Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
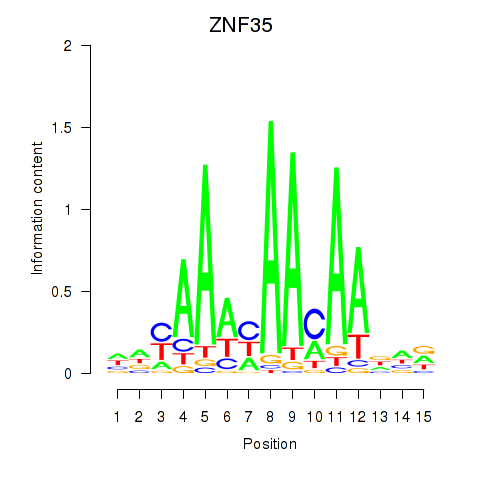
Results for ZNF35
Z-value: 0.70
Transcription factors associated with ZNF35
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF35
|
ENSG00000169981.6 | zinc finger protein 35 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF35 | hg19_v2_chr3_+_44690211_44690267 | 0.60 | 2.5e-22 | Click! |
Activity profile of ZNF35 motif
Sorted Z-values of ZNF35 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_74240610 | 19.75 |
ENST00000578092.1
ENST00000578613.1 ENST00000583578.1 |
LINC00908
|
long intergenic non-protein coding RNA 908 |
| chr13_-_41240717 | 5.70 |
ENST00000379561.5
|
FOXO1
|
forkhead box O1 |
| chr1_+_220701456 | 4.40 |
ENST00000366918.4
ENST00000402574.1 |
MARK1
|
MAP/microtubule affinity-regulating kinase 1 |
| chr14_-_31889782 | 4.23 |
ENST00000543095.2
|
HEATR5A
|
HEAT repeat containing 5A |
| chr4_+_88754069 | 4.08 |
ENST00000395102.4
ENST00000497649.2 |
MEPE
|
matrix extracellular phosphoglycoprotein |
| chr12_+_25205446 | 3.58 |
ENST00000557489.1
ENST00000354454.3 ENST00000536173.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr12_+_25205666 | 3.35 |
ENST00000547044.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr13_-_36429763 | 3.30 |
ENST00000379893.1
|
DCLK1
|
doublecortin-like kinase 1 |
| chr5_-_39203093 | 3.27 |
ENST00000515010.1
|
FYB
|
FYN binding protein |
| chr10_-_52645379 | 3.09 |
ENST00000395489.2
|
A1CF
|
APOBEC1 complementation factor |
| chr12_+_51318513 | 3.07 |
ENST00000332160.4
|
METTL7A
|
methyltransferase like 7A |
| chr4_+_88754113 | 3.02 |
ENST00000560249.1
ENST00000540395.1 ENST00000511670.1 ENST00000361056.3 |
MEPE
|
matrix extracellular phosphoglycoprotein |
| chr12_+_4382917 | 2.58 |
ENST00000261254.3
|
CCND2
|
cyclin D2 |
| chr10_-_52645416 | 2.55 |
ENST00000374001.2
ENST00000373997.3 ENST00000373995.3 ENST00000282641.2 ENST00000395495.1 ENST00000414883.1 |
A1CF
|
APOBEC1 complementation factor |
| chr19_-_13617247 | 2.53 |
ENST00000573710.2
|
CACNA1A
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
| chr2_+_166150541 | 2.53 |
ENST00000283256.6
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chr3_+_29323043 | 2.45 |
ENST00000452462.1
ENST00000456853.1 |
RBMS3
|
RNA binding motif, single stranded interacting protein 3 |
| chr1_-_12677714 | 2.20 |
ENST00000376223.2
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr21_-_42219065 | 2.04 |
ENST00000400454.1
|
DSCAM
|
Down syndrome cell adhesion molecule |
| chr4_+_68424434 | 2.04 |
ENST00000265404.2
ENST00000396225.1 |
STAP1
|
signal transducing adaptor family member 1 |
| chr6_-_133084580 | 1.98 |
ENST00000525270.1
ENST00000530536.1 ENST00000524919.1 |
VNN2
|
vanin 2 |
| chr8_+_85095769 | 1.98 |
ENST00000518566.1
|
RALYL
|
RALY RNA binding protein-like |
| chr12_+_79258444 | 1.96 |
ENST00000261205.4
|
SYT1
|
synaptotagmin I |
| chr12_+_54891495 | 1.95 |
ENST00000293373.6
|
NCKAP1L
|
NCK-associated protein 1-like |
| chr7_+_74072288 | 1.83 |
ENST00000443166.1
|
GTF2I
|
general transcription factor IIi |
| chr17_-_58042075 | 1.75 |
ENST00000305783.8
ENST00000589113.1 ENST00000442346.2 |
RNFT1
|
ring finger protein, transmembrane 1 |
| chrX_-_138724677 | 1.73 |
ENST00000370573.4
ENST00000338585.6 ENST00000370576.4 |
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr12_-_102872317 | 1.70 |
ENST00000424202.2
|
IGF1
|
insulin-like growth factor 1 (somatomedin C) |
| chr4_-_89744365 | 1.64 |
ENST00000513837.1
ENST00000503556.1 |
FAM13A
|
family with sequence similarity 13, member A |
| chr5_-_58571935 | 1.62 |
ENST00000503258.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr7_+_27282319 | 1.49 |
ENST00000222761.3
|
EVX1
|
even-skipped homeobox 1 |
| chr6_-_135271219 | 1.43 |
ENST00000367847.2
ENST00000367845.2 |
ALDH8A1
|
aldehyde dehydrogenase 8 family, member A1 |
| chr12_-_22487618 | 1.39 |
ENST00000404299.3
ENST00000396037.4 |
ST8SIA1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 |
| chr15_+_35270552 | 1.38 |
ENST00000391457.2
|
AC114546.1
|
HCG37415; PRO1914; Uncharacterized protein |
| chrM_+_4431 | 1.32 |
ENST00000361453.3
|
MT-ND2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr7_+_123488124 | 1.24 |
ENST00000476325.1
|
HYAL4
|
hyaluronoglucosaminidase 4 |
| chr3_-_148939835 | 1.16 |
ENST00000264613.6
|
CP
|
ceruloplasmin (ferroxidase) |
| chr1_+_214161272 | 1.16 |
ENST00000498508.2
ENST00000366958.4 |
PROX1
|
prospero homeobox 1 |
| chr2_+_177015122 | 1.15 |
ENST00000468418.3
|
HOXD3
|
homeobox D3 |
| chr1_-_169599353 | 1.13 |
ENST00000367793.2
ENST00000367794.2 ENST00000367792.2 ENST00000367791.2 ENST00000367788.2 |
SELP
|
selectin P (granule membrane protein 140kDa, antigen CD62) |
| chr7_-_87342564 | 1.03 |
ENST00000265724.3
ENST00000416177.1 |
ABCB1
|
ATP-binding cassette, sub-family B (MDR/TAP), member 1 |
| chr17_-_42295870 | 0.98 |
ENST00000526094.1
ENST00000529383.1 ENST00000530828.1 |
UBTF
|
upstream binding transcription factor, RNA polymerase I |
| chr3_+_69985792 | 0.77 |
ENST00000531774.1
|
MITF
|
microphthalmia-associated transcription factor |
| chr12_-_11091862 | 0.67 |
ENST00000537503.1
|
TAS2R14
|
taste receptor, type 2, member 14 |
| chr13_-_53422640 | 0.62 |
ENST00000338862.4
ENST00000377942.3 |
PCDH8
|
protocadherin 8 |
| chr14_-_92198403 | 0.62 |
ENST00000553329.1
ENST00000256343.3 |
CATSPERB
|
catsper channel auxiliary subunit beta |
| chr7_+_80231466 | 0.61 |
ENST00000309881.7
ENST00000534394.1 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr13_+_109248500 | 0.61 |
ENST00000356711.2
|
MYO16
|
myosin XVI |
| chr18_+_39535152 | 0.45 |
ENST00000262039.4
ENST00000398870.3 ENST00000586545.1 |
PIK3C3
|
phosphatidylinositol 3-kinase, catalytic subunit type 3 |
| chr20_+_56964169 | 0.42 |
ENST00000475243.1
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr13_-_47471155 | 0.29 |
ENST00000543956.1
ENST00000542664.1 |
HTR2A
|
5-hydroxytryptamine (serotonin) receptor 2A, G protein-coupled |
| chrX_+_9431324 | 0.25 |
ENST00000407597.2
ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X
|
transducin (beta)-like 1X-linked |
| chr17_+_41561317 | 0.22 |
ENST00000540306.1
ENST00000262415.3 ENST00000605777.1 |
DHX8
|
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
| chr4_+_70796784 | 0.16 |
ENST00000246891.4
ENST00000444405.3 |
CSN1S1
|
casein alpha s1 |
| chr3_-_58523010 | 0.13 |
ENST00000459701.2
ENST00000302819.5 |
ACOX2
|
acyl-CoA oxidase 2, branched chain |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF35
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 5.7 | GO:0035947 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) response to fluoride(GO:1902617) |
| 0.9 | 5.6 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.7 | 2.0 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) |
| 0.7 | 2.0 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.5 | 1.5 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.5 | 2.0 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) actin polymerization-dependent cell motility(GO:0070358) |
| 0.4 | 1.2 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.4 | 1.8 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.3 | 1.7 | GO:1904073 | trophectodermal cell proliferation(GO:0001834) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.3 | 7.1 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.2 | 1.1 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.2 | 2.0 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.2 | 2.5 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.2 | 1.6 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.2 | 1.0 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.2 | 2.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.2 | 2.0 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.1 | 1.4 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 2.6 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.1 | 3.3 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.1 | 0.4 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.6 | GO:0072564 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.1 | 1.1 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.1 | 1.0 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) |
| 0.1 | 2.7 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.3 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.1 | 7.5 | GO:0007338 | single fertilization(GO:0007338) |
| 0.1 | 1.4 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 0.4 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 2.5 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 1.2 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 1.2 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.0 | 4.4 | GO:0030010 | establishment of cell polarity(GO:0030010) |
| 0.0 | 0.7 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 1.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 2.4 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 1.7 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.6 | GO:0001756 | somitogenesis(GO:0001756) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.6 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.4 | 2.6 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.4 | 2.0 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.2 | 1.7 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.2 | 1.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.1 | 0.4 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 2.5 | GO:0033270 | voltage-gated sodium channel complex(GO:0001518) paranode region of axon(GO:0033270) |
| 0.1 | 2.0 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 2.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 0.6 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.1 | 6.9 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.1 | 4.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.3 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 3.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.6 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 1.3 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 8.3 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.1 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 2.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 3.3 | GO:0099572 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 2.0 | GO:0030426 | growth cone(GO:0030426) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 7.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.4 | 2.0 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.4 | 2.0 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.4 | 1.1 | GO:0042806 | fucose binding(GO:0042806) |
| 0.3 | 2.0 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.3 | 2.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.2 | 1.0 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.2 | 4.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) phosphatidic acid binding(GO:0070300) |
| 0.2 | 1.4 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.2 | 5.7 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 1.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 1.0 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.1 | 0.4 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 1.4 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.1 | 1.2 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 2.5 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 1.2 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.1 | 2.5 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 0.6 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 1.6 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.1 | 0.7 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 5.6 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 1.7 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 1.8 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.1 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.0 | 1.3 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 2.4 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.3 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 2.0 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.0 | 1.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 3.3 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 3.1 | GO:0008168 | methyltransferase activity(GO:0008168) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 2.6 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 2.0 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 1.7 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 3.3 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 1.7 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 7.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 2.2 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.1 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 2.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 2.0 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.7 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.7 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 2.0 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 2.0 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 3.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 1.0 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 2.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.2 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 2.6 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 1.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.0 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 1.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 2.5 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.0 | 0.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 1.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |