Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
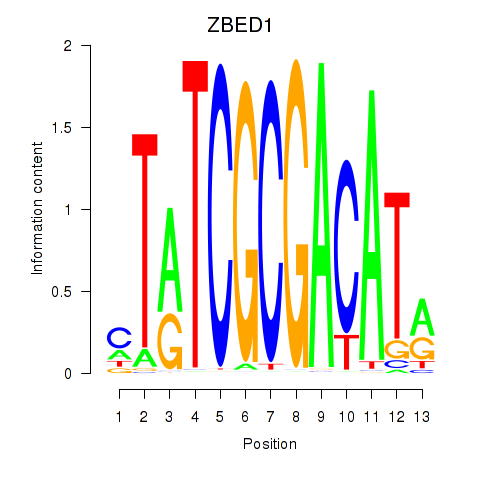
Results for ZBED1
Z-value: 0.60
Transcription factors associated with ZBED1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBED1
|
ENSG00000214717.5 | zinc finger BED-type containing 1 |
|
ZBED1
|
ENSGR0000214717.5 | zinc finger BED-type containing 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBED1 | hg19_v2_chrX_-_2418596_2418621 | 0.44 | 1.6e-11 | Click! |
Activity profile of ZBED1 motif
Sorted Z-values of ZBED1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_151254738 | 10.51 |
ENST00000336715.6
ENST00000324048.5 ENST00000368879.2 |
ZNF687
|
zinc finger protein 687 |
| chr1_+_36023370 | 5.83 |
ENST00000356090.4
ENST00000373243.2 |
NCDN
|
neurochondrin |
| chr7_+_139025875 | 5.65 |
ENST00000297534.6
|
C7orf55
|
chromosome 7 open reading frame 55 |
| chr17_+_4843654 | 5.23 |
ENST00000575111.1
|
RNF167
|
ring finger protein 167 |
| chr17_+_4843413 | 5.08 |
ENST00000572430.1
ENST00000262482.6 |
RNF167
|
ring finger protein 167 |
| chr17_+_4843679 | 4.78 |
ENST00000576229.1
|
RNF167
|
ring finger protein 167 |
| chr17_-_4843316 | 4.34 |
ENST00000544061.2
|
SLC25A11
|
solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11 |
| chr17_+_4843303 | 3.70 |
ENST00000571816.1
|
RNF167
|
ring finger protein 167 |
| chr9_+_33290491 | 3.46 |
ENST00000379540.3
ENST00000379521.4 ENST00000318524.6 |
NFX1
|
nuclear transcription factor, X-box binding 1 |
| chr19_+_54641444 | 3.23 |
ENST00000221232.5
ENST00000358389.3 |
CNOT3
|
CCR4-NOT transcription complex, subunit 3 |
| chr8_+_38089198 | 3.16 |
ENST00000528358.1
ENST00000529642.1 ENST00000532222.1 ENST00000520272.2 |
DDHD2
|
DDHD domain containing 2 |
| chr22_+_19701985 | 2.97 |
ENST00000455784.2
ENST00000406395.1 |
SEPT5
|
septin 5 |
| chrX_-_53711064 | 2.87 |
ENST00000342160.3
ENST00000446750.1 |
HUWE1
|
HECT, UBA and WWE domain containing 1, E3 ubiquitin protein ligase |
| chr17_-_4843206 | 2.37 |
ENST00000576951.1
|
SLC25A11
|
solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11 |
| chrX_-_63615297 | 1.63 |
ENST00000374852.3
ENST00000453546.1 |
MTMR8
|
myotubularin related protein 8 |
| chr22_-_36424458 | 0.17 |
ENST00000438146.2
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr1_-_184723942 | 0.03 |
ENST00000318130.8
|
EDEM3
|
ER degradation enhancer, mannosidase alpha-like 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBED1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 6.7 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.4 | 3.5 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.3 | 3.0 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.1 | 3.2 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.1 | 3.2 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 5.8 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.1 | 21.7 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.1 | 1.6 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.2 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 3.2 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 3.0 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 2.9 | GO:1904813 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.0 | 5.8 | GO:0030424 | axon(GO:0030424) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.7 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.1 | 1.6 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 3.2 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 21.7 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 3.5 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.9 | PID P53 REGULATION PATHWAY | p53 pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.7 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 3.5 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 2.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 2.9 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |