Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
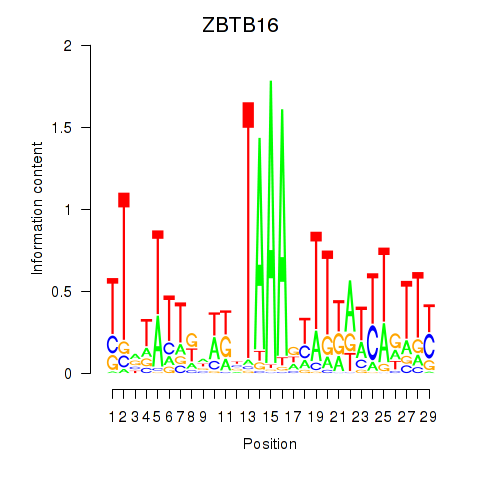
Results for ZBTB16
Z-value: 0.67
Transcription factors associated with ZBTB16
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB16
|
ENSG00000109906.9 | zinc finger and BTB domain containing 16 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB16 | hg19_v2_chr11_+_113930291_113930339 | -0.45 | 1.7e-12 | Click! |
Activity profile of ZBTB16 motif
Sorted Z-values of ZBTB16 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_62538089 | 23.35 |
ENST00000519078.2
ENST00000395284.3 ENST00000316629.4 |
CDK1
|
cyclin-dependent kinase 1 |
| chr5_+_159848807 | 12.29 |
ENST00000352433.5
|
PTTG1
|
pituitary tumor-transforming 1 |
| chr5_+_159848854 | 11.74 |
ENST00000517480.1
ENST00000520452.1 ENST00000393964.1 |
PTTG1
|
pituitary tumor-transforming 1 |
| chr1_+_156095951 | 10.99 |
ENST00000448611.2
ENST00000368297.1 |
LMNA
|
lamin A/C |
| chr3_-_146262488 | 10.50 |
ENST00000487389.1
|
PLSCR1
|
phospholipid scramblase 1 |
| chr15_+_75628232 | 9.26 |
ENST00000267935.8
ENST00000567195.1 |
COMMD4
|
COMM domain containing 4 |
| chr15_+_75628394 | 9.18 |
ENST00000564815.1
ENST00000338995.6 |
COMMD4
|
COMM domain containing 4 |
| chr15_+_75628419 | 9.17 |
ENST00000567377.1
ENST00000562789.1 ENST00000568301.1 |
COMMD4
|
COMM domain containing 4 |
| chr1_-_246357029 | 8.60 |
ENST00000391836.2
|
SMYD3
|
SET and MYND domain containing 3 |
| chr15_-_55562479 | 8.57 |
ENST00000564609.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr16_+_14726672 | 8.00 |
ENST00000261658.2
ENST00000563971.1 |
BFAR
|
bifunctional apoptosis regulator |
| chr7_-_32534850 | 7.62 |
ENST00000409952.3
ENST00000409909.3 |
LSM5
|
LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr2_+_102413726 | 7.56 |
ENST00000350878.4
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr12_+_118454500 | 7.25 |
ENST00000537315.1
ENST00000229043.3 ENST00000484086.2 ENST00000420967.1 ENST00000454402.2 ENST00000392542.2 ENST00000535092.1 |
RFC5
|
replication factor C (activator 1) 5, 36.5kDa |
| chr2_+_242289502 | 7.17 |
ENST00000451310.1
|
SEPT2
|
septin 2 |
| chr21_-_35284635 | 7.10 |
ENST00000429238.1
|
AP000304.12
|
AP000304.12 |
| chr16_-_85722530 | 7.01 |
ENST00000253462.3
|
GINS2
|
GINS complex subunit 2 (Psf2 homolog) |
| chr13_+_76123883 | 6.79 |
ENST00000377595.3
|
UCHL3
|
ubiquitin carboxyl-terminal esterase L3 (ubiquitin thiolesterase) |
| chr19_+_36142147 | 6.54 |
ENST00000590618.1
|
COX6B1
|
cytochrome c oxidase subunit VIb polypeptide 1 (ubiquitous) |
| chrX_+_118602363 | 6.53 |
ENST00000317881.8
|
SLC25A5
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 |
| chr3_-_146262637 | 6.45 |
ENST00000472349.1
ENST00000342435.4 |
PLSCR1
|
phospholipid scramblase 1 |
| chr1_-_115292591 | 6.40 |
ENST00000438362.2
|
CSDE1
|
cold shock domain containing E1, RNA-binding |
| chr19_+_9296279 | 6.24 |
ENST00000344248.2
|
OR7D2
|
olfactory receptor, family 7, subfamily D, member 2 |
| chr1_+_100731749 | 6.21 |
ENST00000370128.4
ENST00000260563.4 |
RTCA
|
RNA 3'-terminal phosphate cyclase |
| chr5_-_34919094 | 6.21 |
ENST00000341754.4
|
RAD1
|
RAD1 homolog (S. pombe) |
| chr4_-_99851766 | 6.12 |
ENST00000450253.2
|
EIF4E
|
eukaryotic translation initiation factor 4E |
| chr16_+_81070792 | 6.11 |
ENST00000564241.1
ENST00000565237.1 |
ATMIN
|
ATM interactor |
| chr20_+_31407692 | 5.89 |
ENST00000375571.5
|
MAPRE1
|
microtubule-associated protein, RP/EB family, member 1 |
| chr4_-_99850243 | 5.87 |
ENST00000280892.6
ENST00000511644.1 ENST00000504432.1 ENST00000505992.1 |
EIF4E
|
eukaryotic translation initiation factor 4E |
| chrX_-_47518527 | 5.83 |
ENST00000333119.3
|
UXT
|
ubiquitously-expressed, prefoldin-like chaperone |
| chr10_+_5135981 | 5.71 |
ENST00000380554.3
|
AKR1C3
|
aldo-keto reductase family 1, member C3 |
| chr19_-_46195029 | 5.66 |
ENST00000588599.1
ENST00000585392.1 ENST00000590212.1 ENST00000587367.1 ENST00000391932.3 |
SNRPD2
|
small nuclear ribonucleoprotein D2 polypeptide 16.5kDa |
| chr1_-_44818599 | 5.62 |
ENST00000537474.1
|
ERI3
|
ERI1 exoribonuclease family member 3 |
| chr17_-_40134339 | 5.61 |
ENST00000587727.1
|
DNAJC7
|
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr15_-_55562582 | 5.60 |
ENST00000396307.2
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr12_-_27090896 | 5.47 |
ENST00000539625.1
ENST00000538727.1 |
ASUN
|
asunder spermatogenesis regulator |
| chr7_+_120591170 | 5.31 |
ENST00000431467.1
|
ING3
|
inhibitor of growth family, member 3 |
| chr6_+_32812568 | 5.23 |
ENST00000414474.1
|
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr3_-_155524049 | 5.22 |
ENST00000534941.1
ENST00000340171.2 |
C3orf33
|
chromosome 3 open reading frame 33 |
| chr8_-_30585439 | 5.14 |
ENST00000221130.5
|
GSR
|
glutathione reductase |
| chr12_+_32832134 | 4.88 |
ENST00000452533.2
|
DNM1L
|
dynamin 1-like |
| chr7_+_7606497 | 4.86 |
ENST00000340080.4
ENST00000405785.1 ENST00000433635.1 |
MIOS
|
missing oocyte, meiosis regulator, homolog (Drosophila) |
| chr12_+_53836339 | 4.71 |
ENST00000549135.1
|
PRR13
|
proline rich 13 |
| chr22_-_36236265 | 4.66 |
ENST00000414461.2
ENST00000416721.2 ENST00000449924.2 ENST00000262829.7 ENST00000397305.3 |
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr1_-_11159887 | 4.64 |
ENST00000544779.1
ENST00000304457.7 ENST00000376936.4 |
EXOSC10
|
exosome component 10 |
| chr12_+_32832203 | 4.55 |
ENST00000553257.1
ENST00000549701.1 ENST00000358214.5 ENST00000266481.6 ENST00000551476.1 ENST00000550154.1 ENST00000547312.1 ENST00000414834.2 ENST00000381000.4 ENST00000548750.1 |
DNM1L
|
dynamin 1-like |
| chr12_-_75905374 | 4.34 |
ENST00000438169.2
ENST00000229214.4 |
KRR1
|
KRR1, small subunit (SSU) processome component, homolog (yeast) |
| chr12_-_71533055 | 4.26 |
ENST00000552128.1
|
TSPAN8
|
tetraspanin 8 |
| chr16_-_20817753 | 4.09 |
ENST00000389345.5
ENST00000300005.3 ENST00000357967.4 ENST00000569729.1 |
ERI2
|
ERI1 exoribonuclease family member 2 |
| chr4_-_83769996 | 3.90 |
ENST00000511338.1
|
SEC31A
|
SEC31 homolog A (S. cerevisiae) |
| chr8_-_74205851 | 3.88 |
ENST00000396467.1
|
RPL7
|
ribosomal protein L7 |
| chr10_+_47894572 | 3.69 |
ENST00000355876.5
|
FAM21B
|
family with sequence similarity 21, member B |
| chr16_+_24549014 | 3.68 |
ENST00000564314.1
ENST00000567686.1 |
RBBP6
|
retinoblastoma binding protein 6 |
| chr17_+_46908350 | 3.61 |
ENST00000258947.3
ENST00000509507.1 ENST00000448105.2 ENST00000570513.1 ENST00000509415.1 ENST00000513119.1 ENST00000416445.2 ENST00000508679.1 ENST00000505071.1 |
CALCOCO2
|
calcium binding and coiled-coil domain 2 |
| chr9_-_110540419 | 3.59 |
ENST00000398726.3
|
AL162389.1
|
Uncharacterized protein |
| chr3_-_93692781 | 3.52 |
ENST00000394236.3
|
PROS1
|
protein S (alpha) |
| chr3_-_165555200 | 3.49 |
ENST00000479451.1
ENST00000540653.1 ENST00000488954.1 ENST00000264381.3 |
BCHE
|
butyrylcholinesterase |
| chr5_-_9630463 | 3.49 |
ENST00000382492.2
|
TAS2R1
|
taste receptor, type 2, member 1 |
| chr11_-_72070206 | 3.41 |
ENST00000544382.1
|
CLPB
|
ClpB caseinolytic peptidase B homolog (E. coli) |
| chr10_+_47894023 | 3.38 |
ENST00000358474.5
|
FAM21B
|
family with sequence similarity 21, member B |
| chr7_-_16872932 | 3.30 |
ENST00000419572.2
ENST00000412973.1 |
AGR2
|
anterior gradient 2 |
| chr12_-_11062161 | 3.20 |
ENST00000390677.2
|
TAS2R13
|
taste receptor, type 2, member 13 |
| chr18_-_21017817 | 3.20 |
ENST00000542162.1
ENST00000383233.3 ENST00000582336.1 ENST00000450466.2 ENST00000578520.1 ENST00000399707.1 |
TMEM241
|
transmembrane protein 241 |
| chr4_+_74606223 | 3.18 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr3_+_44840679 | 3.16 |
ENST00000425755.1
|
KIF15
|
kinesin family member 15 |
| chr12_-_68696652 | 3.10 |
ENST00000539972.1
|
MDM1
|
Mdm1 nuclear protein homolog (mouse) |
| chr8_-_8318847 | 3.05 |
ENST00000521218.1
|
CTA-398F10.2
|
CTA-398F10.2 |
| chr8_+_132952112 | 3.01 |
ENST00000520362.1
ENST00000519656.1 |
EFR3A
|
EFR3 homolog A (S. cerevisiae) |
| chr6_+_26273144 | 3.01 |
ENST00000377733.2
|
HIST1H2BI
|
histone cluster 1, H2bi |
| chr7_+_134464414 | 2.96 |
ENST00000361901.2
|
CALD1
|
caldesmon 1 |
| chr17_-_46623441 | 2.93 |
ENST00000330070.4
|
HOXB2
|
homeobox B2 |
| chr5_+_96212185 | 2.87 |
ENST00000379904.4
|
ERAP2
|
endoplasmic reticulum aminopeptidase 2 |
| chr16_-_18908196 | 2.86 |
ENST00000565324.1
ENST00000561947.1 |
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr10_-_47239738 | 2.78 |
ENST00000413193.2
|
AGAP10
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 10 |
| chr11_+_120255997 | 2.74 |
ENST00000532993.1
|
ARHGEF12
|
Rho guanine nucleotide exchange factor (GEF) 12 |
| chr19_+_21265028 | 2.71 |
ENST00000291770.7
|
ZNF714
|
zinc finger protein 714 |
| chr11_+_67351019 | 2.70 |
ENST00000398606.3
|
GSTP1
|
glutathione S-transferase pi 1 |
| chr5_+_7654057 | 2.69 |
ENST00000537121.1
|
ADCY2
|
adenylate cyclase 2 (brain) |
| chr10_+_8096769 | 2.69 |
ENST00000346208.3
|
GATA3
|
GATA binding protein 3 |
| chrX_-_23926004 | 2.67 |
ENST00000379226.4
ENST00000379220.3 |
APOO
|
apolipoprotein O |
| chr11_-_104972158 | 2.58 |
ENST00000598974.1
ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1
CARD16
CARD17
|
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr5_+_180682720 | 2.55 |
ENST00000599439.1
|
AC008443.1
|
CDNA: FLJ23158 fis, clone LNG09623; Uncharacterized protein |
| chr4_+_74347400 | 2.55 |
ENST00000226355.3
|
AFM
|
afamin |
| chr1_+_33439268 | 2.54 |
ENST00000594612.1
|
FKSG48
|
FKSG48 |
| chr16_-_70239683 | 2.52 |
ENST00000601706.1
|
AC009060.1
|
Uncharacterized protein |
| chr4_+_37962018 | 2.52 |
ENST00000504686.1
|
PTTG2
|
pituitary tumor-transforming 2 |
| chr8_-_122653630 | 2.52 |
ENST00000303924.4
|
HAS2
|
hyaluronan synthase 2 |
| chr1_+_76251879 | 2.36 |
ENST00000535300.1
ENST00000319942.3 |
RABGGTB
|
Rab geranylgeranyltransferase, beta subunit |
| chr6_+_74171301 | 2.26 |
ENST00000415954.2
ENST00000498286.1 ENST00000370305.1 ENST00000370300.4 |
MTO1
|
mitochondrial tRNA translation optimization 1 |
| chr17_-_9479128 | 2.19 |
ENST00000574431.1
|
STX8
|
syntaxin 8 |
| chr2_+_131862872 | 2.16 |
ENST00000439822.2
|
PLEKHB2
|
pleckstrin homology domain containing, family B (evectins) member 2 |
| chr3_-_138312971 | 2.07 |
ENST00000485115.1
ENST00000484888.1 ENST00000468900.1 ENST00000542237.1 ENST00000481834.1 |
CEP70
|
centrosomal protein 70kDa |
| chr4_-_140223614 | 2.03 |
ENST00000394223.1
|
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr12_+_4385230 | 1.93 |
ENST00000536537.1
|
CCND2
|
cyclin D2 |
| chr3_+_150126101 | 1.90 |
ENST00000361875.3
ENST00000361136.2 |
TSC22D2
|
TSC22 domain family, member 2 |
| chr3_-_138313161 | 1.89 |
ENST00000489254.1
ENST00000474781.1 ENST00000462419.1 ENST00000264982.3 |
CEP70
|
centrosomal protein 70kDa |
| chr3_-_37216055 | 1.86 |
ENST00000336686.4
|
LRRFIP2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr12_-_102224704 | 1.78 |
ENST00000299314.7
|
GNPTAB
|
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits |
| chr19_-_19932501 | 1.77 |
ENST00000540806.2
ENST00000590766.1 ENST00000587452.1 ENST00000545006.1 ENST00000590319.1 ENST00000587461.1 ENST00000450683.2 ENST00000443905.2 ENST00000590274.1 |
ZNF506
CTC-559E9.4
|
zinc finger protein 506 CTC-559E9.4 |
| chr4_-_140223670 | 1.76 |
ENST00000394228.1
ENST00000539387.1 |
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr2_-_188419078 | 1.73 |
ENST00000437725.1
ENST00000409676.1 ENST00000339091.4 ENST00000420747.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr3_+_130569592 | 1.71 |
ENST00000533801.2
|
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr3_-_133969673 | 1.67 |
ENST00000427044.2
|
RYK
|
receptor-like tyrosine kinase |
| chr2_-_11606275 | 1.61 |
ENST00000381525.3
ENST00000362009.4 |
E2F6
|
E2F transcription factor 6 |
| chrY_+_26997726 | 1.60 |
ENST00000382296.2
|
DAZ4
|
deleted in azoospermia 4 |
| chrX_+_71401526 | 1.58 |
ENST00000218432.5
ENST00000423432.2 ENST00000373669.2 |
PIN4
|
protein (peptidylprolyl cis/trans isomerase) NIMA-interacting, 4 (parvulin) |
| chr4_+_74301880 | 1.53 |
ENST00000395792.2
ENST00000226359.2 |
AFP
|
alpha-fetoprotein |
| chr12_+_21207503 | 1.49 |
ENST00000545916.1
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr12_+_10460549 | 1.48 |
ENST00000543420.1
ENST00000543777.1 |
KLRD1
|
killer cell lectin-like receptor subfamily D, member 1 |
| chr8_-_33370607 | 1.47 |
ENST00000360742.5
ENST00000523305.1 |
TTI2
|
TELO2 interacting protein 2 |
| chr11_+_67351213 | 1.46 |
ENST00000398603.1
|
GSTP1
|
glutathione S-transferase pi 1 |
| chr6_+_42749759 | 1.43 |
ENST00000314073.5
|
GLTSCR1L
|
GLTSCR1-like |
| chr18_-_64271363 | 1.36 |
ENST00000262150.2
|
CDH19
|
cadherin 19, type 2 |
| chr10_+_48189612 | 1.36 |
ENST00000453919.1
|
AGAP9
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 9 |
| chr17_-_37607497 | 1.36 |
ENST00000394287.3
ENST00000300651.6 |
MED1
|
mediator complex subunit 1 |
| chr11_-_327537 | 1.33 |
ENST00000602735.1
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr2_+_131862900 | 1.29 |
ENST00000438882.2
ENST00000538982.1 ENST00000404460.1 |
PLEKHB2
|
pleckstrin homology domain containing, family B (evectins) member 2 |
| chr12_-_10007448 | 1.18 |
ENST00000538152.1
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr9_-_36400213 | 1.17 |
ENST00000259605.6
ENST00000353739.4 |
RNF38
|
ring finger protein 38 |
| chr14_+_39703084 | 1.13 |
ENST00000553728.1
|
RP11-407N17.3
|
cTAGE family member 5 isoform 4 |
| chr3_+_51663407 | 1.12 |
ENST00000432863.1
ENST00000296477.3 |
RAD54L2
|
RAD54-like 2 (S. cerevisiae) |
| chr5_+_140560980 | 1.10 |
ENST00000361016.2
|
PCDHB16
|
protocadherin beta 16 |
| chr10_+_111765562 | 1.03 |
ENST00000360162.3
|
ADD3
|
adducin 3 (gamma) |
| chr18_+_616672 | 0.99 |
ENST00000338387.7
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr10_+_8096631 | 0.97 |
ENST00000379328.3
|
GATA3
|
GATA binding protein 3 |
| chr19_+_13135386 | 0.96 |
ENST00000360105.4
ENST00000588228.1 ENST00000591028.1 |
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr3_-_120365866 | 0.95 |
ENST00000475447.2
|
HGD
|
homogentisate 1,2-dioxygenase |
| chrX_+_115301975 | 0.94 |
ENST00000371906.4
|
AGTR2
|
angiotensin II receptor, type 2 |
| chr10_+_5005598 | 0.89 |
ENST00000442997.1
|
AKR1C1
|
aldo-keto reductase family 1, member C1 |
| chr17_-_18266797 | 0.89 |
ENST00000316694.3
ENST00000539052.1 |
SHMT1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr22_-_36236623 | 0.87 |
ENST00000405409.2
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr10_+_5005445 | 0.81 |
ENST00000380872.4
|
AKR1C1
|
aldo-keto reductase family 1, member C1 |
| chr19_-_29704448 | 0.79 |
ENST00000304863.4
|
UQCRFS1
|
ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 |
| chr1_+_170633047 | 0.77 |
ENST00000239461.6
ENST00000497230.2 |
PRRX1
|
paired related homeobox 1 |
| chr12_+_26164645 | 0.77 |
ENST00000542004.1
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr6_-_49712123 | 0.75 |
ENST00000263045.4
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr1_+_210501589 | 0.74 |
ENST00000413764.2
ENST00000541565.1 |
HHAT
|
hedgehog acyltransferase |
| chr17_-_39041479 | 0.73 |
ENST00000167588.3
|
KRT20
|
keratin 20 |
| chr7_-_38289173 | 0.73 |
ENST00000436911.2
|
TRGC2
|
T cell receptor gamma constant 2 |
| chr15_-_85197501 | 0.72 |
ENST00000434634.2
|
WDR73
|
WD repeat domain 73 |
| chr19_+_45281118 | 0.71 |
ENST00000270279.3
ENST00000341505.4 |
CBLC
|
Cbl proto-oncogene C, E3 ubiquitin protein ligase |
| chr3_+_179065474 | 0.68 |
ENST00000471841.1
ENST00000280653.7 |
MFN1
|
mitofusin 1 |
| chr2_+_172544011 | 0.66 |
ENST00000508530.1
|
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr12_+_10460417 | 0.65 |
ENST00000381908.3
ENST00000336164.4 ENST00000350274.5 |
KLRD1
|
killer cell lectin-like receptor subfamily D, member 1 |
| chr5_+_35856951 | 0.63 |
ENST00000303115.3
ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr3_+_160394940 | 0.61 |
ENST00000320767.2
|
ARL14
|
ADP-ribosylation factor-like 14 |
| chr3_-_126327398 | 0.56 |
ENST00000383572.2
|
TXNRD3NB
|
thioredoxin reductase 3 neighbor |
| chr2_+_172543919 | 0.54 |
ENST00000452242.1
ENST00000340296.4 |
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr4_-_70518941 | 0.53 |
ENST00000286604.4
ENST00000505512.1 ENST00000514019.1 |
UGT2A1
UGT2A1
|
UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus |
| chr18_+_616711 | 0.53 |
ENST00000579494.1
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr19_-_44079611 | 0.51 |
ENST00000594107.1
ENST00000543982.1 |
XRCC1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr2_+_191334212 | 0.50 |
ENST00000444317.1
ENST00000535751.1 |
MFSD6
|
major facilitator superfamily domain containing 6 |
| chr1_+_32608566 | 0.47 |
ENST00000545542.1
|
KPNA6
|
karyopherin alpha 6 (importin alpha 7) |
| chrX_+_65382381 | 0.46 |
ENST00000519389.1
|
HEPH
|
hephaestin |
| chr18_+_39535152 | 0.43 |
ENST00000262039.4
ENST00000398870.3 ENST00000586545.1 |
PIK3C3
|
phosphatidylinositol 3-kinase, catalytic subunit type 3 |
| chr4_+_70796784 | 0.43 |
ENST00000246891.4
ENST00000444405.3 |
CSN1S1
|
casein alpha s1 |
| chr17_+_32597232 | 0.39 |
ENST00000378569.2
ENST00000200307.4 ENST00000394627.1 ENST00000394630.3 |
CCL7
|
chemokine (C-C motif) ligand 7 |
| chr4_+_88532028 | 0.36 |
ENST00000282478.7
|
DSPP
|
dentin sialophosphoprotein |
| chr21_-_30391636 | 0.27 |
ENST00000493196.1
|
RWDD2B
|
RWD domain containing 2B |
| chr5_+_67586465 | 0.27 |
ENST00000336483.5
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr12_+_133657461 | 0.26 |
ENST00000412146.2
ENST00000544426.1 ENST00000440984.2 ENST00000319849.3 ENST00000440550.2 |
ZNF140
|
zinc finger protein 140 |
| chr2_+_172543967 | 0.25 |
ENST00000534253.2
ENST00000263811.4 ENST00000397119.3 ENST00000410079.3 ENST00000438879.1 |
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr13_+_77564795 | 0.23 |
ENST00000377453.3
|
CLN5
|
ceroid-lipofuscinosis, neuronal 5 |
| chr6_+_25963020 | 0.23 |
ENST00000357085.3
|
TRIM38
|
tripartite motif containing 38 |
| chr10_+_52094298 | 0.14 |
ENST00000595931.1
|
AC069547.1
|
HCG1745369; PRO3073; Uncharacterized protein |
| chr21_+_37692481 | 0.12 |
ENST00000400485.1
|
MORC3
|
MORC family CW-type zinc finger 3 |
| chr15_-_50406680 | 0.11 |
ENST00000559829.1
|
ATP8B4
|
ATPase, class I, type 8B, member 4 |
| chr19_-_14992264 | 0.10 |
ENST00000327462.2
|
OR7A17
|
olfactory receptor, family 7, subfamily A, member 17 |
| chr9_+_79074068 | 0.03 |
ENST00000444201.2
ENST00000376730.4 |
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB16
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 23.4 | GO:0014038 | regulation of Schwann cell differentiation(GO:0014038) |
| 3.1 | 9.4 | GO:0090149 | synaptic vesicle recycling via endosome(GO:0036466) mitochondrial membrane fission(GO:0090149) |
| 2.4 | 14.2 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 2.2 | 11.0 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 2.1 | 16.9 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 2.0 | 5.9 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 1.9 | 5.7 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 1.6 | 6.2 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 1.5 | 4.6 | GO:0071048 | nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 1.5 | 7.3 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 1.2 | 3.7 | GO:1901536 | regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) |
| 1.2 | 3.5 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 1.1 | 8.0 | GO:1903895 | negative regulation of IRE1-mediated unfolded protein response(GO:1903895) |
| 1.1 | 6.5 | GO:1901029 | adenine transport(GO:0015853) negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 1.1 | 3.2 | GO:0015783 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 1.1 | 6.4 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 1.0 | 4.2 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 1.0 | 2.9 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.9 | 5.5 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.9 | 7.0 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.6 | 2.6 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.6 | 3.6 | GO:1901098 | regulation of autophagosome maturation(GO:1901096) positive regulation of autophagosome maturation(GO:1901098) |
| 0.6 | 26.5 | GO:0045143 | homologous chromosome segregation(GO:0045143) negative regulation of mitotic sister chromatid separation(GO:2000816) |
| 0.5 | 3.3 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.5 | 2.5 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.5 | 1.4 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.4 | 21.5 | GO:0071549 | cellular response to dexamethasone stimulus(GO:0071549) |
| 0.4 | 1.7 | GO:0061643 | chemorepulsion of dopaminergic neuron axon(GO:0036518) chemorepulsion of axon(GO:0061643) |
| 0.4 | 5.6 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.3 | 1.7 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.3 | 7.6 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.3 | 3.9 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.3 | 6.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.3 | 0.9 | GO:1904482 | response to tetrahydrofolate(GO:1904481) cellular response to tetrahydrofolate(GO:1904482) |
| 0.3 | 2.4 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.3 | 1.7 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.3 | 1.7 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.3 | 6.1 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.3 | 2.3 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.2 | 6.7 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.2 | 3.1 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.2 | 5.8 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.2 | 7.6 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.2 | 2.7 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.2 | 2.9 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.2 | 3.5 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.2 | 3.9 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.2 | 5.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.2 | 1.4 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.2 | 4.9 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.2 | 0.8 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 0.9 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.5 | GO:0006288 | base-excision repair, DNA ligation(GO:0006288) |
| 0.1 | 4.3 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 0.4 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 1.9 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.1 | 1.5 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.1 | 5.1 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.1 | 4.2 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 0.8 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.1 | 0.8 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 2.7 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 4.3 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.1 | 1.6 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 | 2.2 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.1 | 0.7 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 1.3 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.1 | 0.6 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.1 | 0.4 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.1 | 2.6 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 7.9 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.1 | 2.5 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.1 | 5.2 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.1 | 3.8 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 3.7 | GO:0061053 | somite development(GO:0061053) |
| 0.0 | 1.6 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 1.1 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 5.0 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 1.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.7 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 3.2 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 2.1 | GO:0002228 | natural killer cell mediated immunity(GO:0002228) |
| 0.0 | 0.2 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 6.3 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.2 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.6 | GO:0007264 | small GTPase mediated signal transduction(GO:0007264) |
| 0.0 | 0.5 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 3.4 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.5 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.5 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 4.3 | GO:0051090 | regulation of sequence-specific DNA binding transcription factor activity(GO:0051090) |
| 0.0 | 1.7 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 23.4 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 1.8 | 7.0 | GO:0000811 | GINS complex(GO:0000811) |
| 1.6 | 6.2 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 1.4 | 4.2 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 1.3 | 5.3 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 1.2 | 11.0 | GO:0005638 | lamin filament(GO:0005638) |
| 1.2 | 7.3 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 1.2 | 14.2 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 1.1 | 12.0 | GO:0033391 | chromatoid body(GO:0033391) |
| 1.1 | 7.6 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 1.1 | 6.5 | GO:0071817 | MMXD complex(GO:0071817) |
| 1.1 | 3.2 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 1.0 | 5.9 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.7 | 5.7 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.7 | 4.9 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.7 | 5.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.6 | 6.4 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.6 | 2.4 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.5 | 2.6 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.5 | 7.2 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.4 | 4.6 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.4 | 2.7 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.3 | 1.9 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.2 | 3.5 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.2 | 5.8 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.2 | 3.0 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 3.9 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.4 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 4.3 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 2.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 9.4 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 3.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 0.5 | GO:0070522 | ERCC4-ERCC1 complex(GO:0070522) |
| 0.1 | 0.8 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 6.5 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 0.7 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.1 | 0.3 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 3.5 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 8.0 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 2.2 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 2.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 1.6 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 2.3 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 16.4 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 1.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 3.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 3.5 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 3.1 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 7.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.2 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 1.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.3 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.7 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 1.0 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.4 | GO:0047718 | androsterone dehydrogenase activity(GO:0047023) indanol dehydrogenase activity(GO:0047718) |
| 1.7 | 5.1 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 1.6 | 6.5 | GO:0015207 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 1.5 | 16.9 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 1.5 | 12.0 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 1.1 | 3.2 | GO:0036080 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 1.0 | 7.3 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 1.0 | 6.2 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 1.0 | 23.4 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 1.0 | 8.6 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.9 | 7.6 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.9 | 10.6 | GO:0089720 | caspase binding(GO:0089720) |
| 0.9 | 3.5 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.8 | 2.5 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.8 | 4.2 | GO:0070026 | nitric oxide binding(GO:0070026) |
| 0.7 | 6.8 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.6 | 2.5 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.6 | 3.7 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.6 | 6.7 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.5 | 14.2 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.5 | 7.0 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.5 | 6.7 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.5 | 2.4 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.5 | 9.4 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.5 | 1.4 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.4 | 2.1 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.3 | 1.7 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.3 | 22.5 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.3 | 6.1 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.2 | 6.5 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 5.9 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 0.6 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.2 | 2.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.2 | 3.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.2 | 0.9 | GO:0070905 | serine binding(GO:0070905) |
| 0.2 | 5.2 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.2 | 2.7 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.2 | 5.6 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.2 | 4.6 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.2 | 5.8 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 2.9 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.8 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 3.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 1.6 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 4.2 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 5.3 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 3.8 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 1.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 3.5 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 5.6 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.1 | 1.9 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.1 | 2.9 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 0.9 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.1 | 0.4 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 1.7 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.5 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 3.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 3.9 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.9 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 9.4 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.8 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.3 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 1.6 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.7 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.0 | 2.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 4.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 5.6 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 3.1 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.4 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 0.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 5.5 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 1.4 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 23.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.3 | 12.0 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.2 | 13.5 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.2 | 6.9 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 5.9 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 7.8 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 13.6 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 3.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 3.7 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 2.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 7.9 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 7.2 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 1.9 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 1.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 5.7 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.7 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 2.7 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 2.7 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 1.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 23.4 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.6 | 12.0 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.5 | 14.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.5 | 7.6 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.5 | 7.3 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.4 | 7.0 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.3 | 29.3 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.2 | 3.5 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.2 | 3.5 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.2 | 14.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.2 | 6.2 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.2 | 2.6 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.2 | 5.1 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 11.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 11.3 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 3.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 4.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 2.5 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 3.2 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.1 | 5.7 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 2.7 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.1 | 9.4 | REACTOME APOPTOTIC EXECUTION PHASE | Genes involved in Apoptotic execution phase |
| 0.1 | 2.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 2.7 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 3.0 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 2.6 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.1 | 1.7 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.1 | 3.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.9 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 6.7 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 1.9 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 1.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.5 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 1.2 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.5 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.5 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |