Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
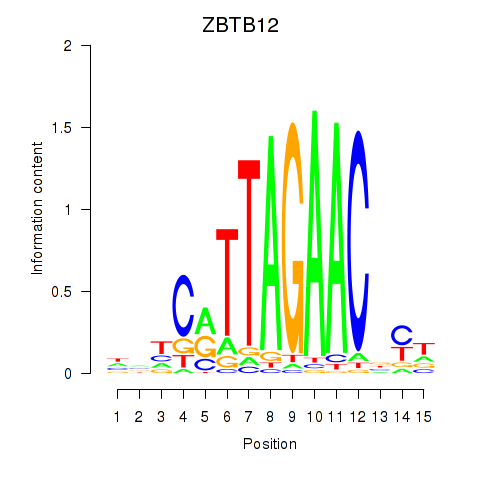
Results for ZBTB12
Z-value: 0.89
Transcription factors associated with ZBTB12
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB12
|
ENSG00000204366.3 | zinc finger and BTB domain containing 12 |
Activity profile of ZBTB12 motif
Sorted Z-values of ZBTB12 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_158979792 | 20.08 |
ENST00000359709.3
ENST00000430894.2 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr1_+_158979686 | 19.25 |
ENST00000368132.3
ENST00000295809.7 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr1_+_158979680 | 19.19 |
ENST00000368131.4
ENST00000340979.6 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr1_+_32757668 | 18.59 |
ENST00000373548.3
|
HDAC1
|
histone deacetylase 1 |
| chr16_-_67969888 | 17.65 |
ENST00000574576.2
|
PSMB10
|
proteasome (prosome, macropain) subunit, beta type, 10 |
| chr13_-_46716969 | 16.67 |
ENST00000435666.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr1_+_26644441 | 16.22 |
ENST00000374213.2
|
CD52
|
CD52 molecule |
| chr11_+_65769550 | 15.88 |
ENST00000312175.2
ENST00000445560.2 ENST00000530204.1 |
BANF1
|
barrier to autointegration factor 1 |
| chr11_+_65769946 | 15.79 |
ENST00000533166.1
|
BANF1
|
barrier to autointegration factor 1 |
| chr15_+_40453204 | 15.65 |
ENST00000287598.6
ENST00000412359.3 |
BUB1B
|
BUB1 mitotic checkpoint serine/threonine kinase B |
| chr1_+_155583012 | 14.06 |
ENST00000462250.2
|
MSTO1
|
misato 1, mitochondrial distribution and morphology regulator |
| chr12_-_7077125 | 14.04 |
ENST00000545555.2
|
PHB2
|
prohibitin 2 |
| chr15_+_66797627 | 12.02 |
ENST00000565627.1
ENST00000564179.1 |
ZWILCH
|
zwilch kinetochore protein |
| chr15_-_55563072 | 11.95 |
ENST00000567380.1
ENST00000565972.1 ENST00000569493.1 |
RAB27A
|
RAB27A, member RAS oncogene family |
| chr15_+_66797455 | 11.93 |
ENST00000446801.2
|
ZWILCH
|
zwilch kinetochore protein |
| chr15_-_55562479 | 11.87 |
ENST00000564609.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr10_+_76969909 | 11.69 |
ENST00000298468.5
ENST00000543351.1 |
VDAC2
|
voltage-dependent anion channel 2 |
| chr20_+_36373032 | 11.16 |
ENST00000373473.1
|
CTNNBL1
|
catenin, beta like 1 |
| chr14_-_74959978 | 10.79 |
ENST00000541064.1
|
NPC2
|
Niemann-Pick disease, type C2 |
| chr1_+_45212074 | 10.57 |
ENST00000372217.1
|
KIF2C
|
kinesin family member 2C |
| chr22_+_22930626 | 10.41 |
ENST00000390302.2
|
IGLV2-33
|
immunoglobulin lambda variable 2-33 (non-functional) |
| chr14_-_74960030 | 9.86 |
ENST00000553490.1
ENST00000557510.1 |
NPC2
|
Niemann-Pick disease, type C2 |
| chr12_+_53689309 | 9.74 |
ENST00000351500.3
ENST00000550846.1 ENST00000334478.4 ENST00000549759.1 |
PFDN5
|
prefoldin subunit 5 |
| chr5_-_176778523 | 9.64 |
ENST00000513877.1
ENST00000515209.1 ENST00000514458.1 ENST00000502560.1 |
LMAN2
|
lectin, mannose-binding 2 |
| chr15_-_55562582 | 9.23 |
ENST00000396307.2
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr14_-_54908043 | 9.22 |
ENST00000556113.1
ENST00000553660.1 ENST00000395573.4 ENST00000557690.1 ENST00000216416.4 |
CNIH1
|
cornichon family AMPA receptor auxiliary protein 1 |
| chr11_-_104972158 | 9.13 |
ENST00000598974.1
ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1
CARD16
CARD17
|
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr15_-_66797172 | 9.10 |
ENST00000569438.1
ENST00000569696.1 ENST00000307961.6 |
RPL4
|
ribosomal protein L4 |
| chr4_-_10118348 | 9.01 |
ENST00000502702.1
|
WDR1
|
WD repeat domain 1 |
| chr12_+_53848549 | 8.64 |
ENST00000439930.3
ENST00000548933.1 ENST00000562264.1 |
PCBP2
|
poly(rC) binding protein 2 |
| chr14_-_74959994 | 8.52 |
ENST00000238633.2
ENST00000434013.2 |
NPC2
|
Niemann-Pick disease, type C2 |
| chr19_-_13227534 | 8.51 |
ENST00000588229.1
ENST00000357720.4 |
TRMT1
|
tRNA methyltransferase 1 homolog (S. cerevisiae) |
| chr17_+_36908984 | 8.49 |
ENST00000225426.4
ENST00000579088.1 |
PSMB3
|
proteasome (prosome, macropain) subunit, beta type, 3 |
| chr1_+_45212051 | 8.22 |
ENST00000372222.3
|
KIF2C
|
kinesin family member 2C |
| chr4_-_99851766 | 8.14 |
ENST00000450253.2
|
EIF4E
|
eukaryotic translation initiation factor 4E |
| chr1_-_94344754 | 7.99 |
ENST00000436063.2
|
DNTTIP2
|
deoxynucleotidyltransferase, terminal, interacting protein 2 |
| chr1_+_46016703 | 7.96 |
ENST00000481885.1
ENST00000351829.4 ENST00000471651.1 |
AKR1A1
|
aldo-keto reductase family 1, member A1 (aldehyde reductase) |
| chr20_-_33543546 | 7.67 |
ENST00000216951.2
|
GSS
|
glutathione synthetase |
| chr5_-_148929848 | 7.64 |
ENST00000504676.1
ENST00000515435.1 |
CSNK1A1
|
casein kinase 1, alpha 1 |
| chr12_-_46766577 | 7.55 |
ENST00000256689.5
|
SLC38A2
|
solute carrier family 38, member 2 |
| chr18_-_47018769 | 6.96 |
ENST00000583637.1
ENST00000578528.1 ENST00000578532.1 ENST00000580387.1 ENST00000579248.1 ENST00000581373.1 |
RPL17
|
ribosomal protein L17 |
| chr6_-_114292449 | 6.56 |
ENST00000519065.1
|
HDAC2
|
histone deacetylase 2 |
| chr5_-_96518907 | 6.55 |
ENST00000508447.1
ENST00000283109.3 |
RIOK2
|
RIO kinase 2 |
| chrX_+_149861836 | 6.50 |
ENST00000542156.1
ENST00000370390.3 ENST00000490316.2 ENST00000445323.2 ENST00000544228.1 ENST00000451863.2 |
MTMR1
|
myotubularin related protein 1 |
| chr17_+_73257945 | 6.36 |
ENST00000579002.1
|
MRPS7
|
mitochondrial ribosomal protein S7 |
| chr17_-_46178527 | 6.35 |
ENST00000393408.3
|
CBX1
|
chromobox homolog 1 |
| chr11_-_104905840 | 6.03 |
ENST00000526568.1
ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1
|
caspase 1, apoptosis-related cysteine peptidase |
| chrX_-_102942961 | 5.98 |
ENST00000434230.1
ENST00000418819.1 ENST00000360458.1 |
MORF4L2
|
mortality factor 4 like 2 |
| chr10_-_12084770 | 5.37 |
ENST00000357604.5
|
UPF2
|
UPF2 regulator of nonsense transcripts homolog (yeast) |
| chr5_-_176778803 | 4.99 |
ENST00000303127.7
|
LMAN2
|
lectin, mannose-binding 2 |
| chr7_-_93520259 | 4.85 |
ENST00000222543.5
|
TFPI2
|
tissue factor pathway inhibitor 2 |
| chr19_-_11688500 | 4.82 |
ENST00000433365.2
|
ACP5
|
acid phosphatase 5, tartrate resistant |
| chr14_-_91526462 | 4.74 |
ENST00000536315.2
|
RPS6KA5
|
ribosomal protein S6 kinase, 90kDa, polypeptide 5 |
| chr7_-_102985035 | 4.65 |
ENST00000426036.2
ENST00000249270.7 ENST00000454277.1 ENST00000412522.1 |
DNAJC2
|
DnaJ (Hsp40) homolog, subfamily C, member 2 |
| chr12_+_54378923 | 4.48 |
ENST00000303460.4
|
HOXC10
|
homeobox C10 |
| chr11_+_74303575 | 4.40 |
ENST00000263681.2
|
POLD3
|
polymerase (DNA-directed), delta 3, accessory subunit |
| chr13_-_37633743 | 4.40 |
ENST00000497318.1
ENST00000475892.1 ENST00000356185.3 ENST00000350612.6 ENST00000542180.1 ENST00000360252.4 |
SUPT20H
|
suppressor of Ty 20 homolog (S. cerevisiae) |
| chr17_-_5487768 | 4.31 |
ENST00000269280.4
ENST00000345221.3 ENST00000262467.5 |
NLRP1
|
NLR family, pyrin domain containing 1 |
| chr6_+_2988847 | 4.18 |
ENST00000380472.3
ENST00000605901.1 ENST00000454015.1 |
NQO2
LINC01011
|
NAD(P)H dehydrogenase, quinone 2 long intergenic non-protein coding RNA 1011 |
| chr7_-_93520191 | 3.94 |
ENST00000545378.1
|
TFPI2
|
tissue factor pathway inhibitor 2 |
| chr2_+_190722119 | 3.79 |
ENST00000452382.1
|
PMS1
|
PMS1 postmeiotic segregation increased 1 (S. cerevisiae) |
| chr7_-_142247606 | 3.75 |
ENST00000390361.3
|
TRBV7-3
|
T cell receptor beta variable 7-3 |
| chr1_+_46049706 | 3.64 |
ENST00000527470.1
ENST00000525515.1 ENST00000537798.1 ENST00000402363.3 ENST00000528238.1 ENST00000350030.3 ENST00000470768.1 ENST00000372052.4 ENST00000351223.3 |
NASP
|
nuclear autoantigenic sperm protein (histone-binding) |
| chr10_+_5454505 | 3.48 |
ENST00000355029.4
|
NET1
|
neuroepithelial cell transforming 1 |
| chr17_-_27405875 | 3.35 |
ENST00000359450.6
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1 |
| chr14_-_50154921 | 3.30 |
ENST00000553805.2
ENST00000554396.1 ENST00000216367.5 ENST00000539565.2 |
POLE2
|
polymerase (DNA directed), epsilon 2, accessory subunit |
| chr8_+_1993173 | 3.20 |
ENST00000523438.1
|
MYOM2
|
myomesin 2 |
| chr11_+_8704748 | 3.13 |
ENST00000526562.1
ENST00000525981.1 |
RPL27A
|
ribosomal protein L27a |
| chr2_+_89999259 | 3.10 |
ENST00000558026.1
|
IGKV2D-28
|
immunoglobulin kappa variable 2D-28 |
| chr8_+_1993152 | 2.76 |
ENST00000262113.4
|
MYOM2
|
myomesin 2 |
| chr17_-_39677971 | 2.71 |
ENST00000393976.2
|
KRT15
|
keratin 15 |
| chr1_+_84873913 | 2.60 |
ENST00000370662.3
|
DNASE2B
|
deoxyribonuclease II beta |
| chr11_-_62323702 | 2.44 |
ENST00000530285.1
|
AHNAK
|
AHNAK nucleoprotein |
| chr20_-_8000426 | 2.37 |
ENST00000527925.1
ENST00000246024.2 |
TMX4
|
thioredoxin-related transmembrane protein 4 |
| chr3_-_52804872 | 2.07 |
ENST00000535191.1
ENST00000461689.1 ENST00000383721.4 ENST00000233027.5 |
NEK4
|
NIMA-related kinase 4 |
| chr22_-_42084863 | 2.05 |
ENST00000401959.1
ENST00000355257.3 |
NHP2L1
|
NHP2 non-histone chromosome protein 2-like 1 (S. cerevisiae) |
| chr15_+_81475047 | 1.98 |
ENST00000559388.1
|
IL16
|
interleukin 16 |
| chr1_-_200992827 | 1.93 |
ENST00000332129.2
ENST00000422435.2 |
KIF21B
|
kinesin family member 21B |
| chr2_+_11674213 | 1.78 |
ENST00000381486.2
|
GREB1
|
growth regulation by estrogen in breast cancer 1 |
| chr6_-_11232891 | 1.69 |
ENST00000379433.5
ENST00000379446.5 |
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr22_-_36236265 | 1.59 |
ENST00000414461.2
ENST00000416721.2 ENST00000449924.2 ENST00000262829.7 ENST00000397305.3 |
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr6_+_26158343 | 1.44 |
ENST00000377777.4
ENST00000289316.2 |
HIST1H2BD
|
histone cluster 1, H2bd |
| chrX_-_24690771 | 1.38 |
ENST00000379145.1
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta |
| chr17_-_61781750 | 1.25 |
ENST00000582026.1
|
STRADA
|
STE20-related kinase adaptor alpha |
| chr20_-_36793663 | 1.25 |
ENST00000536701.1
ENST00000536724.1 |
TGM2
|
transglutaminase 2 |
| chr4_-_168155417 | 1.08 |
ENST00000511269.1
ENST00000506697.1 ENST00000512042.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr5_-_88178964 | 1.06 |
ENST00000513252.1
ENST00000508569.1 ENST00000510942.1 ENST00000506554.1 |
MEF2C
|
myocyte enhancer factor 2C |
| chr5_+_54320078 | 1.06 |
ENST00000231009.2
|
GZMK
|
granzyme K (granzyme 3; tryptase II) |
| chrX_-_122866874 | 1.00 |
ENST00000245838.8
ENST00000355725.4 |
THOC2
|
THO complex 2 |
| chr14_-_23877474 | 0.98 |
ENST00000405093.3
|
MYH6
|
myosin, heavy chain 6, cardiac muscle, alpha |
| chr3_-_151047327 | 0.95 |
ENST00000325602.5
|
P2RY13
|
purinergic receptor P2Y, G-protein coupled, 13 |
| chr8_+_110098850 | 0.88 |
ENST00000518632.1
|
TRHR
|
thyrotropin-releasing hormone receptor |
| chr1_-_157811588 | 0.81 |
ENST00000368174.4
|
CD5L
|
CD5 molecule-like |
| chr6_+_36097992 | 0.80 |
ENST00000211287.4
|
MAPK13
|
mitogen-activated protein kinase 13 |
| chrX_+_101478829 | 0.74 |
ENST00000372763.1
ENST00000372758.1 |
NXF2
|
nuclear RNA export factor 2 |
| chr15_-_34880646 | 0.72 |
ENST00000543376.1
|
GOLGA8A
|
golgin A8 family, member A |
| chr17_+_11501748 | 0.71 |
ENST00000262442.4
ENST00000579828.1 |
DNAH9
|
dynein, axonemal, heavy chain 9 |
| chr19_+_52264449 | 0.67 |
ENST00000599326.1
ENST00000598953.1 |
FPR2
|
formyl peptide receptor 2 |
| chrX_+_70338525 | 0.57 |
ENST00000374102.1
|
MED12
|
mediator complex subunit 12 |
| chr1_+_113010056 | 0.57 |
ENST00000369686.5
|
WNT2B
|
wingless-type MMTV integration site family, member 2B |
| chr19_-_46148820 | 0.48 |
ENST00000587152.1
|
EML2
|
echinoderm microtubule associated protein like 2 |
| chr17_+_46184911 | 0.48 |
ENST00000580219.1
ENST00000452859.2 ENST00000393405.2 ENST00000439357.2 ENST00000359238.2 |
SNX11
|
sorting nexin 11 |
| chr1_+_200993071 | 0.38 |
ENST00000446333.1
ENST00000458003.1 |
RP11-168O16.1
|
RP11-168O16.1 |
| chr14_-_23876801 | 0.33 |
ENST00000356287.3
|
MYH6
|
myosin, heavy chain 6, cardiac muscle, alpha |
| chr1_-_117210290 | 0.22 |
ENST00000369483.1
ENST00000369486.3 |
IGSF3
|
immunoglobulin superfamily, member 3 |
| chr5_-_88179017 | 0.18 |
ENST00000514028.1
ENST00000514015.1 ENST00000503075.1 ENST00000437473.2 |
MEF2C
|
myocyte enhancer factor 2C |
| chr10_+_180405 | 0.08 |
ENST00000439456.1
ENST00000397962.3 ENST00000309776.4 ENST00000381602.4 |
ZMYND11
|
zinc finger, MYND-type containing 11 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB12
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.3 | 29.2 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 5.5 | 33.0 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 5.0 | 25.1 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 3.9 | 15.6 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 3.8 | 15.2 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 2.7 | 8.0 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 2.6 | 31.7 | GO:0015074 | DNA integration(GO:0015074) |
| 2.3 | 9.0 | GO:0042247 | morphogenesis of follicular epithelium(GO:0016333) establishment or maintenance of polarity of follicular epithelium(GO:0016334) establishment of planar polarity of follicular epithelium(GO:0042247) |
| 2.2 | 6.6 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 2.0 | 14.0 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 1.9 | 18.8 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 1.6 | 4.7 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 1.3 | 7.5 | GO:0032328 | alanine transport(GO:0032328) |
| 1.2 | 62.8 | GO:0032731 | positive regulation of interleukin-1 beta production(GO:0032731) |
| 1.1 | 7.6 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 1.0 | 16.7 | GO:0051639 | actin filament network formation(GO:0051639) |
| 1.0 | 4.8 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.9 | 4.7 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) histone H2A phosphorylation(GO:1990164) |
| 0.8 | 14.1 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.7 | 6.0 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.5 | 8.6 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.4 | 16.2 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.4 | 1.3 | GO:0018153 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.4 | 3.6 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.3 | 7.7 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.3 | 6.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.3 | 3.5 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.3 | 26.1 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.2 | 7.7 | GO:0032201 | telomere maintenance via semi-conservative replication(GO:0032201) |
| 0.2 | 7.7 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.2 | 14.6 | GO:0050766 | positive regulation of phagocytosis(GO:0050766) |
| 0.2 | 11.7 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.2 | 6.5 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.2 | 5.9 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.2 | 6.0 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.2 | 0.6 | GO:0060492 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.2 | 23.9 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.2 | 1.0 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.2 | 4.5 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.2 | 1.3 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) atrial cardiac muscle tissue morphogenesis(GO:0055009) |
| 0.2 | 8.1 | GO:0071549 | cellular response to dexamethasone stimulus(GO:0071549) |
| 0.2 | 11.2 | GO:0016445 | somatic diversification of immunoglobulins(GO:0016445) |
| 0.2 | 3.8 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.1 | 4.2 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.1 | 2.1 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.1 | 19.2 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 1.4 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.1 | 0.6 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.1 | 10.4 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 9.2 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 2.6 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.1 | 0.7 | GO:0032930 | positive regulation of superoxide anion generation(GO:0032930) |
| 0.1 | 2.0 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.1 | 5.4 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 9.7 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.1 | 0.9 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 1.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 2.4 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.7 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.5 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 2.4 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 3.1 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.0 | 0.2 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 4.4 | GO:0007369 | gastrulation(GO:0007369) |
| 0.0 | 2.7 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.7 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 1.3 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 1.4 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 3.4 | GO:0007249 | I-kappaB kinase/NF-kappaB signaling(GO:0007249) |
| 0.0 | 0.8 | GO:0050729 | positive regulation of inflammatory response(GO:0050729) |
| 0.0 | 1.7 | GO:0051017 | actin filament bundle assembly(GO:0051017) actin filament bundle organization(GO:0061572) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.0 | 23.9 | GO:1990423 | RZZ complex(GO:1990423) |
| 3.2 | 19.5 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 2.8 | 33.0 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 2.2 | 17.7 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 1.8 | 9.0 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 1.4 | 9.7 | GO:0016272 | prefoldin complex(GO:0016272) |
| 1.3 | 15.6 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 1.3 | 6.3 | GO:0001940 | male pronucleus(GO:0001940) |
| 1.1 | 4.4 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 1.0 | 25.1 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.9 | 8.5 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.8 | 16.7 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.8 | 6.6 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.7 | 8.1 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.7 | 3.3 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.6 | 7.6 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.6 | 18.8 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.5 | 11.2 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.5 | 4.5 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.5 | 11.7 | GO:0046930 | pore complex(GO:0046930) |
| 0.5 | 3.8 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.3 | 4.4 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.3 | 7.3 | GO:0032982 | myosin filament(GO:0032982) |
| 0.3 | 29.2 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.3 | 23.8 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.2 | 6.4 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.2 | 6.0 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.2 | 5.4 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 0.7 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.1 | 31.7 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.1 | 1.0 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 27.2 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 51.1 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 11.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 16.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 2.4 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 7.5 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 1.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 2.7 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 9.4 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.7 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 8.2 | GO:0005925 | focal adhesion(GO:0005925) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 8.5 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 1.9 | 18.8 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 1.8 | 25.1 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 1.4 | 8.6 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 1.3 | 11.7 | GO:0015288 | porin activity(GO:0015288) |
| 1.3 | 33.0 | GO:0031489 | myosin V binding(GO:0031489) |
| 1.1 | 15.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 1.0 | 4.2 | GO:1904408 | dihydronicotinamide riboside quinone reductase activity(GO:0001512) melatonin binding(GO:1904408) |
| 1.0 | 8.1 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.9 | 8.0 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.9 | 26.1 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.9 | 2.6 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.8 | 4.7 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.7 | 14.6 | GO:0005537 | mannose binding(GO:0005537) |
| 0.7 | 6.3 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.5 | 7.7 | GO:0016594 | glycine binding(GO:0016594) |
| 0.5 | 29.2 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.5 | 6.5 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.4 | 4.8 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.4 | 8.4 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.3 | 48.8 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.3 | 1.4 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.3 | 45.7 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.3 | 7.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.2 | 3.8 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.2 | 1.3 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.2 | 3.5 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.2 | 0.7 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.2 | 1.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 25.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 2.4 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 25.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 5.4 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 8.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 2.0 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 7.5 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.1 | 3.6 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 31.1 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.1 | 4.3 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.9 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 1.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.7 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.6 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 5.0 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 1.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 9.8 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 3.1 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.8 | GO:0005044 | scavenger receptor activity(GO:0005044) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 18.6 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.6 | 15.8 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.3 | 18.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.3 | 7.6 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.3 | 15.6 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.2 | 4.7 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.2 | 8.1 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.2 | 15.8 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 6.6 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 0.8 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 9.7 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 3.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.3 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.7 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 31.7 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 1.3 | 33.0 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.8 | 15.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.8 | 7.6 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.8 | 18.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.6 | 18.6 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.5 | 15.6 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.5 | 7.7 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.4 | 8.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.4 | 26.1 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.4 | 4.3 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.3 | 7.5 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.2 | 9.7 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.2 | 20.2 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.2 | 7.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 8.6 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 6.5 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 24.6 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.1 | 4.7 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 6.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 4.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 9.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 3.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 1.3 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 1.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 0.8 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.9 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 1.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.7 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |