Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
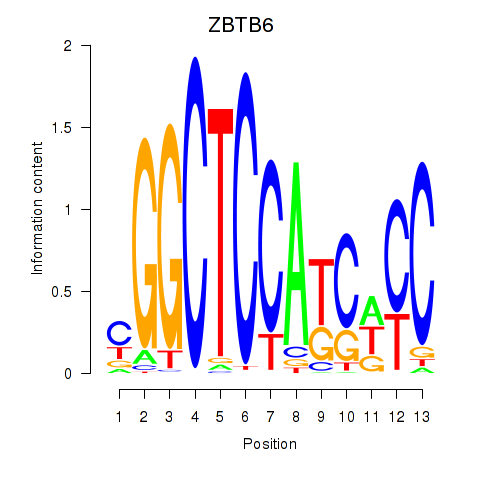
Results for ZBTB6
Z-value: 0.92
Transcription factors associated with ZBTB6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB6
|
ENSG00000186130.4 | zinc finger and BTB domain containing 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB6 | hg19_v2_chr9_-_125675576_125675612 | 0.21 | 2.2e-03 | Click! |
Activity profile of ZBTB6 motif
Sorted Z-values of ZBTB6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB6
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.4 | 33.8 | GO:1902613 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 6.7 | 20.1 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 6.6 | 19.8 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 4.7 | 37.8 | GO:0007506 | gonadal mesoderm development(GO:0007506) |
| 4.4 | 4.4 | GO:0032058 | positive regulation of translation in response to stress(GO:0032056) positive regulation of translational initiation in response to stress(GO:0032058) |
| 4.3 | 39.0 | GO:0003190 | atrioventricular valve formation(GO:0003190) |
| 4.2 | 37.9 | GO:1903912 | negative regulation of PERK-mediated unfolded protein response(GO:1903898) negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 3.9 | 11.6 | GO:0045643 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 3.8 | 11.5 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 3.8 | 19.1 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 3.8 | 11.3 | GO:0051710 | regulation of cytolysis in other organism(GO:0051710) |
| 3.5 | 10.5 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 3.5 | 10.4 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 3.4 | 10.2 | GO:0007412 | axon target recognition(GO:0007412) |
| 3.1 | 12.6 | GO:1904978 | regulation of endosome organization(GO:1904978) |
| 3.1 | 40.3 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 2.9 | 8.8 | GO:0018282 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 2.9 | 2.9 | GO:0051084 | 'de novo' posttranslational protein folding(GO:0051084) chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 2.8 | 14.0 | GO:0099540 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 2.8 | 8.3 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 2.6 | 7.9 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 2.6 | 7.9 | GO:1901624 | negative regulation of lymphocyte chemotaxis(GO:1901624) |
| 2.6 | 23.5 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 2.4 | 12.0 | GO:0044026 | DNA hypermethylation(GO:0044026) |
| 2.3 | 6.8 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 2.2 | 8.8 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 2.1 | 6.2 | GO:1900138 | negative regulation of phospholipase A2 activity(GO:1900138) |
| 2.0 | 6.0 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 1.9 | 7.7 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 1.9 | 5.7 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 1.9 | 11.3 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 1.8 | 5.5 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 1.8 | 23.2 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 1.7 | 21.0 | GO:1905146 | lysosomal protein catabolic process(GO:1905146) |
| 1.7 | 8.7 | GO:2001074 | regulation of metanephric ureteric bud development(GO:2001074) positive regulation of metanephric ureteric bud development(GO:2001076) |
| 1.7 | 5.2 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 1.6 | 11.1 | GO:1900107 | regulation of nodal signaling pathway(GO:1900107) |
| 1.6 | 7.8 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 1.5 | 3.0 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 1.5 | 4.5 | GO:1903465 | vacuolar phosphate transport(GO:0007037) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 1.5 | 7.4 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 1.5 | 4.4 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 1.4 | 17.1 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 1.4 | 5.7 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 1.4 | 8.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 1.4 | 4.1 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 1.3 | 9.2 | GO:0006477 | protein sulfation(GO:0006477) |
| 1.3 | 10.4 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 1.3 | 3.8 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 1.3 | 10.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 1.3 | 3.8 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 1.2 | 18.5 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 1.2 | 8.5 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 1.2 | 8.4 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 1.2 | 7.1 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 1.1 | 3.4 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 1.1 | 9.1 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 1.1 | 4.5 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 1.1 | 16.8 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 1.1 | 4.4 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 1.1 | 14.3 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 1.1 | 5.3 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) growth plate cartilage chondrocyte development(GO:0003431) Harderian gland development(GO:0070384) |
| 1.0 | 4.2 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 1.0 | 4.2 | GO:0019086 | late viral transcription(GO:0019086) |
| 1.0 | 6.2 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 1.0 | 7.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 1.0 | 3.9 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 1.0 | 2.0 | GO:0070459 | prolactin secretion(GO:0070459) |
| 1.0 | 10.7 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 1.0 | 6.8 | GO:0048104 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 1.0 | 7.6 | GO:0070235 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.9 | 7.3 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.9 | 6.4 | GO:1903433 | regulation of constitutive secretory pathway(GO:1903433) |
| 0.9 | 4.6 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.9 | 10.9 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.9 | 19.6 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.9 | 2.7 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.9 | 13.1 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.9 | 2.6 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.9 | 6.8 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.9 | 14.5 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.8 | 2.5 | GO:1903348 | cellular response to lead ion(GO:0071284) positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.8 | 6.6 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.8 | 9.1 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.8 | 4.9 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.8 | 3.2 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.8 | 45.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.8 | 3.9 | GO:1900020 | embryonic genitalia morphogenesis(GO:0030538) regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.8 | 2.3 | GO:0010519 | negative regulation of phospholipase activity(GO:0010519) |
| 0.8 | 6.2 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.8 | 16.0 | GO:0070977 | bone maturation(GO:0070977) |
| 0.7 | 2.9 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.7 | 2.9 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.7 | 12.3 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.7 | 2.9 | GO:0042335 | cuticle development(GO:0042335) |
| 0.7 | 12.9 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.7 | 5.0 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.7 | 28.2 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.7 | 8.4 | GO:0010566 | regulation of ketone biosynthetic process(GO:0010566) |
| 0.7 | 2.1 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.7 | 12.9 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.7 | 5.3 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.7 | 7.2 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.7 | 4.6 | GO:0090650 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.7 | 10.5 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.6 | 3.2 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 0.6 | 11.5 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.6 | 4.5 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.6 | 3.2 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.6 | 1.9 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.6 | 7.4 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.6 | 1.8 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.6 | 6.1 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.6 | 23.0 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.6 | 1.2 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.6 | 4.7 | GO:0010666 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.6 | 4.7 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.6 | 6.4 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.6 | 2.9 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.6 | 7.2 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.5 | 3.8 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.5 | 1.1 | GO:1904779 | regulation of protein localization to centrosome(GO:1904779) |
| 0.5 | 1.6 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.5 | 1.1 | GO:0061526 | acetylcholine secretion(GO:0061526) |
| 0.5 | 1.6 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.5 | 5.8 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.5 | 1.0 | GO:0036022 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.5 | 6.1 | GO:1903799 | negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.5 | 4.1 | GO:0048505 | regulation of development, heterochronic(GO:0040034) regulation of timing of cell differentiation(GO:0048505) |
| 0.5 | 2.0 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.5 | 0.5 | GO:0030514 | regulation of BMP signaling pathway(GO:0030510) negative regulation of BMP signaling pathway(GO:0030514) |
| 0.5 | 2.5 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.5 | 2.5 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.5 | 6.0 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.5 | 3.5 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.5 | 10.9 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.5 | 2.0 | GO:0032431 | activation of phospholipase A2 activity(GO:0032431) |
| 0.5 | 4.9 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.5 | 1.4 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) skeletal muscle fiber differentiation(GO:0098528) regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.5 | 2.4 | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 0.5 | 7.1 | GO:0021756 | striatum development(GO:0021756) |
| 0.5 | 3.2 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.5 | 1.8 | GO:0033076 | alkaloid catabolic process(GO:0009822) isoquinoline alkaloid metabolic process(GO:0033076) cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 0.4 | 2.2 | GO:0002218 | activation of innate immune response(GO:0002218) innate immune response activating cell surface receptor signaling pathway(GO:0002220) stimulatory C-type lectin receptor signaling pathway(GO:0002223) innate immune response-activating signal transduction(GO:0002758) |
| 0.4 | 8.3 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.4 | 7.4 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.4 | 3.9 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.4 | 6.4 | GO:0010629 | negative regulation of gene expression(GO:0010629) |
| 0.4 | 2.5 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.4 | 16.3 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.4 | 9.2 | GO:0032402 | melanosome transport(GO:0032402) |
| 0.4 | 5.8 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.4 | 2.5 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) |
| 0.4 | 19.0 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.4 | 3.2 | GO:0008212 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.4 | 32.9 | GO:0009301 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.4 | 5.8 | GO:0021681 | cerebellar granular layer development(GO:0021681) |
| 0.4 | 5.0 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.4 | 7.6 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.4 | 1.9 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.4 | 3.4 | GO:0051005 | plasma membrane to endosome transport(GO:0048227) negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.4 | 23.9 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.4 | 9.7 | GO:0003299 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.4 | 3.9 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.4 | 3.9 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.4 | 6.4 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.3 | 2.4 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.3 | 6.8 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.3 | 2.0 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.3 | 1.0 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.3 | 3.0 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.3 | 6.6 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.3 | 11.4 | GO:0043278 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.3 | 4.6 | GO:0007409 | axonogenesis(GO:0007409) |
| 0.3 | 5.5 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.3 | 2.6 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.3 | 2.8 | GO:0032226 | positive regulation of synaptic transmission, dopaminergic(GO:0032226) positive regulation of cell adhesion molecule production(GO:0060355) positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.3 | 13.9 | GO:1901998 | toxin transport(GO:1901998) |
| 0.3 | 0.9 | GO:1902256 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) positive regulation of vascular wound healing(GO:0035470) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.3 | 9.5 | GO:0001510 | RNA methylation(GO:0001510) |
| 0.3 | 7.6 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.3 | 2.1 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.3 | 1.2 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.3 | 9.6 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.3 | 17.0 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.3 | 0.9 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.3 | 2.0 | GO:0008595 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.3 | 1.4 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.3 | 3.1 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.3 | 3.3 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.3 | 3.3 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.3 | 1.3 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.3 | 2.6 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.3 | 1.8 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.2 | 1.2 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.2 | 5.6 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.2 | 3.9 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.2 | 4.5 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.2 | 12.5 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.2 | 10.8 | GO:0051646 | mitochondrion localization(GO:0051646) |
| 0.2 | 6.7 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.2 | 0.9 | GO:0042428 | serotonin metabolic process(GO:0042428) |
| 0.2 | 2.3 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.2 | 0.7 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.2 | 6.3 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.2 | 4.0 | GO:0048535 | lymph node development(GO:0048535) |
| 0.2 | 5.4 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.2 | 3.0 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.2 | 1.3 | GO:1990822 | basic amino acid transmembrane transport(GO:1990822) |
| 0.2 | 5.1 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.2 | 1.7 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.2 | 4.7 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.2 | 1.9 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.2 | 2.1 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.2 | 3.5 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.2 | 5.4 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.2 | 0.4 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.2 | 0.4 | GO:0042713 | sperm ejaculation(GO:0042713) |
| 0.2 | 3.8 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.2 | 2.4 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.2 | 6.3 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.2 | 3.3 | GO:0009452 | 7-methylguanosine mRNA capping(GO:0006370) 7-methylguanosine RNA capping(GO:0009452) RNA capping(GO:0036260) |
| 0.2 | 8.2 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.2 | 1.0 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.2 | 4.0 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.2 | 1.3 | GO:0008585 | female gonad development(GO:0008585) |
| 0.2 | 2.1 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.2 | 3.2 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.2 | 1.3 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.2 | 7.9 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.2 | 5.3 | GO:0009081 | branched-chain amino acid metabolic process(GO:0009081) branched-chain amino acid catabolic process(GO:0009083) |
| 0.2 | 4.2 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.2 | 3.2 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.2 | 2.0 | GO:0043116 | negative regulation of vascular permeability(GO:0043116) |
| 0.2 | 2.5 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.2 | 1.3 | GO:1902744 | negative regulation of lamellipodium assembly(GO:0010593) negative regulation of lamellipodium organization(GO:1902744) |
| 0.2 | 1.0 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.2 | 10.9 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.2 | 1.3 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.2 | 0.6 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.2 | 5.0 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.2 | 4.5 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.1 | 0.6 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.1 | 10.0 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 5.1 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.1 | 7.1 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.1 | 1.0 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.1 | 0.9 | GO:1904646 | positive regulation of superoxide anion generation(GO:0032930) cellular response to beta-amyloid(GO:1904646) |
| 0.1 | 2.1 | GO:0098856 | intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.1 | 4.1 | GO:0006693 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.1 | 1.1 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.1 | 2.0 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 3.4 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 0.4 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) |
| 0.1 | 0.4 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.1 | 3.2 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.1 | 11.2 | GO:0010921 | regulation of phosphatase activity(GO:0010921) |
| 0.1 | 2.5 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.1 | 1.6 | GO:0007625 | grooming behavior(GO:0007625) |
| 0.1 | 2.1 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 2.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 6.2 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.1 | 2.2 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 2.5 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.1 | 1.4 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.1 | 2.6 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.1 | 3.3 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 15.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 2.6 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 2.8 | GO:0044803 | multi-organism membrane organization(GO:0044803) |
| 0.1 | 14.6 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 0.7 | GO:0023014 | signal transduction by protein phosphorylation(GO:0023014) |
| 0.1 | 1.7 | GO:1902895 | positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.1 | 0.4 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.1 | 0.9 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 0.7 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.1 | 0.7 | GO:0050654 | chondroitin sulfate metabolic process(GO:0030204) chondroitin sulfate catabolic process(GO:0030207) chondroitin sulfate proteoglycan metabolic process(GO:0050654) |
| 0.1 | 5.5 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.1 | 1.7 | GO:0070734 | peptidyl-lysine dimethylation(GO:0018027) histone H3-K27 methylation(GO:0070734) |
| 0.1 | 3.4 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.1 | 1.3 | GO:0021904 | dorsal/ventral neural tube patterning(GO:0021904) |
| 0.1 | 4.7 | GO:0021954 | central nervous system neuron development(GO:0021954) |
| 0.1 | 3.7 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.1 | 3.9 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.1 | 0.3 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.1 | 1.0 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.1 | 1.8 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.1 | 1.3 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.1 | 3.4 | GO:0034113 | heterotypic cell-cell adhesion(GO:0034113) |
| 0.1 | 2.5 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 1.0 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.1 | 3.1 | GO:0030317 | sperm motility(GO:0030317) |
| 0.1 | 1.8 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 0.4 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 4.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 4.0 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 2.9 | GO:0043647 | inositol phosphate metabolic process(GO:0043647) |
| 0.1 | 1.7 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.1 | 1.5 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.1 | 0.8 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.1 | 5.6 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.1 | 0.3 | GO:0043524 | negative regulation of neuron apoptotic process(GO:0043524) |
| 0.1 | 1.7 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.1 | 5.2 | GO:0006024 | aminoglycan biosynthetic process(GO:0006023) glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.1 | 0.9 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.6 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.1 | 0.3 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 0.8 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
| 0.1 | 0.9 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.1 | 7.5 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.1 | 3.5 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.4 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 4.0 | GO:0003014 | renal system process(GO:0003014) |
| 0.0 | 0.2 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) histone H2A phosphorylation(GO:1990164) |
| 0.0 | 1.8 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 1.6 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 2.2 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 2.9 | GO:0009306 | protein secretion(GO:0009306) |
| 0.0 | 0.9 | GO:0001895 | retina homeostasis(GO:0001895) |
| 0.0 | 2.5 | GO:1901184 | regulation of ERBB signaling pathway(GO:1901184) |
| 0.0 | 0.2 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.7 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.9 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 1.7 | GO:0000271 | polysaccharide biosynthetic process(GO:0000271) |
| 0.0 | 1.2 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.0 | 1.2 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 0.2 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 2.1 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 1.7 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 1.2 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 0.5 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 1.2 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 0.5 | GO:0030218 | erythrocyte differentiation(GO:0030218) |
| 0.0 | 5.1 | GO:0016311 | dephosphorylation(GO:0016311) |
| 0.0 | 2.3 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 | 0.3 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.2 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.0 | 0.1 | GO:0038089 | positive regulation of cell migration by vascular endothelial growth factor signaling pathway(GO:0038089) |
| 0.0 | 0.2 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.9 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.0 | 1.0 | GO:0051291 | protein heterooligomerization(GO:0051291) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 32.9 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 3.5 | 10.5 | GO:0019034 | viral replication complex(GO:0019034) |
| 2.4 | 7.3 | GO:0034657 | GID complex(GO:0034657) |
| 2.3 | 11.3 | GO:0071797 | LUBAC complex(GO:0071797) |
| 2.1 | 6.4 | GO:0031251 | PAN complex(GO:0031251) |
| 2.1 | 19.0 | GO:0005638 | lamin filament(GO:0005638) |
| 2.1 | 35.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 1.7 | 6.7 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 1.6 | 4.7 | GO:0060187 | cell pole(GO:0060187) |
| 1.4 | 14.5 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 1.4 | 11.5 | GO:0060203 | clathrin-sculpted glutamate transport vesicle(GO:0060199) clathrin-sculpted glutamate transport vesicle membrane(GO:0060203) |
| 1.3 | 6.6 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 1.3 | 5.2 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 1.3 | 10.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 1.2 | 2.3 | GO:0033648 | host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) |
| 1.2 | 13.9 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 1.1 | 12.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 1.1 | 9.9 | GO:0030897 | HOPS complex(GO:0030897) |
| 1.1 | 5.5 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 1.1 | 5.3 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 1.0 | 5.8 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 1.0 | 10.5 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.9 | 8.4 | GO:0034464 | BBSome(GO:0034464) |
| 0.9 | 6.5 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.9 | 7.2 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.9 | 21.0 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.9 | 11.3 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.8 | 8.5 | GO:0045180 | basal cortex(GO:0045180) |
| 0.8 | 3.3 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.8 | 18.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.8 | 8.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.7 | 9.6 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.7 | 4.1 | GO:0002177 | manchette(GO:0002177) |
| 0.7 | 7.4 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.6 | 1.8 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.6 | 2.4 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.6 | 3.5 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.6 | 2.3 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.6 | 5.5 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.5 | 11.8 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.5 | 6.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.5 | 8.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.5 | 27.1 | GO:0030118 | clathrin coat(GO:0030118) |
| 0.5 | 7.7 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) |
| 0.4 | 12.4 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.4 | 5.6 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.4 | 3.9 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.4 | 19.8 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.4 | 2.1 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.4 | 1.7 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.4 | 2.5 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.4 | 4.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.4 | 2.9 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.4 | 6.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.4 | 34.2 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.4 | 30.6 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.4 | 3.2 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.4 | 8.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.4 | 4.1 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.4 | 4.0 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.3 | 1.7 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.3 | 19.3 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.3 | 6.0 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.3 | 2.0 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.3 | 2.6 | GO:0032009 | early phagosome(GO:0032009) |
| 0.3 | 4.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.3 | 2.7 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.3 | 14.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.3 | 3.5 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.3 | 10.2 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.3 | 5.0 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.3 | 9.0 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.2 | 7.0 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.2 | 3.9 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.2 | 16.0 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.2 | 3.1 | GO:0000800 | lateral element(GO:0000800) |
| 0.2 | 4.2 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.2 | 3.2 | GO:0001741 | XY body(GO:0001741) |
| 0.2 | 3.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.2 | 12.8 | GO:0005844 | polysome(GO:0005844) |
| 0.2 | 1.0 | GO:0044308 | axonal spine(GO:0044308) |
| 0.2 | 0.8 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.2 | 20.6 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.2 | 24.2 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.2 | 2.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.2 | 2.7 | GO:0036038 | MKS complex(GO:0036038) |
| 0.2 | 10.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.2 | 5.1 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.2 | 6.0 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.2 | 6.2 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.2 | 25.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.2 | 5.2 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.2 | 3.9 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.2 | 4.2 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.2 | 1.0 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.2 | 0.6 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.1 | 4.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 2.5 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 13.0 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 2.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 6.0 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.1 | 7.6 | GO:0009986 | cell surface(GO:0009986) |
| 0.1 | 4.1 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 1.4 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 6.8 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 3.2 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 11.6 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 5.2 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 8.4 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 12.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 1.6 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 5.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 5.7 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 1.6 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 1.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 1.0 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 5.7 | GO:0043235 | receptor complex(GO:0043235) |
| 0.1 | 25.1 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.1 | 4.7 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 2.2 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 115.7 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 6.3 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.1 | 0.4 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 1.7 | GO:0098793 | presynapse(GO:0098793) |
| 0.1 | 3.4 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 2.2 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 1.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 2.4 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 2.0 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.3 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 3.3 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.0 | 2.5 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.3 | GO:0031082 | BLOC complex(GO:0031082) |
| 0.0 | 3.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 4.6 | GO:0030016 | myofibril(GO:0030016) |
| 0.0 | 47.7 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 1.0 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.9 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 0.1 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.7 | GO:1904724 | tertiary granule lumen(GO:1904724) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.7 | 20.1 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 5.1 | 15.2 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 4.3 | 17.1 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 3.9 | 11.6 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 3.8 | 19.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 3.5 | 10.4 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 3.4 | 16.8 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 3.1 | 12.6 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 3.1 | 12.5 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 2.9 | 11.5 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 2.9 | 8.6 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 2.9 | 28.6 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 2.8 | 8.5 | GO:0034584 | piRNA binding(GO:0034584) |
| 2.8 | 14.0 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 2.8 | 8.3 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 2.7 | 16.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 2.6 | 10.2 | GO:0052812 | phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 2.5 | 10.2 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 2.5 | 45.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 2.5 | 12.4 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 2.4 | 7.1 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 2.3 | 13.6 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 1.9 | 5.7 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 1.9 | 3.8 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 1.8 | 10.5 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 1.7 | 5.2 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 1.7 | 13.8 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 1.7 | 3.4 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 1.7 | 3.4 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 1.7 | 16.9 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 1.6 | 6.6 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 1.6 | 7.9 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 1.6 | 7.8 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 1.5 | 10.7 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 1.5 | 10.4 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 1.5 | 7.3 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 1.3 | 6.6 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 1.3 | 4.0 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 1.3 | 15.5 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 1.3 | 33.1 | GO:0031489 | myosin V binding(GO:0031489) |
| 1.2 | 5.0 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 1.2 | 7.4 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 1.2 | 6.0 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 1.2 | 5.9 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 1.1 | 4.6 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 1.1 | 3.3 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 1.1 | 6.4 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 1.1 | 3.2 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) steroid delta-isomerase activity(GO:0004769) |
| 1.0 | 16.6 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 1.0 | 8.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 1.0 | 4.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 1.0 | 11.4 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.9 | 9.3 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.9 | 37.9 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.9 | 10.5 | GO:0048185 | activin binding(GO:0048185) |
| 0.8 | 7.6 | GO:0043426 | MRF binding(GO:0043426) |
| 0.8 | 4.9 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.8 | 4.0 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.8 | 5.5 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.8 | 4.6 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.8 | 16.6 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.7 | 10.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.7 | 4.4 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.7 | 10.1 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.7 | 10.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.7 | 7.9 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.7 | 2.9 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.7 | 4.2 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.7 | 2.1 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.7 | 4.7 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.6 | 14.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.6 | 3.2 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.6 | 2.3 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.6 | 10.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.6 | 1.7 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.6 | 3.3 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.6 | 8.8 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.5 | 2.7 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.5 | 14.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.5 | 3.2 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.5 | 3.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.5 | 4.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.5 | 10.9 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.5 | 6.6 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.5 | 3.0 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.5 | 12.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.5 | 2.4 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.5 | 13.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.5 | 2.4 | GO:0086077 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 0.5 | 7.9 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.5 | 2.3 | GO:0070905 | serine binding(GO:0070905) |
| 0.5 | 6.0 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.5 | 7.3 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.5 | 4.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.4 | 7.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.4 | 6.6 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.4 | 2.6 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.4 | 4.7 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.4 | 3.4 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.4 | 10.9 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.4 | 8.4 | GO:0034979 | NAD-dependent protein deacetylase activity(GO:0034979) |
| 0.4 | 2.5 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.4 | 13.8 | GO:0008066 | glutamate receptor activity(GO:0008066) |
| 0.4 | 6.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.4 | 8.4 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.4 | 3.2 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.4 | 6.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.4 | 18.8 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.4 | 36.3 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.4 | 1.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.4 | 2.2 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.4 | 1.8 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.4 | 2.9 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.4 | 2.5 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.4 | 6.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.4 | 5.3 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.3 | 6.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.3 | 1.7 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.3 | 2.9 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.3 | 1.6 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.3 | 7.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.3 | 6.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.3 | 3.8 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.3 | 9.9 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.3 | 24.1 | GO:0032813 | tumor necrosis factor receptor superfamily binding(GO:0032813) |
| 0.3 | 1.3 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.3 | 4.6 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.3 | 8.3 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.3 | 11.7 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.3 | 8.1 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.3 | 1.8 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.3 | 3.5 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.3 | 11.2 | GO:0015144 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.3 | 5.1 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.3 | 2.8 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.3 | 6.6 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.3 | 0.3 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.3 | 1.9 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.3 | 4.1 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.3 | 3.2 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.3 | 6.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.3 | 3.7 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.3 | 1.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.3 | 4.4 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.3 | 2.6 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.3 | 5.1 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.2 | 3.5 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.2 | 1.0 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.2 | 9.5 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.2 | 0.9 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.2 | 0.9 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.2 | 1.6 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.2 | 3.8 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.2 | 2.9 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.2 | 0.2 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.2 | 5.4 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.2 | 4.4 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.2 | 10.9 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.2 | 5.5 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.2 | 10.0 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 9.2 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.2 | 3.2 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.2 | 5.3 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.2 | 7.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 22.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.2 | 2.9 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.2 | 1.0 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.2 | 3.9 | GO:0031005 | filamin binding(GO:0031005) |
| 0.2 | 3.5 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.2 | 2.6 | GO:0035325 | Toll-like receptor binding(GO:0035325) |
| 0.2 | 7.7 | GO:0030551 | cyclic nucleotide binding(GO:0030551) |
| 0.2 | 3.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.2 | 28.5 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.2 | 2.2 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.2 | 2.1 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.2 | 3.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 4.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.2 | 15.3 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.2 | 3.3 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.2 | 1.7 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.2 | 6.6 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.2 | 0.6 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.1 | 2.7 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 1.3 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 8.2 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.1 | 3.1 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 0.7 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 0.8 | GO:0031432 | titin binding(GO:0031432) |
| 0.1 | 8.6 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 2.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.4 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 5.3 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 2.8 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 1.0 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 1.3 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.1 | 6.1 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.1 | 1.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 21.9 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.1 | 0.9 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 1.9 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.1 | 2.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 5.1 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 5.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 1.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 3.4 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 6.2 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 2.9 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 24.0 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.1 | 2.3 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 2.8 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 0.4 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.1 | 10.4 | GO:0046906 | heme binding(GO:0020037) tetrapyrrole binding(GO:0046906) |
| 0.1 | 4.6 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.1 | 0.6 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 2.3 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 3.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.9 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.7 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 2.6 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 1.2 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.4 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 1.3 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.4 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.4 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 10.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 4.0 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 2.8 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.7 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.4 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 2.5 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.3 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 6.5 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 1.3 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 1.1 | GO:0005230 | extracellular ligand-gated ion channel activity(GO:0005230) |
| 0.0 | 7.1 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 0.2 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.8 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.6 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.4 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.5 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.1 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 42.4 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.9 | 15.8 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.8 | 18.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.7 | 12.9 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.6 | 49.9 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.5 | 6.7 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.5 | 13.1 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.5 | 22.1 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.5 | 11.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.4 | 7.8 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.4 | 12.2 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.4 | 6.7 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.3 | 11.2 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.3 | 16.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.3 | 13.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.3 | 7.0 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.3 | 3.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.3 | 11.6 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.3 | 15.3 | PID FGF PATHWAY | FGF signaling pathway |
| 0.3 | 5.8 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.2 | 8.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.2 | 5.8 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.2 | 10.2 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.2 | 10.2 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.2 | 18.1 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.2 | 14.1 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.2 | 4.8 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.2 | 5.8 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.2 | 7.1 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.2 | 7.5 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.2 | 4.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.2 | 5.9 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.2 | 2.8 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.1 | 5.0 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 9.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 7.9 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 3.8 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 2.3 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 19.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 2.0 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 0.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 2.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 3.2 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 3.2 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 8.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 3.4 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.1 | 2.1 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 2.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 1.6 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 1.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 3.5 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 2.9 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.9 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 1.0 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.4 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.4 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 1.9 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.4 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 28.9 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 1.2 | 16.0 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 1.2 | 2.4 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 1.0 | 18.5 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.9 | 4.5 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.9 | 11.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.8 | 11.6 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.8 | 26.9 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.6 | 10.2 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.6 | 5.5 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.5 | 4.4 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.5 | 10.6 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.5 | 10.5 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.5 | 7.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.5 | 8.8 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.5 | 30.6 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.5 | 5.0 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.5 | 10.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.5 | 21.8 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.5 | 14.1 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.5 | 10.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.5 | 6.0 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.5 | 2.7 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.5 | 9.5 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.4 | 52.1 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.4 | 11.4 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.4 | 13.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.4 | 33.1 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.4 | 25.3 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.4 | 11.4 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.3 | 4.8 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.3 | 4.4 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.3 | 6.6 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.3 | 16.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.3 | 5.0 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.3 | 10.8 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.3 | 2.9 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.3 | 6.8 | REACTOME PROLONGED ERK ACTIVATION EVENTS | Genes involved in Prolonged ERK activation events |
| 0.2 | 6.0 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.2 | 5.1 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.2 | 10.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 10.1 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.2 | 3.2 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.2 | 6.1 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.2 | 2.6 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.2 | 2.6 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.2 | 9.9 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.2 | 6.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 3.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.2 | 5.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.2 | 16.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.2 | 5.5 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.2 | 4.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 2.4 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.2 | 2.5 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.2 | 3.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 2.5 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.2 | 2.1 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 4.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.6 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 6.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 0.4 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 9.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 2.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 1.1 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 3.4 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 2.4 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 5.4 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 27.4 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.1 | 2.3 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 1.6 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.1 | 9.3 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.1 | 1.7 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 7.0 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 1.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 4.2 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.1 | 1.9 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 1.6 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.1 | 2.5 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.1 | 13.1 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 0.5 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 3.4 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 1.8 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.8 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 1.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.4 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.4 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.2 | REACTOME BILE ACID AND BILE SALT METABOLISM | Genes involved in Bile acid and bile salt metabolism |
| 0.0 | 2.3 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.4 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 1.4 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |