Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
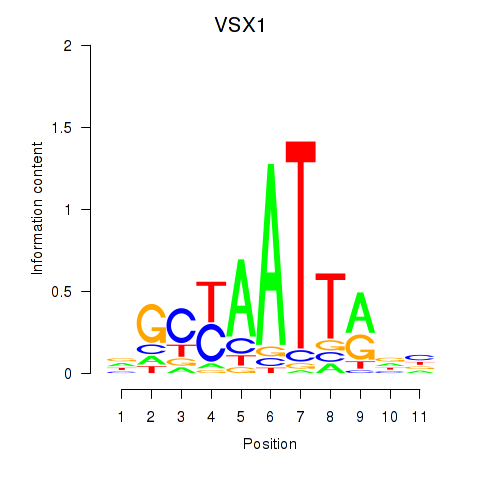
Results for VSX1
Z-value: 1.16
Transcription factors associated with VSX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
VSX1
|
ENSG00000100987.10 | visual system homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| VSX1 | hg19_v2_chr20_-_25062767_25062779 | -0.37 | 1.5e-08 | Click! |
Activity profile of VSX1 motif
Sorted Z-values of VSX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_64673630 | 32.29 |
ENST00000558008.1
ENST00000559519.1 ENST00000380258.2 |
KIAA0101
|
KIAA0101 |
| chr19_+_50180317 | 30.16 |
ENST00000534465.1
|
PRMT1
|
protein arginine methyltransferase 1 |
| chr1_+_154947148 | 28.36 |
ENST00000368436.1
ENST00000308987.5 |
CKS1B
|
CDC28 protein kinase regulatory subunit 1B |
| chr1_+_154947126 | 26.83 |
ENST00000368439.1
|
CKS1B
|
CDC28 protein kinase regulatory subunit 1B |
| chr1_-_197115818 | 26.59 |
ENST00000367409.4
ENST00000294732.7 |
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr14_+_58711539 | 24.98 |
ENST00000216455.4
ENST00000412908.2 ENST00000557508.1 |
PSMA3
|
proteasome (prosome, macropain) subunit, alpha type, 3 |
| chr17_+_1733276 | 24.60 |
ENST00000254719.5
|
RPA1
|
replication protein A1, 70kDa |
| chr7_-_99699538 | 23.13 |
ENST00000343023.6
ENST00000303887.5 |
MCM7
|
minichromosome maintenance complex component 7 |
| chr5_+_33440802 | 22.91 |
ENST00000502553.1
ENST00000514259.1 ENST00000265112.3 |
TARS
|
threonyl-tRNA synthetase |
| chr15_-_91537723 | 22.90 |
ENST00000394249.3
ENST00000559811.1 ENST00000442656.2 ENST00000557905.1 ENST00000361919.3 |
PRC1
|
protein regulator of cytokinesis 1 |
| chrX_+_19373700 | 22.72 |
ENST00000379804.1
|
PDHA1
|
pyruvate dehydrogenase (lipoamide) alpha 1 |
| chr5_+_177631523 | 22.45 |
ENST00000506339.1
ENST00000355836.5 ENST00000514633.1 ENST00000515193.1 ENST00000506259.1 ENST00000504898.1 |
HNRNPAB
|
heterogeneous nuclear ribonucleoprotein A/B |
| chr5_+_33441053 | 22.35 |
ENST00000541634.1
ENST00000455217.2 ENST00000414361.2 |
TARS
|
threonyl-tRNA synthetase |
| chr20_+_44441304 | 21.45 |
ENST00000352551.5
|
UBE2C
|
ubiquitin-conjugating enzyme E2C |
| chr5_+_177631497 | 21.02 |
ENST00000358344.3
|
HNRNPAB
|
heterogeneous nuclear ribonucleoprotein A/B |
| chr20_+_44441215 | 20.59 |
ENST00000356455.4
ENST00000405520.1 |
UBE2C
|
ubiquitin-conjugating enzyme E2C |
| chr5_+_44809027 | 19.89 |
ENST00000507110.1
|
MRPS30
|
mitochondrial ribosomal protein S30 |
| chr18_-_33702078 | 19.40 |
ENST00000586829.1
|
SLC39A6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr18_-_33709268 | 18.83 |
ENST00000269187.5
ENST00000590986.1 ENST00000440549.2 |
SLC39A6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr2_-_153574480 | 18.12 |
ENST00000410080.1
|
PRPF40A
|
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr19_+_50180409 | 18.01 |
ENST00000391851.4
|
PRMT1
|
protein arginine methyltransferase 1 |
| chr19_+_50180507 | 17.25 |
ENST00000454376.2
ENST00000524771.1 |
PRMT1
|
protein arginine methyltransferase 1 |
| chrX_-_151999269 | 17.18 |
ENST00000370277.3
|
CETN2
|
centrin, EF-hand protein, 2 |
| chr1_-_149900122 | 17.06 |
ENST00000271628.8
|
SF3B4
|
splicing factor 3b, subunit 4, 49kDa |
| chr2_-_70520539 | 16.86 |
ENST00000482975.2
ENST00000438261.1 |
SNRPG
|
small nuclear ribonucleoprotein polypeptide G |
| chr2_-_70520832 | 16.81 |
ENST00000454893.1
ENST00000272348.2 |
SNRPG
|
small nuclear ribonucleoprotein polypeptide G |
| chr15_+_80351910 | 16.72 |
ENST00000261749.6
ENST00000561060.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr2_-_10952832 | 16.27 |
ENST00000540494.1
|
PDIA6
|
protein disulfide isomerase family A, member 6 |
| chr12_-_49075941 | 16.19 |
ENST00000553086.1
ENST00000548304.1 |
KANSL2
|
KAT8 regulatory NSL complex subunit 2 |
| chr7_-_86849883 | 16.19 |
ENST00000433078.1
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr14_-_74960030 | 16.11 |
ENST00000553490.1
ENST00000557510.1 |
NPC2
|
Niemann-Pick disease, type C2 |
| chr14_-_74959978 | 16.05 |
ENST00000541064.1
|
NPC2
|
Niemann-Pick disease, type C2 |
| chr19_-_46195029 | 15.76 |
ENST00000588599.1
ENST00000585392.1 ENST00000590212.1 ENST00000587367.1 ENST00000391932.3 |
SNRPD2
|
small nuclear ribonucleoprotein D2 polypeptide 16.5kDa |
| chr7_+_77428149 | 15.67 |
ENST00000415251.2
ENST00000275575.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr14_-_74959994 | 15.51 |
ENST00000238633.2
ENST00000434013.2 |
NPC2
|
Niemann-Pick disease, type C2 |
| chr7_+_12727250 | 15.44 |
ENST00000404894.1
|
ARL4A
|
ADP-ribosylation factor-like 4A |
| chr17_-_47785504 | 15.43 |
ENST00000514907.1
ENST00000503334.1 ENST00000508520.1 |
SLC35B1
|
solute carrier family 35, member B1 |
| chrX_+_70503037 | 15.18 |
ENST00000535149.1
|
NONO
|
non-POU domain containing, octamer-binding |
| chr17_-_5322786 | 15.00 |
ENST00000225696.4
|
NUP88
|
nucleoporin 88kDa |
| chr22_+_24309089 | 14.94 |
ENST00000215770.5
|
DDTL
|
D-dopachrome tautomerase-like |
| chr2_-_10952922 | 14.90 |
ENST00000272227.3
|
PDIA6
|
protein disulfide isomerase family A, member 6 |
| chr17_-_5323480 | 14.72 |
ENST00000573584.1
|
NUP88
|
nucleoporin 88kDa |
| chr7_-_137686791 | 14.41 |
ENST00000452463.1
ENST00000330387.6 ENST00000456390.1 |
CREB3L2
|
cAMP responsive element binding protein 3-like 2 |
| chr7_-_44613494 | 14.35 |
ENST00000431640.1
ENST00000258772.5 |
DDX56
|
DEAD (Asp-Glu-Ala-Asp) box helicase 56 |
| chr5_+_151151504 | 14.28 |
ENST00000356245.3
ENST00000507878.2 |
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr5_+_151151471 | 13.88 |
ENST00000394123.3
ENST00000543466.1 |
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr13_-_50367057 | 13.51 |
ENST00000261667.3
|
KPNA3
|
karyopherin alpha 3 (importin alpha 4) |
| chr2_+_65454926 | 13.51 |
ENST00000542850.1
ENST00000377982.4 |
ACTR2
|
ARP2 actin-related protein 2 homolog (yeast) |
| chr7_-_148725733 | 12.70 |
ENST00000286091.4
|
PDIA4
|
protein disulfide isomerase family A, member 4 |
| chr1_-_167905225 | 12.61 |
ENST00000367846.4
|
MPC2
|
mitochondrial pyruvate carrier 2 |
| chr15_-_64673665 | 12.51 |
ENST00000300035.4
|
KIAA0101
|
KIAA0101 |
| chr2_-_175113088 | 12.47 |
ENST00000409546.1
ENST00000428402.2 |
OLA1
|
Obg-like ATPase 1 |
| chr2_-_175113301 | 12.18 |
ENST00000344357.5
ENST00000284719.3 |
OLA1
|
Obg-like ATPase 1 |
| chr22_-_24316648 | 11.89 |
ENST00000403754.3
ENST00000430101.2 ENST00000398344.4 |
DDT
|
D-dopachrome tautomerase |
| chr15_+_80351977 | 11.89 |
ENST00000559157.1
ENST00000561012.1 ENST00000564367.1 ENST00000558494.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr11_-_66056596 | 11.86 |
ENST00000471387.2
ENST00000359461.6 ENST00000376901.4 |
YIF1A
|
Yip1 interacting factor homolog A (S. cerevisiae) |
| chr1_-_150669500 | 11.74 |
ENST00000271732.3
|
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr4_-_143227088 | 11.70 |
ENST00000511838.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chrX_-_16887963 | 11.66 |
ENST00000380084.4
|
RBBP7
|
retinoblastoma binding protein 7 |
| chr12_+_28410128 | 11.53 |
ENST00000381259.1
ENST00000381256.1 |
CCDC91
|
coiled-coil domain containing 91 |
| chr15_-_66816853 | 11.48 |
ENST00000568588.1
|
RPL4
|
ribosomal protein L4 |
| chr12_-_56694142 | 11.30 |
ENST00000550655.1
ENST00000548567.1 ENST00000551430.2 ENST00000351328.3 |
CS
|
citrate synthase |
| chr17_-_47785265 | 11.29 |
ENST00000511763.1
ENST00000515850.1 ENST00000415270.2 ENST00000240333.6 |
SLC35B1
|
solute carrier family 35, member B1 |
| chr3_+_179280668 | 11.22 |
ENST00000429709.2
ENST00000450518.2 ENST00000392662.1 ENST00000490364.1 |
ACTL6A
|
actin-like 6A |
| chr2_-_61697862 | 10.97 |
ENST00000398571.2
|
USP34
|
ubiquitin specific peptidase 34 |
| chr12_-_49076002 | 10.95 |
ENST00000357861.3
ENST00000550347.1 ENST00000420613.2 ENST00000550931.1 ENST00000550870.1 |
KANSL2
|
KAT8 regulatory NSL complex subunit 2 |
| chr1_-_85742773 | 10.89 |
ENST00000370580.1
|
BCL10
|
B-cell CLL/lymphoma 10 |
| chr16_+_69458537 | 10.89 |
ENST00000515314.1
ENST00000561792.1 ENST00000568237.1 |
CYB5B
|
cytochrome b5 type B (outer mitochondrial membrane) |
| chr4_+_147096837 | 10.89 |
ENST00000296581.5
ENST00000502781.1 |
LSM6
|
LSM6 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr7_+_6048856 | 10.55 |
ENST00000223029.3
ENST00000400479.2 ENST00000395236.2 |
AIMP2
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 2 |
| chrX_+_154444643 | 10.53 |
ENST00000286428.5
|
VBP1
|
von Hippel-Lindau binding protein 1 |
| chr18_-_71959159 | 10.52 |
ENST00000494131.2
ENST00000397914.4 ENST00000340533.4 |
CYB5A
|
cytochrome b5 type A (microsomal) |
| chr7_+_77428066 | 10.13 |
ENST00000422959.2
ENST00000307305.8 ENST00000424760.1 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chrX_+_70503433 | 10.08 |
ENST00000276079.8
ENST00000373856.3 ENST00000373841.1 ENST00000420903.1 |
NONO
|
non-POU domain containing, octamer-binding |
| chr12_-_6960407 | 10.02 |
ENST00000540683.1
ENST00000229265.6 ENST00000535406.1 ENST00000422785.3 |
CDCA3
|
cell division cycle associated 3 |
| chrX_+_151999511 | 9.96 |
ENST00000370274.3
ENST00000440023.1 ENST00000432467.1 |
NSDHL
|
NAD(P) dependent steroid dehydrogenase-like |
| chr15_-_37393406 | 9.76 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr19_+_54619125 | 9.33 |
ENST00000445811.1
ENST00000419967.1 ENST00000445124.1 ENST00000447810.1 |
PRPF31
|
pre-mRNA processing factor 31 |
| chr3_+_138340049 | 9.27 |
ENST00000464668.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr11_+_63754294 | 8.79 |
ENST00000543988.1
|
OTUB1
|
OTU domain, ubiquitin aldehyde binding 1 |
| chr5_+_145826867 | 8.62 |
ENST00000296702.5
ENST00000394421.2 |
TCERG1
|
transcription elongation regulator 1 |
| chr4_+_166248775 | 8.59 |
ENST00000261507.6
ENST00000507013.1 ENST00000393766.2 ENST00000504317.1 |
MSMO1
|
methylsterol monooxygenase 1 |
| chr1_+_43148059 | 8.56 |
ENST00000321358.7
ENST00000332220.6 |
YBX1
|
Y box binding protein 1 |
| chr6_+_160211481 | 8.54 |
ENST00000367034.4
|
MRPL18
|
mitochondrial ribosomal protein L18 |
| chr2_+_131100423 | 8.44 |
ENST00000409935.1
ENST00000409649.1 ENST00000428740.1 |
IMP4
|
IMP4, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
| chr2_-_86422523 | 8.39 |
ENST00000442664.2
ENST00000409051.2 ENST00000449247.2 |
IMMT
|
inner membrane protein, mitochondrial |
| chr10_-_1095050 | 8.37 |
ENST00000381344.3
|
IDI1
|
isopentenyl-diphosphate delta isomerase 1 |
| chr14_+_23790690 | 8.30 |
ENST00000556821.1
|
PABPN1
|
poly(A) binding protein, nuclear 1 |
| chr19_+_13049413 | 8.27 |
ENST00000316448.5
ENST00000588454.1 |
CALR
|
calreticulin |
| chr19_+_45394477 | 8.27 |
ENST00000252487.5
ENST00000405636.2 ENST00000592434.1 ENST00000426677.2 ENST00000589649.1 |
TOMM40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr3_-_52567792 | 8.19 |
ENST00000307092.4
ENST00000422318.2 ENST00000459839.1 |
NT5DC2
|
5'-nucleotidase domain containing 2 |
| chr11_+_57480046 | 8.09 |
ENST00000378312.4
ENST00000278422.4 |
TMX2
|
thioredoxin-related transmembrane protein 2 |
| chr20_+_36322408 | 8.02 |
ENST00000361383.6
ENST00000447935.1 ENST00000405275.2 |
CTNNBL1
|
catenin, beta like 1 |
| chr19_+_19303720 | 8.01 |
ENST00000392324.4
|
RFXANK
|
regulatory factor X-associated ankyrin-containing protein |
| chr11_+_67798363 | 7.97 |
ENST00000525628.1
|
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr5_+_159436120 | 7.93 |
ENST00000522793.1
ENST00000231238.5 |
TTC1
|
tetratricopeptide repeat domain 1 |
| chr19_+_17420340 | 7.91 |
ENST00000359866.4
|
DDA1
|
DET1 and DDB1 associated 1 |
| chr11_-_3818932 | 7.75 |
ENST00000324932.7
ENST00000359171.4 |
NUP98
|
nucleoporin 98kDa |
| chr2_-_69664586 | 7.70 |
ENST00000303698.3
ENST00000394305.1 ENST00000410022.2 |
NFU1
|
NFU1 iron-sulfur cluster scaffold homolog (S. cerevisiae) |
| chr4_-_143226979 | 7.53 |
ENST00000514525.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr7_+_55433131 | 7.53 |
ENST00000254770.2
|
LANCL2
|
LanC lantibiotic synthetase component C-like 2 (bacterial) |
| chr1_-_35658736 | 7.43 |
ENST00000357214.5
|
SFPQ
|
splicing factor proline/glutamine-rich |
| chr15_-_77363441 | 7.42 |
ENST00000346495.2
ENST00000424443.3 ENST00000561277.1 |
TSPAN3
|
tetraspanin 3 |
| chr2_-_96874553 | 7.42 |
ENST00000337288.5
ENST00000443962.1 |
STARD7
|
StAR-related lipid transfer (START) domain containing 7 |
| chr10_+_85899196 | 7.41 |
ENST00000372134.3
|
GHITM
|
growth hormone inducible transmembrane protein |
| chr17_-_7307358 | 7.37 |
ENST00000576017.1
ENST00000302422.3 ENST00000535512.1 |
TMEM256
TMEM256-PLSCR3
|
transmembrane protein 256 TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr6_+_30687978 | 7.36 |
ENST00000327892.8
ENST00000435534.1 |
TUBB
|
tubulin, beta class I |
| chr13_-_41593425 | 7.36 |
ENST00000239882.3
|
ELF1
|
E74-like factor 1 (ets domain transcription factor) |
| chr1_-_23670813 | 7.26 |
ENST00000374612.1
|
HNRNPR
|
heterogeneous nuclear ribonucleoprotein R |
| chr17_+_79650962 | 7.20 |
ENST00000329138.4
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate |
| chr10_+_70661014 | 7.19 |
ENST00000373585.3
|
DDX50
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 50 |
| chr1_-_23670817 | 7.19 |
ENST00000478691.1
|
HNRNPR
|
heterogeneous nuclear ribonucleoprotein R |
| chr1_+_24286287 | 7.14 |
ENST00000334351.7
ENST00000374468.1 |
PNRC2
|
proline-rich nuclear receptor coactivator 2 |
| chr17_+_8191815 | 7.13 |
ENST00000226105.6
ENST00000407006.4 ENST00000580434.1 ENST00000439238.3 |
RANGRF
|
RAN guanine nucleotide release factor |
| chr14_-_34931458 | 7.11 |
ENST00000298130.4
|
SPTSSA
|
serine palmitoyltransferase, small subunit A |
| chr12_+_98909351 | 7.09 |
ENST00000343315.5
ENST00000266732.4 ENST00000393053.2 |
TMPO
|
thymopoietin |
| chr9_+_140135665 | 7.01 |
ENST00000340384.4
|
TUBB4B
|
tubulin, beta 4B class IVb |
| chr11_-_3818688 | 6.96 |
ENST00000355260.3
ENST00000397004.4 ENST00000397007.4 ENST00000532475.1 |
NUP98
|
nucleoporin 98kDa |
| chr5_+_179125907 | 6.95 |
ENST00000247461.4
ENST00000452673.2 ENST00000502498.1 ENST00000507307.1 ENST00000513246.1 ENST00000502673.1 ENST00000506654.1 ENST00000512607.2 ENST00000510810.1 |
CANX
|
calnexin |
| chr4_+_1283639 | 6.93 |
ENST00000303400.4
ENST00000505177.2 ENST00000503653.1 ENST00000264750.6 ENST00000502558.1 ENST00000452175.2 ENST00000514708.1 |
MAEA
|
macrophage erythroblast attacher |
| chr19_+_19303572 | 6.82 |
ENST00000407360.3
ENST00000540981.1 |
RFXANK
|
regulatory factor X-associated ankyrin-containing protein |
| chr1_-_20987851 | 6.78 |
ENST00000464364.1
ENST00000602624.2 |
DDOST
|
dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit (non-catalytic) |
| chr1_-_154946825 | 6.71 |
ENST00000368453.4
ENST00000368450.1 ENST00000366442.2 |
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr14_-_78083112 | 6.55 |
ENST00000216484.2
|
SPTLC2
|
serine palmitoyltransferase, long chain base subunit 2 |
| chr1_-_20987889 | 6.52 |
ENST00000415136.2
|
DDOST
|
dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit (non-catalytic) |
| chr9_+_12693336 | 6.51 |
ENST00000381137.2
ENST00000388918.5 |
TYRP1
|
tyrosinase-related protein 1 |
| chr16_-_11036300 | 6.44 |
ENST00000331808.4
|
DEXI
|
Dexi homolog (mouse) |
| chr6_-_111927062 | 6.43 |
ENST00000359831.4
|
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr19_+_19496624 | 6.36 |
ENST00000494516.2
ENST00000360315.3 ENST00000252577.5 |
GATAD2A
|
GATA zinc finger domain containing 2A |
| chr16_+_69458428 | 6.33 |
ENST00000512062.1
ENST00000307892.8 |
CYB5B
|
cytochrome b5 type B (outer mitochondrial membrane) |
| chr3_-_33481835 | 6.33 |
ENST00000283629.3
|
UBP1
|
upstream binding protein 1 (LBP-1a) |
| chr12_+_21654714 | 6.22 |
ENST00000542038.1
ENST00000540141.1 ENST00000229314.5 |
GOLT1B
|
golgi transport 1B |
| chr11_+_95523621 | 6.21 |
ENST00000325542.5
ENST00000325486.5 ENST00000544522.1 ENST00000541365.1 |
CEP57
|
centrosomal protein 57kDa |
| chr12_-_6961050 | 6.14 |
ENST00000538862.2
|
CDCA3
|
cell division cycle associated 3 |
| chr7_-_25019760 | 6.10 |
ENST00000352860.1
ENST00000353930.1 ENST00000431825.2 ENST00000313367.2 |
OSBPL3
|
oxysterol binding protein-like 3 |
| chr13_+_103249322 | 6.07 |
ENST00000376065.4
ENST00000376052.3 |
TPP2
|
tripeptidyl peptidase II |
| chr15_-_77363513 | 6.03 |
ENST00000267970.4
|
TSPAN3
|
tetraspanin 3 |
| chr19_-_13227463 | 6.03 |
ENST00000437766.1
ENST00000221504.8 |
TRMT1
|
tRNA methyltransferase 1 homolog (S. cerevisiae) |
| chr16_+_68119247 | 5.99 |
ENST00000575270.1
|
NFATC3
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 |
| chr2_-_201828356 | 5.96 |
ENST00000234296.2
|
ORC2
|
origin recognition complex, subunit 2 |
| chr19_+_16222439 | 5.94 |
ENST00000300935.3
|
RAB8A
|
RAB8A, member RAS oncogene family |
| chr6_+_32146131 | 5.92 |
ENST00000375094.3
|
RNF5
|
ring finger protein 5, E3 ubiquitin protein ligase |
| chr11_+_67798114 | 5.84 |
ENST00000453471.2
ENST00000528492.1 ENST00000526339.1 ENST00000525419.1 |
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr10_-_126107482 | 5.83 |
ENST00000368845.5
ENST00000539214.1 |
OAT
|
ornithine aminotransferase |
| chr22_-_42486747 | 5.71 |
ENST00000602404.1
|
NDUFA6
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 6, 14kDa |
| chr15_-_77363375 | 5.67 |
ENST00000559494.1
|
TSPAN3
|
tetraspanin 3 |
| chr19_-_13227534 | 5.62 |
ENST00000588229.1
ENST00000357720.4 |
TRMT1
|
tRNA methyltransferase 1 homolog (S. cerevisiae) |
| chr4_-_105416039 | 5.61 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr15_+_89631381 | 5.58 |
ENST00000352732.5
|
ABHD2
|
abhydrolase domain containing 2 |
| chr11_+_67798090 | 5.51 |
ENST00000313468.5
|
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chrX_-_77150985 | 5.51 |
ENST00000358075.6
|
MAGT1
|
magnesium transporter 1 |
| chr17_-_74733404 | 5.45 |
ENST00000508921.3
ENST00000583836.1 ENST00000358156.6 ENST00000392485.2 ENST00000359995.5 |
SRSF2
|
serine/arginine-rich splicing factor 2 |
| chr19_+_39421556 | 5.40 |
ENST00000407800.2
ENST00000402029.3 |
MRPS12
|
mitochondrial ribosomal protein S12 |
| chr2_-_191878874 | 5.33 |
ENST00000392322.3
ENST00000392323.2 ENST00000424722.1 ENST00000361099.3 |
STAT1
|
signal transducer and activator of transcription 1, 91kDa |
| chr3_-_165555200 | 5.32 |
ENST00000479451.1
ENST00000540653.1 ENST00000488954.1 ENST00000264381.3 |
BCHE
|
butyrylcholinesterase |
| chr9_-_99801592 | 5.21 |
ENST00000259470.5
|
CTSV
|
cathepsin V |
| chr6_+_26199737 | 5.09 |
ENST00000359985.1
|
HIST1H2BF
|
histone cluster 1, H2bf |
| chr14_+_23790655 | 5.05 |
ENST00000397276.2
|
PABPN1
|
poly(A) binding protein, nuclear 1 |
| chr3_+_120461484 | 5.00 |
ENST00000484715.1
ENST00000469772.1 ENST00000283875.5 ENST00000492959.1 |
GTF2E1
|
general transcription factor IIE, polypeptide 1, alpha 56kDa |
| chr19_-_47616992 | 4.98 |
ENST00000253048.5
|
ZC3H4
|
zinc finger CCCH-type containing 4 |
| chr4_+_169013666 | 4.91 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chr16_+_69345243 | 4.87 |
ENST00000254950.11
|
VPS4A
|
vacuolar protein sorting 4 homolog A (S. cerevisiae) |
| chr22_-_32651326 | 4.80 |
ENST00000266086.4
|
SLC5A4
|
solute carrier family 5 (glucose activated ion channel), member 4 |
| chr15_-_43802769 | 4.51 |
ENST00000263801.3
|
TP53BP1
|
tumor protein p53 binding protein 1 |
| chr1_-_20987982 | 4.50 |
ENST00000375048.3
|
DDOST
|
dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit (non-catalytic) |
| chr15_+_41523335 | 4.47 |
ENST00000334660.5
|
CHP1
|
calcineurin-like EF-hand protein 1 |
| chr3_-_119813264 | 4.41 |
ENST00000264235.8
|
GSK3B
|
glycogen synthase kinase 3 beta |
| chr22_-_30960876 | 4.32 |
ENST00000401975.1
ENST00000428682.1 ENST00000423299.1 |
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr6_+_33172407 | 4.25 |
ENST00000374662.3
|
HSD17B8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr1_-_23670752 | 4.25 |
ENST00000302271.6
ENST00000426846.2 ENST00000427764.2 ENST00000606561.1 ENST00000374616.3 |
HNRNPR
|
heterogeneous nuclear ribonucleoprotein R |
| chr3_-_195538760 | 4.21 |
ENST00000475231.1
|
MUC4
|
mucin 4, cell surface associated |
| chr8_+_120428546 | 4.21 |
ENST00000259526.3
|
NOV
|
nephroblastoma overexpressed |
| chr7_+_138145145 | 4.11 |
ENST00000415680.2
|
TRIM24
|
tripartite motif containing 24 |
| chr1_-_47655686 | 4.08 |
ENST00000294338.2
|
PDZK1IP1
|
PDZK1 interacting protein 1 |
| chr4_-_39979576 | 4.05 |
ENST00000303538.8
ENST00000503396.1 |
PDS5A
|
PDS5, regulator of cohesion maintenance, homolog A (S. cerevisiae) |
| chr10_+_1095416 | 4.03 |
ENST00000358220.1
|
WDR37
|
WD repeat domain 37 |
| chr2_+_162087577 | 3.97 |
ENST00000439442.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr12_-_21654479 | 3.94 |
ENST00000421138.2
ENST00000444129.2 ENST00000539672.1 ENST00000542432.1 ENST00000536964.1 ENST00000536240.1 ENST00000396093.3 ENST00000314748.6 |
RECQL
|
RecQ protein-like (DNA helicase Q1-like) |
| chr5_+_179125368 | 3.85 |
ENST00000502296.1
ENST00000504734.1 ENST00000415618.2 |
CANX
|
calnexin |
| chrX_-_15872914 | 3.84 |
ENST00000380291.1
ENST00000545766.1 ENST00000421527.2 ENST00000329235.2 |
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr16_+_68279207 | 3.83 |
ENST00000413021.2
ENST00000565744.1 ENST00000219345.5 |
PLA2G15
|
phospholipase A2, group XV |
| chr9_+_34646624 | 3.80 |
ENST00000450095.2
ENST00000556278.1 |
GALT
GALT
|
galactose-1-phosphate uridylyltransferase Uncharacterized protein |
| chr2_-_192711968 | 3.78 |
ENST00000304141.4
|
SDPR
|
serum deprivation response |
| chr10_-_127505167 | 3.78 |
ENST00000368786.1
|
UROS
|
uroporphyrinogen III synthase |
| chr17_+_46970134 | 3.75 |
ENST00000503641.1
ENST00000514808.1 |
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr3_-_105588231 | 3.72 |
ENST00000545639.1
ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr17_+_45727204 | 3.70 |
ENST00000290158.4
|
KPNB1
|
karyopherin (importin) beta 1 |
| chr11_+_64018955 | 3.69 |
ENST00000279230.6
ENST00000540288.1 ENST00000325234.5 |
PLCB3
|
phospholipase C, beta 3 (phosphatidylinositol-specific) |
| chr16_+_46723552 | 3.66 |
ENST00000219097.2
ENST00000568364.2 |
ORC6
|
origin recognition complex, subunit 6 |
| chr17_-_47841485 | 3.64 |
ENST00000506156.1
ENST00000240364.2 |
FAM117A
|
family with sequence similarity 117, member A |
| chr19_+_41770236 | 3.64 |
ENST00000392006.3
|
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr11_-_122932730 | 3.63 |
ENST00000532182.1
ENST00000524590.1 ENST00000528292.1 ENST00000533540.1 ENST00000525463.1 |
HSPA8
|
heat shock 70kDa protein 8 |
| chr17_-_79980734 | 3.63 |
ENST00000584600.1
ENST00000584347.1 ENST00000580435.1 ENST00000306704.6 ENST00000392359.3 |
STRA13
|
stimulated by retinoic acid 13 |
| chr12_+_3000037 | 3.57 |
ENST00000544943.1
ENST00000448120.2 |
TULP3
|
tubby like protein 3 |
| chr15_-_43785303 | 3.52 |
ENST00000382039.3
ENST00000450115.2 ENST00000382044.4 |
TP53BP1
|
tumor protein p53 binding protein 1 |
| chr2_+_234637754 | 3.50 |
ENST00000482026.1
ENST00000609767.1 |
UGT1A3
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A3 UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr1_+_167905894 | 3.46 |
ENST00000367843.3
ENST00000432587.2 ENST00000312263.6 |
DCAF6
|
DDB1 and CUL4 associated factor 6 |
| chr15_+_89631647 | 3.43 |
ENST00000569550.1
ENST00000565066.1 ENST00000565973.1 |
ABHD2
|
abhydrolase domain containing 2 |
| chr3_+_179322481 | 3.38 |
ENST00000259037.3
|
NDUFB5
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa |
| chr11_+_3819049 | 3.37 |
ENST00000396986.2
ENST00000300730.6 ENST00000532535.1 ENST00000396993.4 ENST00000396991.2 ENST00000532523.1 ENST00000459679.1 ENST00000464261.1 ENST00000464906.2 ENST00000464441.1 |
PGAP2
|
post-GPI attachment to proteins 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of VSX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 15.1 | 45.3 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 11.9 | 47.7 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 10.9 | 65.4 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 8.9 | 26.7 | GO:0072334 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 5.3 | 42.0 | GO:0010994 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) positive regulation of exit from mitosis(GO:0031536) |
| 5.1 | 25.3 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 4.5 | 13.5 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 4.4 | 13.3 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 4.2 | 29.7 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 3.9 | 35.3 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 3.8 | 38.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 3.6 | 10.9 | GO:0032765 | lymphotoxin A production(GO:0032641) positive regulation of mast cell cytokine production(GO:0032765) lymphotoxin A biosynthetic process(GO:0042109) |
| 3.5 | 10.6 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 3.3 | 26.6 | GO:0090306 | spindle assembly involved in meiosis(GO:0090306) |
| 2.9 | 14.7 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 2.8 | 8.5 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 2.3 | 23.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 1.9 | 5.7 | GO:0006258 | UDP-glucose catabolic process(GO:0006258) galactose catabolic process via UDP-galactose(GO:0033499) |
| 1.8 | 27.1 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 1.8 | 7.1 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 1.8 | 5.3 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 1.6 | 6.5 | GO:1904502 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 1.6 | 4.9 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 1.6 | 9.3 | GO:0048254 | snoRNA localization(GO:0048254) |
| 1.5 | 10.6 | GO:1900045 | negative regulation of histone ubiquitination(GO:0033183) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) negative regulation of protein polyubiquitination(GO:1902915) |
| 1.5 | 13.3 | GO:1901419 | regulation of response to alcohol(GO:1901419) |
| 1.5 | 4.4 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 1.4 | 5.7 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 1.4 | 4.2 | GO:1990523 | bone regeneration(GO:1990523) |
| 1.4 | 8.3 | GO:0002501 | peptide antigen assembly with MHC protein complex(GO:0002501) |
| 1.3 | 22.9 | GO:0000022 | mitotic spindle elongation(GO:0000022) |
| 1.3 | 5.3 | GO:2000697 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of nephron tubule epithelial cell differentiation(GO:0072183) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) negative regulation of epithelial cell differentiation involved in kidney development(GO:2000697) |
| 1.3 | 6.3 | GO:0061738 | late endosomal microautophagy(GO:0061738) negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 1.2 | 7.4 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 1.2 | 8.6 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 1.2 | 17.1 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 1.2 | 3.7 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 1.1 | 55.2 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 1.1 | 28.6 | GO:0006625 | protein targeting to peroxisome(GO:0006625) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 1.1 | 3.2 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 1.1 | 2.1 | GO:0060434 | bronchus morphogenesis(GO:0060434) |
| 1.1 | 49.4 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 1.0 | 41.8 | GO:0006294 | nucleotide-excision repair, preincision complex assembly(GO:0006294) |
| 1.0 | 5.2 | GO:1990834 | response to odorant(GO:1990834) |
| 1.0 | 8.0 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 1.0 | 6.9 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 1.0 | 5.8 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 1.0 | 11.5 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.9 | 3.8 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) uroporphyrinogen III metabolic process(GO:0046502) |
| 0.9 | 2.8 | GO:0071934 | thiamine transport(GO:0015888) thiamine transmembrane transport(GO:0071934) |
| 0.9 | 7.1 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.9 | 0.9 | GO:1903179 | regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) |
| 0.8 | 10.5 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.8 | 10.8 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.8 | 2.3 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 0.8 | 19.1 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.7 | 6.5 | GO:0048023 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.7 | 1.4 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.7 | 3.5 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.7 | 5.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.7 | 4.0 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.7 | 5.3 | GO:0070885 | positive regulation of sodium:proton antiporter activity(GO:0032417) negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.6 | 32.1 | GO:0032508 | DNA duplex unwinding(GO:0032508) |
| 0.6 | 2.5 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.6 | 3.7 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.6 | 13.5 | GO:0019054 | modulation by virus of host process(GO:0019054) |
| 0.6 | 10.5 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.6 | 7.2 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.6 | 8.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.5 | 21.5 | GO:0042407 | cristae formation(GO:0042407) |
| 0.5 | 44.0 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.5 | 11.2 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.5 | 4.3 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.5 | 11.0 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.5 | 10.0 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.5 | 5.2 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.5 | 11.9 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.5 | 2.3 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.5 | 1.4 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.4 | 7.2 | GO:1903543 | positive regulation of exosomal secretion(GO:1903543) |
| 0.4 | 11.6 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.4 | 11.7 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.4 | 8.5 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.4 | 7.3 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.4 | 5.9 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.4 | 7.7 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.3 | 25.3 | GO:0070125 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 0.3 | 6.2 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.3 | 10.9 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.3 | 1.7 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.3 | 17.8 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.3 | 21.0 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.3 | 5.9 | GO:0098969 | neurotransmitter receptor transport to plasma membrane(GO:0098877) neurotransmitter receptor transport to postsynaptic membrane(GO:0098969) establishment of protein localization to postsynaptic membrane(GO:1903540) |
| 0.3 | 4.3 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.3 | 7.4 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.3 | 9.6 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.3 | 5.4 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.3 | 7.0 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.3 | 0.3 | GO:1900158 | negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.3 | 25.6 | GO:0051436 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) |
| 0.3 | 3.8 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.2 | 1.5 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.2 | 14.4 | GO:0042267 | natural killer cell mediated cytotoxicity(GO:0042267) |
| 0.2 | 9.0 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.2 | 9.2 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.2 | 2.3 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.2 | 16.4 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.2 | 6.2 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.2 | 0.6 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.2 | 13.9 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.2 | 0.7 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.2 | 27.1 | GO:0001837 | epithelial to mesenchymal transition(GO:0001837) |
| 0.2 | 7.1 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.2 | 6.1 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.1 | 4.0 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.1 | 1.5 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 1.9 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.1 | 6.7 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.1 | 0.8 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 | 34.2 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.1 | 0.9 | GO:0042421 | norepinephrine biosynthetic process(GO:0042421) |
| 0.1 | 9.8 | GO:0045638 | negative regulation of myeloid cell differentiation(GO:0045638) |
| 0.1 | 6.7 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.1 | 0.4 | GO:0061743 | motor learning(GO:0061743) |
| 0.1 | 1.5 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.1 | 3.2 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 1.4 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 17.2 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.1 | 2.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 3.4 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 3.7 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.1 | 3.8 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.1 | 7.3 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.1 | 0.8 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 0.5 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.1 | 4.2 | GO:0016445 | somatic diversification of immunoglobulins(GO:0016445) |
| 0.1 | 1.5 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 2.0 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 2.0 | GO:0006397 | mRNA processing(GO:0006397) |
| 0.0 | 3.1 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.0 | 6.3 | GO:0019079 | viral genome replication(GO:0019079) |
| 0.0 | 0.7 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.5 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 11.9 | GO:0007265 | Ras protein signal transduction(GO:0007265) |
| 0.0 | 5.1 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.8 | GO:0019228 | neuronal action potential(GO:0019228) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.4 | 114.9 | GO:0034709 | methylosome(GO:0034709) |
| 4.1 | 32.7 | GO:0042382 | paraspeckles(GO:0042382) |
| 3.8 | 26.6 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 3.4 | 13.7 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 3.0 | 29.8 | GO:0070938 | contractile ring(GO:0070938) |
| 2.5 | 14.7 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 2.4 | 7.2 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 2.2 | 10.9 | GO:0032449 | CBM complex(GO:0032449) lipopolysaccharide receptor complex(GO:0046696) |
| 2.1 | 25.0 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 2.0 | 6.0 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 1.9 | 23.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 1.8 | 34.7 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 1.8 | 23.1 | GO:0042555 | MCM complex(GO:0042555) |
| 1.8 | 24.6 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 1.7 | 8.4 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 1.6 | 10.9 | GO:0005688 | U6 snRNP(GO:0005688) |
| 1.5 | 25.6 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 1.5 | 10.5 | GO:0016272 | prefoldin complex(GO:0016272) |
| 1.5 | 14.6 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 1.4 | 42.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 1.4 | 5.4 | GO:0035061 | interchromatin granule(GO:0035061) |
| 1.3 | 18.1 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 1.2 | 8.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 1.2 | 8.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 1.2 | 9.3 | GO:0005687 | U4 snRNP(GO:0005687) |
| 1.1 | 5.7 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 1.1 | 6.7 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 1.1 | 13.3 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 1.1 | 10.8 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 1.1 | 17.2 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 1.0 | 8.3 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 1.0 | 38.9 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 1.0 | 55.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 1.0 | 38.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.9 | 12.7 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.9 | 13.5 | GO:0030478 | actin cap(GO:0030478) |
| 0.9 | 6.3 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.8 | 2.5 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.7 | 9.0 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.7 | 4.0 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.6 | 10.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.6 | 16.6 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.5 | 5.9 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.5 | 49.0 | GO:0043657 | host(GO:0018995) host cell(GO:0043657) |
| 0.5 | 52.3 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.5 | 10.6 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.5 | 10.3 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.5 | 4.9 | GO:0090543 | Flemming body(GO:0090543) |
| 0.5 | 5.0 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.4 | 28.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.4 | 3.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.4 | 6.5 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.4 | 8.0 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.4 | 4.4 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.4 | 3.7 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.4 | 1.4 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.4 | 2.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.3 | 6.1 | GO:0097038 | filopodium tip(GO:0032433) perinuclear endoplasmic reticulum(GO:0097038) |
| 0.3 | 21.0 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.3 | 0.9 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.3 | 1.9 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.2 | 25.1 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.2 | 1.7 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.2 | 22.6 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.2 | 11.5 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.2 | 5.4 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.2 | 8.5 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.2 | 0.8 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.2 | 3.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.2 | 3.7 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.2 | 5.9 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.2 | 9.8 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.1 | 1.5 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 10.4 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 10.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 4.2 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 16.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 1.4 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 28.1 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 2.5 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 7.9 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 5.9 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 12.7 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 16.4 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 7.5 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 0.9 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 27.5 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 1.2 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 5.2 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 1.9 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 7.3 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 3.8 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.1 | 1.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 4.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 1.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 2.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 6.9 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 26.5 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 0.8 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 4.0 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 4.3 | GO:0005740 | mitochondrial envelope(GO:0005740) |
| 0.0 | 2.0 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 2.3 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 2.0 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 15.1 | 45.3 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 10.9 | 65.4 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 8.9 | 26.7 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 7.9 | 55.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 5.7 | 22.7 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 4.6 | 13.7 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 3.8 | 19.2 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 3.4 | 17.2 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 3.3 | 10.0 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 3.0 | 11.9 | GO:0033981 | D-dopachrome decarboxylase activity(GO:0033981) |
| 2.9 | 11.6 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 2.8 | 8.4 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 2.3 | 23.3 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 2.3 | 27.1 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 2.3 | 11.3 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 2.2 | 24.6 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 2.1 | 43.9 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 2.0 | 38.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 1.8 | 7.3 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 1.7 | 5.2 | GO:0030984 | kininogen binding(GO:0030984) |
| 1.6 | 12.6 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 1.3 | 8.0 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 1.3 | 9.3 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 1.3 | 5.3 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 1.2 | 55.2 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 1.2 | 4.8 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 1.2 | 8.3 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 1.1 | 10.6 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 1.0 | 7.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 1.0 | 6.1 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 1.0 | 9.8 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 1.0 | 6.7 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.9 | 2.8 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.9 | 8.4 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.9 | 8.3 | GO:0015288 | porin activity(GO:0015288) |
| 0.9 | 42.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.9 | 49.2 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.9 | 34.0 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.9 | 4.3 | GO:0070404 | NADH binding(GO:0070404) |
| 0.8 | 25.0 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.8 | 25.5 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.7 | 8.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.7 | 9.0 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.7 | 14.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.7 | 4.0 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.6 | 3.7 | GO:0047115 | phenanthrene 9,10-monooxygenase activity(GO:0018636) ketosteroid monooxygenase activity(GO:0047086) trans-1,2-dihydrobenzene-1,2-diol dehydrogenase activity(GO:0047115) |
| 0.6 | 4.3 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.6 | 8.5 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.6 | 9.6 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.6 | 27.0 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.5 | 10.8 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.5 | 5.7 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.5 | 5.7 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.5 | 3.1 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.5 | 5.9 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.4 | 9.4 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.4 | 10.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.4 | 3.8 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.4 | 22.9 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.4 | 41.2 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.4 | 11.4 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.4 | 10.5 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.3 | 14.8 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.3 | 5.8 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.2 | 3.5 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.2 | 6.8 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.2 | 0.7 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.2 | 3.9 | GO:0005521 | lamin binding(GO:0005521) |
| 0.2 | 5.9 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.2 | 3.7 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.2 | 6.2 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.2 | 6.3 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.2 | 38.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.2 | 4.4 | GO:0050321 | protein kinase A catalytic subunit binding(GO:0034236) tau-protein kinase activity(GO:0050321) |
| 0.2 | 4.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.2 | 8.6 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.2 | 50.8 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.2 | 11.0 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.2 | 2.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.2 | 3.2 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.2 | 9.9 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.2 | 5.3 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.2 | 10.6 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 1.5 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 2.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.8 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.1 | 0.5 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.1 | 3.7 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.1 | 14.9 | GO:0016829 | lyase activity(GO:0016829) |
| 0.1 | 4.9 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.1 | 2.0 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 13.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 3.5 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 4.9 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 1.5 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.1 | 2.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 8.1 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.1 | 5.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.4 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 3.8 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 20.2 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 2.1 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 3.1 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 1.7 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.8 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 3.5 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.9 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 3.9 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 0.6 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.3 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 2.7 | GO:0005516 | calmodulin binding(GO:0005516) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 25.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.8 | 47.7 | PID ATR PATHWAY | ATR signaling pathway |
| 0.6 | 45.7 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.5 | 10.9 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.4 | 22.9 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.4 | 22.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.4 | 5.3 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.3 | 13.3 | PID MYC PATHWAY | C-MYC pathway |
| 0.3 | 60.2 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.3 | 14.1 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.3 | 13.5 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.2 | 8.0 | PID ATM PATHWAY | ATM pathway |
| 0.2 | 5.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.2 | 12.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.2 | 8.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.2 | 8.6 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 5.7 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.1 | 9.0 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 3.7 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 7.4 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 12.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 6.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 2.5 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.8 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 1.0 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.7 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 38.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 2.1 | 33.7 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 2.1 | 24.6 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 1.6 | 55.8 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 1.5 | 42.6 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 1.4 | 23.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 1.3 | 19.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 1.1 | 19.1 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 1.0 | 12.1 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 1.0 | 13.3 | REACTOME PROCESSING OF CAPPED INTRONLESS PRE MRNA | Genes involved in Processing of Capped Intronless Pre-mRNA |
| 0.9 | 15.8 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.9 | 35.5 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.9 | 80.2 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.9 | 14.6 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.7 | 10.9 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.7 | 53.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.6 | 17.8 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.6 | 12.1 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.6 | 35.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.5 | 23.9 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.5 | 15.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.5 | 10.9 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.4 | 8.0 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.4 | 7.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.4 | 31.6 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.3 | 28.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.3 | 3.7 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.3 | 28.8 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.3 | 13.8 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.3 | 5.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.3 | 11.1 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.3 | 7.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 14.0 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.2 | 8.5 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.2 | 11.1 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.2 | 5.8 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.2 | 13.3 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 1.7 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 4.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 3.5 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.1 | 4.3 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 1.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 2.3 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 11.4 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.9 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 2.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.2 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.7 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.7 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 2.6 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 3.2 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.4 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |