Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for VAX2_RHOXF2
Z-value: 1.28
Transcription factors associated with VAX2_RHOXF2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
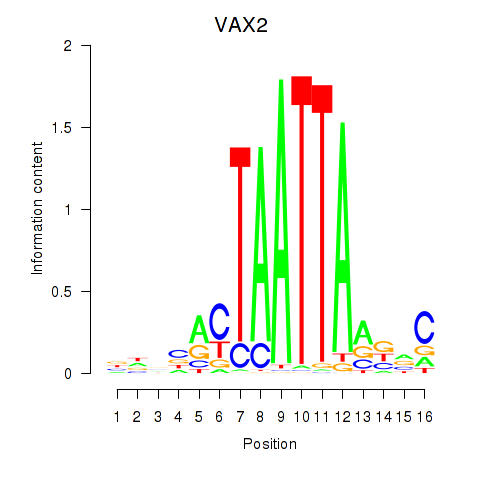
VAX2
|
ENSG00000116035.2 | ventral anterior homeobox 2 |
|
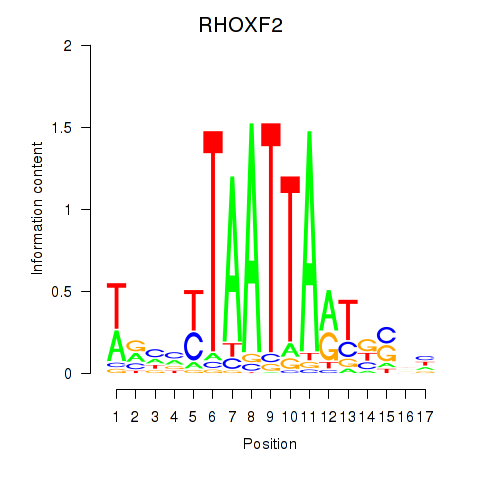
RHOXF2
|
ENSG00000131721.4 | Rhox homeobox family member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| VAX2 | hg19_v2_chr2_+_71127699_71127744 | 0.41 | 2.1e-10 | Click! |
Activity profile of VAX2_RHOXF2 motif
Sorted Z-values of VAX2_RHOXF2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_57045228 | 15.68 |
ENST00000371250.3
|
PPAP2B
|
phosphatidic acid phosphatase type 2B |
| chr12_-_6233828 | 11.91 |
ENST00000572068.1
ENST00000261405.5 |
VWF
|
von Willebrand factor |
| chr2_+_90248739 | 11.02 |
ENST00000468879.1
|
IGKV1D-43
|
immunoglobulin kappa variable 1D-43 |
| chr2_-_89340242 | 9.97 |
ENST00000480492.1
|
IGKV1-12
|
immunoglobulin kappa variable 1-12 |
| chr5_+_66300446 | 9.82 |
ENST00000261569.7
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr11_-_117747607 | 9.63 |
ENST00000540359.1
ENST00000539526.1 |
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr11_-_117747434 | 9.58 |
ENST00000529335.2
ENST00000530956.1 ENST00000260282.4 |
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr11_-_117748138 | 8.61 |
ENST00000527717.1
|
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr11_-_111794446 | 7.35 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr17_-_41466555 | 6.96 |
ENST00000586231.1
|
LINC00910
|
long intergenic non-protein coding RNA 910 |
| chr4_-_186696425 | 6.71 |
ENST00000430503.1
ENST00000319454.6 ENST00000450341.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr12_+_26348246 | 5.32 |
ENST00000422622.2
|
SSPN
|
sarcospan |
| chr3_-_112127981 | 4.71 |
ENST00000486726.2
|
RP11-231E6.1
|
RP11-231E6.1 |
| chr2_-_89292422 | 4.42 |
ENST00000495489.1
|
IGKV1-8
|
immunoglobulin kappa variable 1-8 |
| chr1_-_211307404 | 4.37 |
ENST00000367007.4
|
KCNH1
|
potassium voltage-gated channel, subfamily H (eag-related), member 1 |
| chr6_+_26402517 | 4.06 |
ENST00000414912.2
|
BTN3A1
|
butyrophilin, subfamily 3, member A1 |
| chr16_-_55866997 | 3.51 |
ENST00000360526.3
ENST00000361503.4 |
CES1
|
carboxylesterase 1 |
| chr11_+_71900572 | 3.38 |
ENST00000312293.4
|
FOLR1
|
folate receptor 1 (adult) |
| chr19_+_50016610 | 3.27 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr14_-_53258314 | 3.21 |
ENST00000216410.3
ENST00000557604.1 |
GNPNAT1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr6_+_26402465 | 3.13 |
ENST00000476549.2
ENST00000289361.6 ENST00000450085.2 ENST00000425234.2 ENST00000427334.1 ENST00000506698.1 |
BTN3A1
|
butyrophilin, subfamily 3, member A1 |
| chr11_+_71900703 | 3.10 |
ENST00000393681.2
|
FOLR1
|
folate receptor 1 (adult) |
| chrX_+_43515467 | 2.99 |
ENST00000338702.3
ENST00000542639.1 |
MAOA
|
monoamine oxidase A |
| chr9_+_131062367 | 2.95 |
ENST00000601297.1
|
AL359091.2
|
CDNA: FLJ21673 fis, clone COL09042; HCG2036511; Uncharacterized protein |
| chr20_+_56136136 | 2.72 |
ENST00000319441.4
ENST00000543666.1 |
PCK1
|
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr1_+_35258592 | 2.62 |
ENST00000342280.4
ENST00000450137.1 |
GJA4
|
gap junction protein, alpha 4, 37kDa |
| chr9_-_95166841 | 2.43 |
ENST00000262551.4
|
OGN
|
osteoglycin |
| chr17_+_40610862 | 2.42 |
ENST00000393829.2
ENST00000546249.1 ENST00000537728.1 ENST00000264649.6 ENST00000585525.1 ENST00000343619.4 ENST00000544137.1 ENST00000589727.1 ENST00000587824.1 |
ATP6V0A1
|
ATPase, H+ transporting, lysosomal V0 subunit a1 |
| chr2_+_113763031 | 2.36 |
ENST00000259211.6
|
IL36A
|
interleukin 36, alpha |
| chr12_-_118796910 | 2.31 |
ENST00000541186.1
ENST00000539872.1 |
TAOK3
|
TAO kinase 3 |
| chr3_+_115342349 | 2.05 |
ENST00000393780.3
|
GAP43
|
growth associated protein 43 |
| chr4_+_88571429 | 1.97 |
ENST00000339673.6
ENST00000282479.7 |
DMP1
|
dentin matrix acidic phosphoprotein 1 |
| chr1_+_62439037 | 1.94 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr4_+_88754113 | 1.89 |
ENST00000560249.1
ENST00000540395.1 ENST00000511670.1 ENST00000361056.3 |
MEPE
|
matrix extracellular phosphoglycoprotein |
| chr10_-_17171817 | 1.87 |
ENST00000377833.4
|
CUBN
|
cubilin (intrinsic factor-cobalamin receptor) |
| chr7_+_50348268 | 1.83 |
ENST00000438033.1
ENST00000439701.1 |
IKZF1
|
IKAROS family zinc finger 1 (Ikaros) |
| chr16_+_72088376 | 1.80 |
ENST00000570083.1
ENST00000355906.5 ENST00000398131.2 ENST00000569639.1 ENST00000564499.1 ENST00000357763.4 ENST00000562526.1 ENST00000565574.1 ENST00000568417.2 ENST00000356967.5 |
HP
HPR
|
haptoglobin haptoglobin-related protein |
| chr1_-_92952433 | 1.80 |
ENST00000294702.5
|
GFI1
|
growth factor independent 1 transcription repressor |
| chr12_+_26348429 | 1.80 |
ENST00000242729.2
|
SSPN
|
sarcospan |
| chr4_+_80584903 | 1.68 |
ENST00000506460.1
|
RP11-452C8.1
|
RP11-452C8.1 |
| chr1_-_24151903 | 1.66 |
ENST00000436439.2
ENST00000374490.3 |
HMGCL
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase |
| chr12_+_81110684 | 1.65 |
ENST00000228644.3
|
MYF5
|
myogenic factor 5 |
| chr2_+_169926047 | 1.62 |
ENST00000428522.1
ENST00000450153.1 ENST00000421653.1 |
DHRS9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr6_+_26440700 | 1.57 |
ENST00000494393.1
ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3
|
butyrophilin, subfamily 3, member A3 |
| chr2_-_89597542 | 1.55 |
ENST00000465170.1
|
IGKV1-37
|
immunoglobulin kappa variable 1-37 (non-functional) |
| chr5_-_20575959 | 1.51 |
ENST00000507958.1
|
CDH18
|
cadherin 18, type 2 |
| chrX_+_135730373 | 1.47 |
ENST00000370628.2
|
CD40LG
|
CD40 ligand |
| chr5_+_129083772 | 1.45 |
ENST00000564719.1
|
KIAA1024L
|
KIAA1024-like |
| chr9_-_5339873 | 1.40 |
ENST00000223862.1
ENST00000223858.4 |
RLN1
|
relaxin 1 |
| chr12_-_10022735 | 1.38 |
ENST00000228438.2
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr11_-_62521614 | 1.38 |
ENST00000527994.1
ENST00000394807.3 |
ZBTB3
|
zinc finger and BTB domain containing 3 |
| chrX_+_153533275 | 1.36 |
ENST00000426989.1
ENST00000426203.1 ENST00000369912.2 |
TKTL1
|
transketolase-like 1 |
| chr12_-_9760482 | 1.36 |
ENST00000229402.3
|
KLRB1
|
killer cell lectin-like receptor subfamily B, member 1 |
| chr1_+_46972668 | 1.32 |
ENST00000371956.4
ENST00000360032.3 |
DMBX1
|
diencephalon/mesencephalon homeobox 1 |
| chr14_+_103851712 | 1.31 |
ENST00000440884.3
ENST00000416682.2 ENST00000429436.2 ENST00000303622.9 |
MARK3
|
MAP/microtubule affinity-regulating kinase 3 |
| chr15_+_58702742 | 1.29 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chr17_-_40337470 | 1.24 |
ENST00000293330.1
|
HCRT
|
hypocretin (orexin) neuropeptide precursor |
| chr7_-_100860851 | 1.17 |
ENST00000223127.3
|
PLOD3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr3_+_152552685 | 1.13 |
ENST00000305097.3
|
P2RY1
|
purinergic receptor P2Y, G-protein coupled, 1 |
| chr9_-_5304432 | 1.05 |
ENST00000416837.1
ENST00000308420.3 |
RLN2
|
relaxin 2 |
| chr17_-_18430160 | 0.97 |
ENST00000392176.3
|
FAM106A
|
family with sequence similarity 106, member A |
| chr16_-_28608424 | 0.95 |
ENST00000335715.4
|
SULT1A2
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 |
| chr21_-_37914898 | 0.93 |
ENST00000399136.1
|
CLDN14
|
claudin 14 |
| chr12_-_22063787 | 0.89 |
ENST00000544039.1
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chrX_+_135730297 | 0.82 |
ENST00000370629.2
|
CD40LG
|
CD40 ligand |
| chr10_+_18549645 | 0.78 |
ENST00000396576.2
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr14_+_32798547 | 0.75 |
ENST00000557354.1
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr11_+_33061543 | 0.74 |
ENST00000432887.1
ENST00000528898.1 ENST00000531632.2 |
TCP11L1
|
t-complex 11, testis-specific-like 1 |
| chr1_+_153747746 | 0.72 |
ENST00000368661.3
|
SLC27A3
|
solute carrier family 27 (fatty acid transporter), member 3 |
| chr9_-_95166884 | 0.71 |
ENST00000375561.5
|
OGN
|
osteoglycin |
| chr4_+_169418255 | 0.70 |
ENST00000505667.1
ENST00000511948.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr6_-_133035185 | 0.69 |
ENST00000367928.4
|
VNN1
|
vanin 1 |
| chr18_-_67624160 | 0.66 |
ENST00000581982.1
ENST00000280200.4 |
CD226
|
CD226 molecule |
| chr2_+_103089756 | 0.65 |
ENST00000295269.4
|
SLC9A4
|
solute carrier family 9, subfamily A (NHE4, cation proton antiporter 4), member 4 |
| chr2_-_214016314 | 0.64 |
ENST00000434687.1
ENST00000374319.4 |
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr14_+_32798462 | 0.61 |
ENST00000280979.4
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr14_-_20923195 | 0.61 |
ENST00000206542.4
|
OSGEP
|
O-sialoglycoprotein endopeptidase |
| chr19_+_12175504 | 0.59 |
ENST00000439326.3
|
ZNF844
|
zinc finger protein 844 |
| chr7_-_44580861 | 0.57 |
ENST00000546276.1
ENST00000289547.4 ENST00000381160.3 ENST00000423141.1 |
NPC1L1
|
NPC1-like 1 |
| chr4_-_74486347 | 0.56 |
ENST00000342081.3
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr5_+_140557371 | 0.51 |
ENST00000239444.2
|
PCDHB8
|
protocadherin beta 8 |
| chr17_-_10372875 | 0.47 |
ENST00000255381.2
|
MYH4
|
myosin, heavy chain 4, skeletal muscle |
| chr14_+_72399833 | 0.46 |
ENST00000553530.1
ENST00000556437.1 |
RGS6
|
regulator of G-protein signaling 6 |
| chr9_-_21239978 | 0.41 |
ENST00000380222.2
|
IFNA14
|
interferon, alpha 14 |
| chr16_-_28608364 | 0.38 |
ENST00000533150.1
|
SULT1A2
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 |
| chr13_-_36050819 | 0.37 |
ENST00000379919.4
|
MAB21L1
|
mab-21-like 1 (C. elegans) |
| chr5_+_174151536 | 0.35 |
ENST00000239243.6
ENST00000507785.1 |
MSX2
|
msh homeobox 2 |
| chr15_-_89755034 | 0.33 |
ENST00000563254.1
|
RLBP1
|
retinaldehyde binding protein 1 |
| chr1_+_225600404 | 0.31 |
ENST00000366845.2
|
AC092811.1
|
AC092811.1 |
| chr20_-_50722183 | 0.31 |
ENST00000371523.4
|
ZFP64
|
ZFP64 zinc finger protein |
| chr6_+_167536230 | 0.29 |
ENST00000341935.5
ENST00000349984.4 |
CCR6
|
chemokine (C-C motif) receptor 6 |
| chr7_+_73245193 | 0.27 |
ENST00000340958.2
|
CLDN4
|
claudin 4 |
| chr1_-_152386732 | 0.26 |
ENST00000271835.3
|
CRNN
|
cornulin |
| chr16_-_30122717 | 0.24 |
ENST00000566613.1
|
GDPD3
|
glycerophosphodiester phosphodiesterase domain containing 3 |
| chrX_-_18690210 | 0.21 |
ENST00000379984.3
|
RS1
|
retinoschisin 1 |
| chr19_-_46088068 | 0.16 |
ENST00000263275.4
ENST00000323060.3 |
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia) |
| chr4_-_74486217 | 0.15 |
ENST00000335049.5
ENST00000307439.5 |
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr5_-_149829244 | 0.13 |
ENST00000312037.5
|
RPS14
|
ribosomal protein S14 |
| chr7_+_148936732 | 0.09 |
ENST00000335870.2
|
ZNF212
|
zinc finger protein 212 |
| chr2_-_85555385 | 0.07 |
ENST00000377386.3
|
TGOLN2
|
trans-golgi network protein 2 |
| chr4_+_70796784 | 0.06 |
ENST00000246891.4
ENST00000444405.3 |
CSN1S1
|
casein alpha s1 |
| chr11_-_102709441 | 0.05 |
ENST00000434103.1
|
MMP3
|
matrix metallopeptidase 3 (stromelysin 1, progelatinase) |
| chr14_+_74034310 | 0.03 |
ENST00000538782.1
|
ACOT2
|
acyl-CoA thioesterase 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of VAX2_RHOXF2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.5 | GO:0061713 | neural crest cell migration involved in heart formation(GO:0003147) cell migration involved in heart formation(GO:0060974) anterior neural tube closure(GO:0061713) |
| 0.7 | 2.7 | GO:0006114 | glycerol biosynthetic process(GO:0006114) response to methionine(GO:1904640) |
| 0.6 | 3.2 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.6 | 4.4 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.6 | 1.8 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.6 | 1.8 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.5 | 11.9 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.5 | 3.3 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.4 | 7.4 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.4 | 1.2 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.4 | 27.8 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.4 | 3.5 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.3 | 1.4 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.3 | 6.5 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.3 | 2.3 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.2 | 1.1 | GO:0060406 | positive regulation of penile erection(GO:0060406) |
| 0.2 | 0.7 | GO:0060369 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.2 | 6.7 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.2 | 1.2 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.2 | 2.0 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.2 | 0.6 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.2 | 2.4 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.2 | 1.4 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.2 | 1.3 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.2 | 3.0 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.2 | 1.9 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.2 | 1.7 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.2 | 3.1 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.2 | 0.8 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.2 | 1.7 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.1 | 1.6 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 1.3 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.1 | 23.1 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 1.3 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 0.7 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.1 | 1.9 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.1 | 2.6 | GO:0048265 | response to pain(GO:0048265) |
| 0.1 | 1.3 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 0.6 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.3 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 1.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.6 | GO:0098856 | intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.0 | 2.4 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) |
| 0.0 | 0.6 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 10.2 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.9 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.2 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.0 | 1.6 | GO:0002456 | T cell mediated immunity(GO:0002456) |
| 0.0 | 7.6 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 1.8 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 2.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.0 | GO:0030819 | positive regulation of cAMP biosynthetic process(GO:0030819) |
| 0.0 | 3.9 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.0 | 0.1 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 11.9 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.5 | 7.4 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.5 | 1.8 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.4 | 6.5 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.3 | 1.9 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.3 | 0.9 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.3 | 2.0 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.3 | 2.4 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.2 | 7.1 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.2 | 1.4 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 2.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 1.8 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.1 | 0.6 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 4.4 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 0.8 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 1.2 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 1.2 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 6.8 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.6 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.5 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 1.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 3.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.3 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 3.2 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 5.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 3.0 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.7 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 20.6 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.7 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 18.6 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.5 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.9 | 2.7 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.8 | 3.3 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.8 | 2.3 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.7 | 4.4 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.7 | 3.5 | GO:0004771 | sterol esterase activity(GO:0004771) methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.6 | 1.7 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.5 | 27.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.5 | 1.4 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.4 | 11.9 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.4 | 1.2 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) procollagen galactosyltransferase activity(GO:0050211) |
| 0.4 | 1.9 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.4 | 3.0 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.3 | 1.8 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.3 | 1.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.2 | 6.7 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 7.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 2.0 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.2 | 1.3 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.2 | 1.9 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.2 | 1.6 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.1 | 0.7 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 2.4 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 25.4 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 0.8 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.1 | 2.4 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 1.4 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 0.3 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.1 | 0.9 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.1 | 1.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.1 | 0.6 | GO:0015386 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.7 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 1.2 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 3.2 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 1.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.3 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 1.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.2 | GO:0070492 | phosphatidylinositol-5-phosphate binding(GO:0010314) oligosaccharide binding(GO:0070492) |
| 0.0 | 1.9 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 11.7 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 3.1 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.5 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 3.5 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 15.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 3.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 2.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 2.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.8 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 11.9 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.5 | 15.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.2 | 2.7 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.2 | 3.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.2 | 3.0 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.2 | 3.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 2.6 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 4.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 3.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 3.2 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.1 | 1.1 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 1.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 1.5 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 1.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 3.0 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.3 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.9 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.8 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 1.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.3 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |