Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
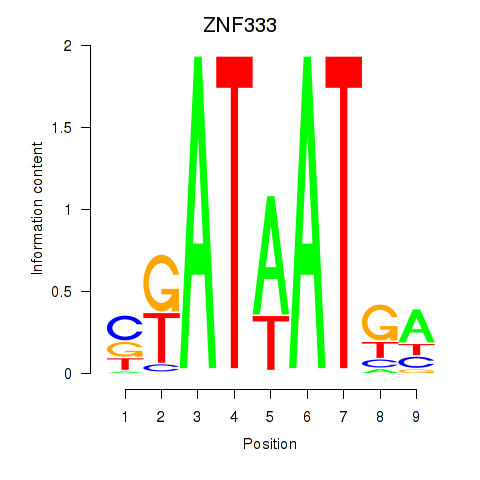
Results for ZNF333
Z-value: 0.53
Transcription factors associated with ZNF333
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF333
|
ENSG00000160961.7 | zinc finger protein 333 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF333 | hg19_v2_chr19_+_14800711_14800917 | 0.08 | 2.2e-01 | Click! |
Activity profile of ZNF333 motif
Sorted Z-values of ZNF333 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_92085262 | 14.29 |
ENST00000298047.6
ENST00000409404.2 ENST00000541502.1 |
FAT3
|
FAT atypical cadherin 3 |
| chr18_+_71815743 | 10.85 |
ENST00000169551.6
ENST00000580087.1 |
TIMM21
|
translocase of inner mitochondrial membrane 21 homolog (yeast) |
| chr6_-_32908765 | 6.95 |
ENST00000416244.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr11_-_85376121 | 5.47 |
ENST00000527447.1
|
CREBZF
|
CREB/ATF bZIP transcription factor |
| chr2_-_127977654 | 5.30 |
ENST00000409327.1
|
CYP27C1
|
cytochrome P450, family 27, subfamily C, polypeptide 1 |
| chr12_-_71031220 | 5.25 |
ENST00000334414.6
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr6_-_52705641 | 4.65 |
ENST00000370989.2
|
GSTA5
|
glutathione S-transferase alpha 5 |
| chrX_-_21776281 | 4.55 |
ENST00000379494.3
|
SMPX
|
small muscle protein, X-linked |
| chr7_-_115670792 | 4.43 |
ENST00000265440.7
ENST00000393485.1 |
TFEC
|
transcription factor EC |
| chr14_-_90798418 | 4.34 |
ENST00000354366.3
|
NRDE2
|
NRDE-2, necessary for RNA interference, domain containing |
| chr7_-_115670804 | 4.02 |
ENST00000320239.7
|
TFEC
|
transcription factor EC |
| chr5_+_137225158 | 4.01 |
ENST00000290431.5
|
PKD2L2
|
polycystic kidney disease 2-like 2 |
| chr6_+_131894284 | 3.92 |
ENST00000368087.3
ENST00000356962.2 |
ARG1
|
arginase 1 |
| chr1_+_19638788 | 3.85 |
ENST00000375155.3
ENST00000375153.3 ENST00000400548.2 |
PQLC2
|
PQ loop repeat containing 2 |
| chr12_-_71031185 | 3.80 |
ENST00000548122.1
ENST00000551525.1 ENST00000550358.1 |
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr19_-_45004556 | 3.79 |
ENST00000587047.1
ENST00000391956.4 ENST00000221327.4 ENST00000586637.1 ENST00000591064.1 ENST00000592529.1 |
ZNF180
|
zinc finger protein 180 |
| chr9_-_21351377 | 3.61 |
ENST00000380210.1
|
IFNA6
|
interferon, alpha 6 |
| chr14_+_76618242 | 3.39 |
ENST00000557542.1
ENST00000557263.1 ENST00000557207.1 ENST00000312858.5 ENST00000261530.7 |
GPATCH2L
|
G patch domain containing 2-like |
| chr16_+_84328252 | 3.36 |
ENST00000219454.5
|
WFDC1
|
WAP four-disulfide core domain 1 |
| chr11_-_6440624 | 3.34 |
ENST00000311051.3
|
APBB1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr5_+_140800638 | 3.11 |
ENST00000398587.2
ENST00000518882.1 |
PCDHGA11
|
protocadherin gamma subfamily A, 11 |
| chr1_+_36789335 | 3.09 |
ENST00000373137.2
|
RP11-268J15.5
|
RP11-268J15.5 |
| chr16_+_84328429 | 3.00 |
ENST00000568638.1
|
WFDC1
|
WAP four-disulfide core domain 1 |
| chr12_+_112563303 | 2.92 |
ENST00000412615.2
|
TRAFD1
|
TRAF-type zinc finger domain containing 1 |
| chr11_-_117698765 | 2.88 |
ENST00000532119.1
|
FXYD2
|
FXYD domain containing ion transport regulator 2 |
| chr8_+_18248755 | 2.72 |
ENST00000286479.3
|
NAT2
|
N-acetyltransferase 2 (arylamine N-acetyltransferase) |
| chr5_+_137225125 | 2.66 |
ENST00000350250.4
ENST00000508638.1 ENST00000502810.1 ENST00000508883.1 |
PKD2L2
|
polycystic kidney disease 2-like 2 |
| chr20_+_58515417 | 2.47 |
ENST00000360816.3
|
FAM217B
|
family with sequence similarity 217, member B |
| chr12_+_112563335 | 2.37 |
ENST00000549358.1
ENST00000257604.5 ENST00000548092.1 ENST00000552896.1 |
TRAFD1
|
TRAF-type zinc finger domain containing 1 |
| chr6_+_160327974 | 2.37 |
ENST00000252660.4
|
MAS1
|
MAS1 oncogene |
| chrX_+_11311533 | 2.35 |
ENST00000380714.3
ENST00000380712.3 ENST00000348912.4 |
AMELX
|
amelogenin, X-linked |
| chr3_+_38323785 | 2.23 |
ENST00000466887.1
ENST00000448498.1 |
SLC22A14
|
solute carrier family 22, member 14 |
| chr20_-_44600810 | 2.18 |
ENST00000322927.2
ENST00000426788.1 |
ZNF335
|
zinc finger protein 335 |
| chr5_+_140535577 | 2.16 |
ENST00000539533.1
|
PCDHB17
|
Protocadherin-psi1; Uncharacterized protein |
| chr17_-_3337135 | 2.11 |
ENST00000248384.1
|
OR1E2
|
olfactory receptor, family 1, subfamily E, member 2 |
| chr14_+_85996507 | 2.09 |
ENST00000554746.1
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr9_-_19786926 | 2.09 |
ENST00000341998.2
ENST00000286344.3 |
SLC24A2
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 2 |
| chr3_+_35721106 | 2.08 |
ENST00000474696.1
ENST00000412048.1 ENST00000396482.2 ENST00000432682.1 |
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr16_-_20702578 | 2.06 |
ENST00000307493.4
ENST00000219151.4 |
ACSM1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr6_+_50786414 | 2.04 |
ENST00000344788.3
ENST00000393655.3 ENST00000263046.4 |
TFAP2B
|
transcription factor AP-2 beta (activating enhancer binding protein 2 beta) |
| chr14_+_85996471 | 2.00 |
ENST00000330753.4
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chrX_-_107975917 | 1.97 |
ENST00000563887.1
|
RP6-24A23.6
|
Uncharacterized protein |
| chr2_+_79412357 | 1.95 |
ENST00000466387.1
|
CTNNA2
|
catenin (cadherin-associated protein), alpha 2 |
| chr2_-_228497888 | 1.94 |
ENST00000264387.4
ENST00000409066.1 |
C2orf83
|
chromosome 2 open reading frame 83 |
| chr12_-_7848364 | 1.90 |
ENST00000329913.3
|
GDF3
|
growth differentiation factor 3 |
| chr14_+_23842018 | 1.88 |
ENST00000397242.2
ENST00000329715.2 |
IL25
|
interleukin 25 |
| chr11_+_18433840 | 1.86 |
ENST00000541669.1
ENST00000280704.4 |
LDHC
|
lactate dehydrogenase C |
| chr5_+_140207536 | 1.74 |
ENST00000529310.1
ENST00000527624.1 |
PCDHA6
|
protocadherin alpha 6 |
| chr2_-_178753465 | 1.71 |
ENST00000389683.3
|
PDE11A
|
phosphodiesterase 11A |
| chr1_+_225600404 | 1.66 |
ENST00000366845.2
|
AC092811.1
|
AC092811.1 |
| chr5_-_27038683 | 1.66 |
ENST00000511822.1
ENST00000231021.4 |
CDH9
|
cadherin 9, type 2 (T1-cadherin) |
| chr3_+_53528659 | 1.65 |
ENST00000350061.5
|
CACNA1D
|
calcium channel, voltage-dependent, L type, alpha 1D subunit |
| chr6_-_135375986 | 1.58 |
ENST00000525067.1
ENST00000367822.5 ENST00000367837.5 |
HBS1L
|
HBS1-like (S. cerevisiae) |
| chr17_-_2966901 | 1.57 |
ENST00000575751.1
|
OR1D5
|
olfactory receptor, family 1, subfamily D, member 5 |
| chr3_-_113918254 | 1.54 |
ENST00000460779.1
|
DRD3
|
dopamine receptor D3 |
| chrX_-_30595959 | 1.54 |
ENST00000378962.3
|
CXorf21
|
chromosome X open reading frame 21 |
| chr6_-_46620522 | 1.47 |
ENST00000275016.2
|
CYP39A1
|
cytochrome P450, family 39, subfamily A, polypeptide 1 |
| chr5_+_137203465 | 1.46 |
ENST00000239926.4
|
MYOT
|
myotilin |
| chrX_-_49121165 | 1.46 |
ENST00000376207.4
ENST00000376199.2 |
FOXP3
|
forkhead box P3 |
| chr7_-_27169801 | 1.45 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr11_-_117698787 | 1.39 |
ENST00000260287.2
|
FXYD2
|
FXYD domain containing ion transport regulator 2 |
| chr17_-_46690839 | 1.36 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chr2_+_233527443 | 1.32 |
ENST00000410095.1
|
EFHD1
|
EF-hand domain family, member D1 |
| chr6_+_166945369 | 1.30 |
ENST00000598601.1
|
Z98049.1
|
CDNA FLJ25492 fis, clone CBR01389; Uncharacterized protein |
| chrX_+_122318318 | 1.25 |
ENST00000371251.1
ENST00000371256.5 |
GRIA3
|
glutamate receptor, ionotropic, AMPA 3 |
| chr7_-_36406750 | 1.21 |
ENST00000453212.1
ENST00000415803.2 ENST00000440378.1 ENST00000431396.1 ENST00000317020.6 ENST00000436884.1 |
KIAA0895
|
KIAA0895 |
| chrY_-_6742068 | 1.19 |
ENST00000215479.5
|
AMELY
|
amelogenin, Y-linked |
| chr11_+_33061543 | 1.19 |
ENST00000432887.1
ENST00000528898.1 ENST00000531632.2 |
TCP11L1
|
t-complex 11, testis-specific-like 1 |
| chr2_-_128432639 | 1.18 |
ENST00000545738.2
ENST00000355119.4 ENST00000409808.2 |
LIMS2
|
LIM and senescent cell antigen-like domains 2 |
| chr3_-_126327398 | 1.14 |
ENST00000383572.2
|
TXNRD3NB
|
thioredoxin reductase 3 neighbor |
| chr5_-_35195338 | 1.11 |
ENST00000509839.1
|
PRLR
|
prolactin receptor |
| chr7_-_83824169 | 1.08 |
ENST00000265362.4
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chrX_+_99839799 | 1.07 |
ENST00000373031.4
|
TNMD
|
tenomodulin |
| chr1_-_169599314 | 0.98 |
ENST00000367786.2
ENST00000458599.2 ENST00000367795.2 ENST00000263686.6 |
SELP
|
selectin P (granule membrane protein 140kDa, antigen CD62) |
| chr4_+_71108300 | 0.94 |
ENST00000304954.3
|
CSN3
|
casein kappa |
| chr1_+_55013889 | 0.91 |
ENST00000343744.2
ENST00000371316.3 |
ACOT11
|
acyl-CoA thioesterase 11 |
| chr16_+_87636474 | 0.89 |
ENST00000284262.2
|
JPH3
|
junctophilin 3 |
| chr8_-_11725549 | 0.85 |
ENST00000505496.2
ENST00000534636.1 ENST00000524500.1 ENST00000345125.3 ENST00000453527.2 ENST00000527215.2 ENST00000532392.1 ENST00000533455.1 ENST00000534510.1 ENST00000530640.2 ENST00000531089.1 ENST00000532656.2 ENST00000531502.1 ENST00000434271.1 ENST00000353047.6 |
CTSB
|
cathepsin B |
| chr12_-_49393092 | 0.83 |
ENST00000421952.2
|
DDN
|
dendrin |
| chr17_-_41623716 | 0.82 |
ENST00000319349.5
|
ETV4
|
ets variant 4 |
| chr4_+_106473768 | 0.79 |
ENST00000265154.2
ENST00000420470.2 |
ARHGEF38
|
Rho guanine nucleotide exchange factor (GEF) 38 |
| chr12_-_10251539 | 0.74 |
ENST00000420265.2
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr11_+_12766583 | 0.71 |
ENST00000361985.2
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr14_+_22748980 | 0.66 |
ENST00000390465.2
|
TRAV38-2DV8
|
T cell receptor alpha variable 38-2/delta variable 8 |
| chr12_+_60058458 | 0.61 |
ENST00000548610.1
|
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr2_+_185463093 | 0.60 |
ENST00000302277.6
|
ZNF804A
|
zinc finger protein 804A |
| chr1_-_169599353 | 0.60 |
ENST00000367793.2
ENST00000367794.2 ENST00000367792.2 ENST00000367791.2 ENST00000367788.2 |
SELP
|
selectin P (granule membrane protein 140kDa, antigen CD62) |
| chr11_-_66313699 | 0.60 |
ENST00000526986.1
ENST00000310442.3 |
ZDHHC24
|
zinc finger, DHHC-type containing 24 |
| chr18_+_46065393 | 0.58 |
ENST00000256413.3
|
CTIF
|
CBP80/20-dependent translation initiation factor |
| chr14_+_24099318 | 0.58 |
ENST00000432832.2
|
DHRS2
|
dehydrogenase/reductase (SDR family) member 2 |
| chr10_+_118350468 | 0.57 |
ENST00000358834.4
ENST00000528052.1 ENST00000442761.1 |
PNLIPRP1
|
pancreatic lipase-related protein 1 |
| chr11_-_18062872 | 0.52 |
ENST00000250018.2
|
TPH1
|
tryptophan hydroxylase 1 |
| chr20_+_56964169 | 0.51 |
ENST00000475243.1
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr5_-_78281603 | 0.50 |
ENST00000264914.4
|
ARSB
|
arylsulfatase B |
| chr10_+_118350522 | 0.47 |
ENST00000530319.1
ENST00000527980.1 ENST00000471549.1 ENST00000534537.1 |
PNLIPRP1
|
pancreatic lipase-related protein 1 |
| chrX_-_114253536 | 0.47 |
ENST00000371936.1
|
IL13RA2
|
interleukin 13 receptor, alpha 2 |
| chr12_-_7656357 | 0.45 |
ENST00000396620.3
ENST00000432237.2 ENST00000359156.4 |
CD163
|
CD163 molecule |
| chr4_-_186696425 | 0.40 |
ENST00000430503.1
ENST00000319454.6 ENST00000450341.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr4_+_110749143 | 0.40 |
ENST00000317735.4
|
RRH
|
retinal pigment epithelium-derived rhodopsin homolog |
| chr6_+_17110726 | 0.39 |
ENST00000354384.5
|
STMND1
|
stathmin domain containing 1 |
| chr4_-_100356551 | 0.32 |
ENST00000209665.4
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr12_+_58176525 | 0.31 |
ENST00000543727.1
ENST00000540550.1 ENST00000323833.8 ENST00000350762.5 ENST00000550559.1 ENST00000548851.1 ENST00000434359.1 ENST00000457189.1 |
TSFM
|
Ts translation elongation factor, mitochondrial |
| chr1_+_159409512 | 0.25 |
ENST00000423932.3
|
OR10J1
|
olfactory receptor, family 10, subfamily J, member 1 |
| chr5_-_147162078 | 0.23 |
ENST00000507386.1
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2 |
| chr17_-_33446735 | 0.21 |
ENST00000460118.2
ENST00000335858.7 |
RAD51D
|
RAD51 paralog D |
| chr13_-_103719196 | 0.19 |
ENST00000245312.3
|
SLC10A2
|
solute carrier family 10 (sodium/bile acid cotransporter), member 2 |
| chr4_-_26492076 | 0.18 |
ENST00000295589.3
|
CCKAR
|
cholecystokinin A receptor |
| chr1_-_51763661 | 0.12 |
ENST00000530004.1
|
TTC39A
|
tetratricopeptide repeat domain 39A |
| chr8_+_40010989 | 0.07 |
ENST00000315792.3
|
C8orf4
|
chromosome 8 open reading frame 4 |
| chr12_-_10959892 | 0.04 |
ENST00000240615.2
|
TAS2R8
|
taste receptor, type 2, member 8 |
| chr7_-_25268104 | 0.03 |
ENST00000222674.2
|
NPVF
|
neuropeptide VF precursor |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF333
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.8 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 1.3 | 3.9 | GO:0090467 | L-arginine import(GO:0043091) arginine import(GO:0090467) L-arginine transport(GO:1902023) |
| 1.3 | 3.8 | GO:1903401 | L-lysine transmembrane transport(GO:1903401) |
| 1.1 | 3.3 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.8 | 10.9 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.7 | 1.5 | GO:0032831 | regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) |
| 0.7 | 2.1 | GO:0018874 | benzoate metabolic process(GO:0018874) butyrate metabolic process(GO:0019605) |
| 0.7 | 4.1 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.6 | 1.9 | GO:0015888 | thiamine transport(GO:0015888) thiamine transmembrane transport(GO:0071934) |
| 0.5 | 1.9 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.4 | 2.3 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.4 | 1.5 | GO:0050883 | negative regulation of sodium:proton antiporter activity(GO:0032416) gastric motility(GO:0035482) gastric emptying(GO:0035483) musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.3 | 5.3 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.3 | 1.9 | GO:0009624 | response to nematode(GO:0009624) |
| 0.3 | 1.9 | GO:0019249 | lactate biosynthetic process(GO:0019249) |
| 0.3 | 2.0 | GO:0072501 | cellular divalent inorganic anion homeostasis(GO:0072501) |
| 0.3 | 1.6 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.3 | 1.1 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.3 | 4.3 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.2 | 1.1 | GO:0048880 | sensory system development(GO:0048880) |
| 0.2 | 3.6 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.2 | 4.3 | GO:0016246 | RNA interference(GO:0016246) |
| 0.2 | 1.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.2 | 0.5 | GO:0042427 | serotonin biosynthetic process(GO:0042427) |
| 0.2 | 6.7 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
| 0.1 | 1.9 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.1 | 2.4 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.1 | 1.5 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 1.6 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.1 | 20.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 1.2 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.1 | 0.3 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.1 | 1.7 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 2.2 | GO:0002052 | positive regulation of neuroblast proliferation(GO:0002052) |
| 0.1 | 0.5 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.1 | 3.9 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 4.2 | GO:0045824 | negative regulation of innate immune response(GO:0045824) |
| 0.1 | 0.6 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 2.7 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.1 | 2.2 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.1 | 1.4 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.1 | 8.2 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 1.2 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.1 | 0.5 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 0.1 | 6.4 | GO:0061045 | negative regulation of wound healing(GO:0061045) |
| 0.1 | 2.1 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.1 | 0.2 | GO:0038188 | cholecystokinin signaling pathway(GO:0038188) |
| 0.1 | 10.2 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 5.5 | GO:0045814 | negative regulation of gene expression, epigenetic(GO:0045814) |
| 0.0 | 4.6 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 1.6 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.9 | GO:0046697 | decidualization(GO:0046697) |
| 0.0 | 0.2 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.9 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.0 | 0.3 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 1.2 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.7 | GO:1902895 | positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.0 | 0.6 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.4 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.9 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.9 | GO:0007595 | lactation(GO:0007595) |
| 0.0 | 0.4 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 2.7 | GO:0006941 | striated muscle contraction(GO:0006941) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 10.9 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.8 | 4.5 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.4 | 3.3 | GO:1990812 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.3 | 5.8 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 0.9 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.2 | 4.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 0.7 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.1 | 1.6 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 0.9 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.1 | 1.6 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.1 | 9.0 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.8 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 1.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 3.9 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 4.3 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.2 | GO:0033063 | DNA recombinase mediator complex(GO:0033061) Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 1.1 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 7.6 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 1.9 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 1.5 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 3.8 | GO:0031301 | integral component of organelle membrane(GO:0031301) |
| 0.0 | 1.9 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 1.7 | GO:0043204 | perikaryon(GO:0043204) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.3 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 1.3 | 3.8 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.8 | 2.3 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.7 | 2.7 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.6 | 1.9 | GO:0015403 | thiamine transmembrane transporter activity(GO:0015234) thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.5 | 1.6 | GO:0042806 | fucose binding(GO:0042806) |
| 0.5 | 2.4 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.4 | 2.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.3 | 5.2 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.3 | 8.2 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.3 | 1.7 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.3 | 3.9 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.3 | 4.3 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.3 | 1.9 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 0.3 | 3.6 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.2 | 1.2 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.2 | 2.1 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.2 | 1.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.2 | 1.5 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.2 | 5.8 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.2 | 3.3 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.2 | 0.6 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.1 | 1.6 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.1 | 1.5 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 2.2 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.1 | 0.6 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.1 | 1.2 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 4.6 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 0.3 | GO:0035276 | aldehyde oxidase activity(GO:0004031) ethanol binding(GO:0035276) |
| 0.1 | 3.9 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 1.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.5 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 7.6 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.1 | 0.2 | GO:0004951 | cholecystokinin receptor activity(GO:0004951) |
| 0.1 | 6.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.9 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.1 | 2.0 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 1.5 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.6 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.0 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 1.5 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.4 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.5 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.7 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 1.9 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 8.4 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.9 | GO:0043394 | proteoglycan binding(GO:0043394) |
| 0.0 | 2.2 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 17.3 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 1.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.9 | GO:0005125 | cytokine activity(GO:0005125) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 6.0 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 1.5 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 8.4 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 3.6 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 3.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.1 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 3.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 3.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 4.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 3.6 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 4.3 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 3.9 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 0.9 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 1.1 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 1.2 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.1 | 1.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 5.8 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 1.7 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.5 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.6 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.4 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 1.5 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 1.1 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 2.1 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.5 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 2.1 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.6 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |