 2.8 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.2 | 8.6 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.2 | 1.9 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.2 | 0.8 | GO:0017198 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.2 | 1.7 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.2 | 1.2 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.2 | 0.4 | GO:0052314 | phytoalexin metabolic process(GO:0052314) |
| 0.2 | 1.2 | GO:1902745 | positive regulation of lamellipodium organization(GO:1902745) |
| 0.2 | 1.8 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.2 | 4.9 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.2 | 0.8 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.2 | 0.8 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.2 | 1.5 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.2 | 5.1 | GO:0036120 | cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.2 | 0.6 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.2 | 4.1 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.2 | 0.9 | GO:0035766 | cell chemotaxis to fibroblast growth factor(GO:0035766) endothelial cell chemotaxis to fibroblast growth factor(GO:0035768) regulation of endothelial tube morphogenesis(GO:1901509) regulation of cell chemotaxis to fibroblast growth factor(GO:1904847) regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000544) |
| 0.2 | 0.9 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.2 | 0.5 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
| 0.2 | 0.5 | GO:0002752 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.2 | 1.0 | GO:0030323 | respiratory tube development(GO:0030323) lung development(GO:0030324) |
| 0.2 | 3.8 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.2 | 2.1 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.2 | 0.8 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.2 | 0.7 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.2 | 3.2 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.2 | 1.2 | GO:0010994 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.2 | 3.0 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.2 | 0.8 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.2 | 1.1 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.2 | 1.4 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.2 | 1.9 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.2 | 0.6 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.2 | 0.5 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.2 | 0.8 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.2 | 0.8 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.2 | 5.9 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.2 | 1.5 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.2 | 0.5 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.2 | 0.8 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.2 | 2.6 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.1 | 1.6 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.1 | 3.1 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 | 1.9 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.1 | 0.4 | GO:0003147 | neural crest cell migration involved in heart formation(GO:0003147) cell migration involved in heart formation(GO:0060974) anterior neural tube closure(GO:0061713) |
| 0.1 | 0.6 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 | 1.6 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.1 | 2.1 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.1 | 1.4 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.1 | 2.3 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.4 | GO:0001694 | histamine biosynthetic process(GO:0001694) |
| 0.1 | 3.4 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 1.7 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.1 | 0.7 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.1 | 0.8 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.1 | 2.7 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 0.8 | GO:1905247 | regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902959) positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) regulation of aspartic-type peptidase activity(GO:1905245) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.1 | 0.4 | GO:1904744 | regulation of single-stranded telomeric DNA binding(GO:0060380) positive regulation of single-stranded telomeric DNA binding(GO:0060381) positive regulation of telomeric DNA binding(GO:1904744) |
| 0.1 | 0.5 | GO:0040034 | regulation of development, heterochronic(GO:0040034) regulation of timing of cell differentiation(GO:0048505) |
| 0.1 | 1.4 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.1 | 0.4 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.1 | 0.8 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.1 | 9.0 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 0.6 | GO:0006041 | glucosamine metabolic process(GO:0006041) glucose 1-phosphate metabolic process(GO:0019255) |
| 0.1 | 1.0 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 | 2.1 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.1 | 0.8 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 1.1 | GO:0021894 | cerebral cortex GABAergic interneuron development(GO:0021894) |
| 0.1 | 1.0 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.1 | 0.4 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.1 | 1.2 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.1 | 0.7 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.1 | 1.7 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.1 | 1.8 | GO:1903204 | negative regulation of oxidative stress-induced neuron death(GO:1903204) |
| 0.1 | 3.0 | GO:0016050 | vesicle organization(GO:0016050) |
| 0.1 | 0.4 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.1 | 0.9 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.1 | 1.2 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.1 | 0.5 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 1.0 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.7 | GO:0052551 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.1 | 1.4 | GO:0035878 | nail development(GO:0035878) |
| 0.1 | 0.7 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 | 0.4 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.1 | 1.3 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.1 | 1.8 | GO:0031424 | keratinization(GO:0031424) |
| 0.1 | 0.6 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.1 | 0.4 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 1.1 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) |
| 0.1 | 4.9 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.1 | 0.3 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.1 | 1.0 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 1.0 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.1 | 0.3 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.1 | 0.7 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.1 | 0.9 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) embryonic neurocranium morphogenesis(GO:0048702) |
| 0.1 | 0.9 | GO:0048569 | post-embryonic organ development(GO:0048569) |
| 0.1 | 0.7 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 0.4 | GO:1900673 | phenylpropanoid catabolic process(GO:0046271) olefin metabolic process(GO:1900673) |
| 0.1 | 1.6 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.1 | 0.4 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.1 | 0.8 | GO:0006983 | ER overload response(GO:0006983) |
| 0.1 | 0.5 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) |
| 0.1 | 0.4 | GO:0060406 | positive regulation of penile erection(GO:0060406) |
| 0.1 | 1.2 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.1 | 1.2 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.1 | 0.3 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.1 | 3.2 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.1 | 1.6 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 1.3 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 6.2 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.1 | 1.4 | GO:0010762 | regulation of fibroblast migration(GO:0010762) |
| 0.1 | 0.2 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 0.1 | 0.3 | GO:0042376 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.1 | 0.7 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.1 | 0.2 | GO:0097021 | lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.1 | 2.6 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.1 | 0.4 | GO:0000819 | sister chromatid segregation(GO:0000819) |
| 0.1 | 0.2 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) negative regulation of urine volume(GO:0035811) |
| 0.1 | 1.5 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 1.3 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 0.9 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.1 | 0.6 | GO:2000507 | cap-dependent translational initiation(GO:0002191) positive regulation of energy homeostasis(GO:2000507) |
| 0.1 | 0.4 | GO:0060157 | urinary bladder development(GO:0060157) |
| 0.1 | 0.2 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.1 | 2.3 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.1 | 0.2 | GO:0070244 | negative regulation of thymocyte apoptotic process(GO:0070244) |
| 0.1 | 0.1 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 | 1.7 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.1 | 0.2 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.1 | 0.3 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.1 | 0.1 | GO:0051710 | regulation of cytolysis in other organism(GO:0051710) |
| 0.1 | 1.2 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.1 | 0.5 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.1 | 0.2 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.1 | 1.6 | GO:0042692 | muscle cell differentiation(GO:0042692) |
| 0.1 | 0.3 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.1 | 0.8 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.1 | 1.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.8 | GO:0046500 | S-adenosylmethionine metabolic process(GO:0046500) |
| 0.1 | 1.3 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.1 | 1.2 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.1 | 3.0 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.1 | 0.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.1 | 1.4 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.1 | 0.1 | GO:0060467 | negative regulation of fertilization(GO:0060467) |
| 0.1 | 0.7 | GO:0019317 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.1 | 0.5 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.1 | 2.6 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.1 | 2.4 | GO:0030835 | negative regulation of actin filament depolymerization(GO:0030835) |
| 0.1 | 0.3 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.1 | 1.2 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.1 | 0.1 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.1 | 1.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 0.2 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.1 | 3.1 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.5 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.1 | 1.3 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.1 | 1.3 | GO:0005977 | glycogen metabolic process(GO:0005977) |
| 0.1 | 1.1 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.1 | 0.4 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 3.3 | GO:0035308 | negative regulation of protein dephosphorylation(GO:0035308) |
| 0.0 | 0.4 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.0 | 1.3 | GO:0043616 | keratinocyte proliferation(GO:0043616) |
| 0.0 | 0.1 | GO:0001575 | globoside metabolic process(GO:0001575) |
| 0.0 | 0.6 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.8 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.5 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 0.5 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.8 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.2 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 0.0 | 0.6 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.3 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.3 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.0 | 0.0 | GO:1901420 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) negative regulation of response to alcohol(GO:1901420) |
| 0.0 | 0.2 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.2 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.0 | 0.3 | GO:0010819 | regulation of T cell chemotaxis(GO:0010819) positive regulation of T cell chemotaxis(GO:0010820) |
| 0.0 | 19.0 | GO:0043687 | post-translational protein modification(GO:0043687) |
| 0.0 | 1.6 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 1.0 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.2 | GO:0007341 | penetration of zona pellucida(GO:0007341) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 1.7 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.0 | 0.4 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:1900076 | regulation of cellular response to insulin stimulus(GO:1900076) |
| 0.0 | 0.1 | GO:0003418 | growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 0.0 | 0.8 | GO:0006312 | mitotic recombination(GO:0006312) |
| 0.0 | 1.1 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 3.3 | GO:0006939 | smooth muscle contraction(GO:0006939) |
| 0.0 | 0.1 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) |
| 0.0 | 1.8 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.4 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.6 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) negative regulation of adherens junction organization(GO:1903392) |
| 0.0 | 0.4 | GO:2000680 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.0 | 0.3 | GO:0015868 | purine nucleotide transport(GO:0015865) purine ribonucleotide transport(GO:0015868) |
| 0.0 | 0.6 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 2.5 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.0 | 2.1 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.0 | 0.6 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 1.2 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.6 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.7 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 1.0 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 1.4 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.0 | 0.6 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.0 | 0.7 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.4 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.8 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 1.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.6 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.3 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.2 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.3 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.8 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.0 | 0.2 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.2 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.1 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) oncogene-induced cell senescence(GO:0090402) |
| 0.0 | 0.1 | GO:1900193 | regulation of oocyte maturation(GO:1900193) negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.4 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 0.1 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 | 0.1 | GO:1902177 | positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902177) |
| 0.0 | 0.2 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 0.3 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.2 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.2 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.2 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.1 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.0 | 0.4 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.2 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.0 | 0.1 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.5 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.1 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.3 | GO:0010644 | cell communication by electrical coupling(GO:0010644) |
| 0.0 | 0.3 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 1.1 | GO:0007422 | peripheral nervous system development(GO:0007422) |
| 0.0 | 0.1 | GO:0051058 | negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.0 | 0.4 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.0 | 0.1 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.6 | GO:0043066 | negative regulation of apoptotic process(GO:0043066) |
| 0.0 | 0.3 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.2 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
| 0.0 | 0.6 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.2 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.2 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.0 | 0.2 | GO:0042177 | negative regulation of protein catabolic process(GO:0042177) |
| 0.0 | 0.5 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.2 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.2 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.3 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.0 | 0.2 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.0 | 0.3 | GO:0051930 | regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.0 | 0.1 | GO:0031125 | rRNA 3'-end processing(GO:0031125) |
| 0.0 | 0.2 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 0.3 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.2 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 | 0.2 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.2 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 12.9 | GO:0097453 | mesaxon(GO:0097453) ensheathing process(GO:1990015) |
| 2.2 | 8.8 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 1.7 | 31.8 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 1.1 | 9.6 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 1.0 | 3.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 1.0 | 5.8 | GO:1990357 | terminal web(GO:1990357) |
| 0.9 | 4.6 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.8 | 2.5 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.8 | 26.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.7 | 3.3 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.6 | 12.6 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.6 | 16.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.6 | 3.0 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.6 | 16.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.6 | 22.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.5 | 3.7 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.5 | 2.6 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.5 | 1.5 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.5 | 7.0 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.5 | 2.4 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.4 | 1.8 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) |
| 0.4 | 1.3 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.4 | 1.2 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.4 | 27.7 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.4 | 3.2 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.4 | 8.5 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.4 | 3.6 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.3 | 2.8 | GO:0098845 | postsynaptic endosome(GO:0098845) |
| 0.3 | 2.2 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.3 | 1.6 | GO:1990761 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.3 | 2.5 | GO:0045179 | apical cortex(GO:0045179) |
| 0.3 | 1.8 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.3 | 1.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.3 | 3.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.3 | 8.1 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.3 | 24.4 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.3 | 2.1 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.2 | 1.0 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.2 | 2.0 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.2 | 8.9 | GO:0032420 | stereocilium(GO:0032420) |
| 0.2 | 0.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.2 | 3.4 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.2 | 0.7 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.2 | 1.4 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.2 | 1.6 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.2 | 2.7 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.2 | 2.9 | GO:0097433 | dense body(GO:0097433) |
| 0.2 | 2.9 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.2 | 0.7 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.2 | 3.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.2 | 1.1 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.2 | 1.8 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 0.6 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.2 | 2.9 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.2 | 18.6 | GO:0005604 | basement membrane(GO:0005604) |
| 0.2 | 11.9 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.2 | 1.2 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.2 | 9.8 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.2 | 0.5 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.2 | 0.6 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.2 | 4.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.2 | 3.4 | GO:0043034 | costamere(GO:0043034) |
| 0.2 | 1.3 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) cytosolic proteasome complex(GO:0031597) |
| 0.2 | 0.6 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 1.6 | GO:0061202 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.1 | 0.7 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 0.8 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.1 | 2.0 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.1 | 0.8 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 14.6 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 2.1 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 1.7 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 1.7 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 27.9 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 0.5 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.1 | 10.1 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 2.2 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 1.7 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.8 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 3.1 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 1.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 7.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 0.7 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 1.9 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 2.0 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 0.3 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.1 | 6.9 | GO:0005761 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.1 | 1.5 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 0.4 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 0.5 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 14.1 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.1 | 2.3 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 1.6 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.1 | 1.9 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 2.7 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 1.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 2.6 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 10.9 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.1 | 1.1 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 7.5 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 1.8 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 6.9 | GO:0098793 | presynapse(GO:0098793) |
| 0.1 | 1.1 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.1 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 1.3 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 0.5 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 0.8 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 1.0 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 3.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 0.5 | GO:0098576 | lumenal side of membrane(GO:0098576) |
| 0.1 | 1.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 1.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 3.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 4.1 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 5.2 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 4.1 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 3.9 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 2.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.2 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.7 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.8 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 1.0 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.4 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 2.1 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.7 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.4 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 5.0 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 1.2 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 1.1 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 1.8 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0000152 | nuclear ubiquitin ligase complex(GO:0000152) |
| 0.0 | 0.8 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 1.9 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.5 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 5.6 | GO:0031252 | cell leading edge(GO:0031252) |
| 0.0 | 0.6 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.5 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 2.2 | GO:0070160 | occluding junction(GO:0070160) |
| 0.0 | 0.5 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.8 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.5 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 0.4 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.0 | 0.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.4 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.2 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.7 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 1.1 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 1.3 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 1.0 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 5.6 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.1 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.6 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.6 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.5 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 1.0 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.5 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 11.3 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.3 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.3 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 0.2 | GO:0035869 | ciliary transition zone(GO:0035869) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 12.9 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 2.9 | 8.8 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 2.2 | 19.6 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 2.0 | 11.7 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 1.4 | 7.2 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 1.3 | 5.4 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 1.1 | 10.9 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 1.1 | 3.2 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 1.0 | 3.0 | GO:0003974 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.9 | 1.8 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.9 | 3.5 | GO:0030760 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.9 | 23.2 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.8 | 2.5 | GO:0004515 | nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.8 | 11.6 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.8 | 3.2 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.7 | 2.2 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.7 | 4.6 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.6 | 6.8 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.6 | 9.2 | GO:0016918 | retinal binding(GO:0016918) |
| 0.6 | 2.9 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.6 | 5.8 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.5 | 2.6 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.5 | 6.6 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.5 | 3.5 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.5 | 22.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.5 | 3.8 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.5 | 6.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.4 | 1.8 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.4 | 2.5 | GO:0008948 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) oxaloacetate decarboxylase activity(GO:0008948) |
| 0.4 | 2.5 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.4 | 5.7 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.4 | 39.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.4 | 3.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.4 | 5.0 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.3 | 1.7 | GO:0004306 | ethanolamine-phosphate cytidylyltransferase activity(GO:0004306) |
| 0.3 | 5.5 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.3 | 1.0 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.3 | 1.7 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.3 | 1.3 | GO:0033906 | protein tyrosine kinase inhibitor activity(GO:0030292) hyaluronoglucuronidase activity(GO:0033906) |
| 0.3 | 1.0 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.3 | 1.3 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) aspartic-type endopeptidase inhibitor activity(GO:0019828) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.3 | 6.5 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.3 | 3.6 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.3 | 1.3 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.3 | 2.6 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.3 | 1.3 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.3 | 15.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.3 | 3.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.3 | 16.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.3 | 2.1 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.3 | 1.2 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.3 | 2.6 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.3 | 7.4 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.3 | 0.8 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.3 | 5.5 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.3 | 1.6 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.3 | 19.4 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.2 | 5.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 1.0 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.2 | 0.5 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.2 | 1.6 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.2 | 0.9 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.2 | 1.6 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.2 | 1.4 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.2 | 0.7 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.2 | 0.9 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.2 | 1.8 | GO:0008430 | selenium binding(GO:0008430) |
| 0.2 | 2.2 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.2 | 1.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.2 | 1.5 | GO:0045502 | dynein binding(GO:0045502) dynein light intermediate chain binding(GO:0051959) |
| 0.2 | 1.7 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.2 | 1.1 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.2 | 0.9 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.2 | 0.8 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.2 | 0.6 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.2 | 9.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.2 | 1.6 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.2 | 1.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.2 | 0.7 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.2 | 4.6 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.2 | 0.7 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.2 | 4.2 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.2 | 1.1 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.2 | 0.5 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.2 | 3.0 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.2 | 1.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 0.9 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.2 | 24.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.2 | 1.5 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.2 | 0.5 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.2 | 2.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.2 | 1.3 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.2 | 0.6 | GO:0042019 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.2 | 0.5 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.2 | 0.3 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.2 | 0.5 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.2 | 2.4 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.2 | 1.6 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.2 | 2.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.2 | 2.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.2 | 10.1 | GO:0016879 | ligase activity, forming carbon-nitrogen bonds(GO:0016879) |
| 0.2 | 0.9 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.2 | 3.0 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.2 | 2.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 1.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 1.5 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 15.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.7 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.1 | 0.4 | GO:0004853 | uroporphyrinogen decarboxylase activity(GO:0004853) |
| 0.1 | 1.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.1 | 1.6 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 1.4 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.1 | 1.0 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 0.7 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.5 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.1 | 2.0 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 0.9 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 1.3 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 1.6 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 0.8 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 1.8 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 0.8 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.1 | 17.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 1.5 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.1 | 0.4 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.1 | 0.9 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 3.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.4 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.1 | 1.0 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 10.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 6.5 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 1.3 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 1.9 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 2.3 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 1.5 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.4 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.1 | 0.4 | GO:0001855 | complement component C4b binding(GO:0001855) |
| 0.1 | 2.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.8 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.1 | 1.4 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 0.4 | GO:0051870 | methotrexate binding(GO:0051870) folic acid receptor activity(GO:0061714) |
| 0.1 | 1.8 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 0.5 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.1 | 0.8 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 4.3 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 6.8 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 0.5 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.1 | 0.4 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 0.4 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.1 | 0.7 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.1 | 1.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 2.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.7 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 0.7 | GO:0042301 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) phosphate ion binding(GO:0042301) |
| 0.1 | 2.9 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 2.9 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.1 | 1.1 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.1 | 4.1 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.3 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 0.2 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.1 | 0.8 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.8 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.1 | 1.3 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 0.3 | GO:0035473 | lipase binding(GO:0035473) |
| 0.1 | 1.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 2.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 1.7 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 7.3 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 0.8 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.9 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 6.8 | GO:0020037 | heme binding(GO:0020037) |
| 0.1 | 0.3 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.1 | 0.7 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.6 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 0.2 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 0.4 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.1 | 0.3 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.1 | 2.1 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 0.4 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 0.2 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.1 | 1.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.3 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 0.6 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.1 | 0.5 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.1 | 29.0 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.1 | 0.3 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 1.5 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 0.2 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.1 | 1.6 | GO:0015464 | acetylcholine receptor activity(GO:0015464) |
| 0.1 | 4.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.9 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 1.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.9 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.0 | 0.7 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.5 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.1 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.0 | 0.5 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.6 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 9.0 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.6 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.9 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.3 | GO:0036310 | annealing helicase activity(GO:0036310) annealing activity(GO:0097617) |
| 0.0 | 0.2 | GO:0019787 | ubiquitin-like protein transferase activity(GO:0019787) |
| 0.0 | 0.1 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.0 | 0.4 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 2.1 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.3 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.3 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.2 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 2.1 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.3 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.8 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 1.2 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 3.4 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.4 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.0 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.3 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 1.5 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 8.6 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.6 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.5 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.0 | 0.9 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.5 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.2 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 1.1 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.3 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 3.9 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.1 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.3 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.1 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.5 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.5 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.7 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 0.4 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.3 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.1 | GO:1901612 | phosphatidylglycerol binding(GO:1901611) cardiolipin binding(GO:1901612) |
| 0.0 | 3.0 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.4 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.3 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 0.1 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.2 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.2 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 10.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.4 | 25.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.4 | 0.4 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.3 | 10.1 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.3 | 18.9 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.3 | 9.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.3 | 18.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.2 | 13.2 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.2 | 0.6 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.2 | 12.1 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.2 | 11.4 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.2 | 2.7 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.2 | 2.7 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.2 | 10.4 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.2 | 7.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 7.3 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.2 | 3.6 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 12.6 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 7.9 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 3.2 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 4.8 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.1 | 4.2 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 3.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.1 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 0.5 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 3.3 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 1.9 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 1.5 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 2.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 11.3 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 2.2 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 2.4 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 0.9 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 2.3 | PID P73PATHWAY | p73 transcription factor network |
| 0.1 | 2.2 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 0.3 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 3.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.5 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.7 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.0 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.3 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 1.2 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.3 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.4 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 20.5 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.7 | 43.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.6 | 6.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.6 | 12.8 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.5 | 22.8 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.5 | 18.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.5 | 23.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.4 | 15.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.4 | 8.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.3 | 5.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.3 | 6.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.3 | 8.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.3 | 2.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.3 | 7.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 15.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.2 | 2.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.2 | 4.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.2 | 5.0 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.2 | 4.9 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.2 | 3.0 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.2 | 11.2 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.2 | 4.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.2 | 2.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 2.6 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 2.0 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 0.6 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.1 | 2.0 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 2.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 4.7 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.1 | 2.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 18.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 1.4 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 6.4 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 1.5 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 1.3 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 1.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 2.2 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.1 | 2.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 2.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 9.3 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.1 | 5.5 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.1 | 1.3 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.1 | 2.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 1.8 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.1 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.1 | 1.3 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 2.4 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.1 | 1.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 1.9 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 1.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.4 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.1 | 1.4 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 13.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 1.7 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.1 | 1.7 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 4.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 1.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 12.7 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 2.7 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.1 | 0.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 0.8 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 6.2 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 3.5 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.8 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.0 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.9 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.4 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 1.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.3 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 1.7 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.6 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.8 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.8 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.3 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.3 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.6 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.6 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.0 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 3.1 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 1.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.3 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.0 | 0.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.1 | REACTOME NCAM SIGNALING FOR NEURITE OUT GROWTH | Genes involved in NCAM signaling for neurite out-growth |
| 0.0 | 0.2 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.6 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 0.2 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |