Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
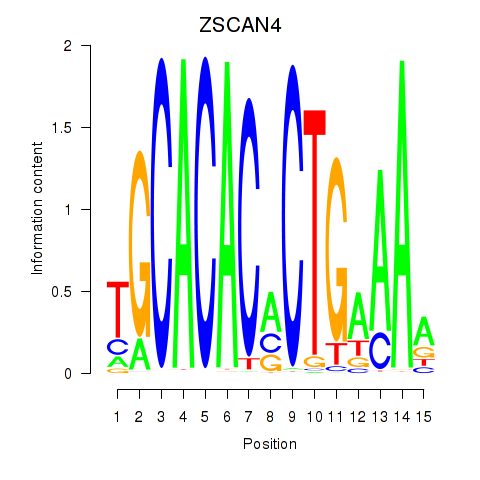
Results for ZSCAN4
Z-value: 0.08
Transcription factors associated with ZSCAN4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZSCAN4
|
ENSG00000180532.6 | zinc finger and SCAN domain containing 4 |
Activity profile of ZSCAN4 motif
Sorted Z-values of ZSCAN4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_58572760 | 11.54 |
ENST00000447756.2
|
FAM107A
|
family with sequence similarity 107, member A |
| chr16_+_23847267 | 9.73 |
ENST00000321728.7
|
PRKCB
|
protein kinase C, beta |
| chr1_+_2036149 | 8.22 |
ENST00000482686.1
ENST00000400920.1 ENST00000486681.1 |
PRKCZ
|
protein kinase C, zeta |
| chr14_-_21493884 | 7.68 |
ENST00000556974.1
ENST00000554419.1 ENST00000298687.5 ENST00000397858.1 ENST00000360463.3 ENST00000350792.3 ENST00000397847.2 |
NDRG2
|
NDRG family member 2 |
| chr14_-_21493649 | 7.68 |
ENST00000553442.1
ENST00000555869.1 ENST00000556457.1 ENST00000397844.2 ENST00000554415.1 |
NDRG2
|
NDRG family member 2 |
| chr1_-_161600990 | 7.12 |
ENST00000531221.1
|
FCGR3B
|
Fc fragment of IgG, low affinity IIIb, receptor (CD16b) |
| chr1_-_154580616 | 7.02 |
ENST00000368474.4
|
ADAR
|
adenosine deaminase, RNA-specific |
| chr8_+_85095769 | 6.18 |
ENST00000518566.1
|
RALYL
|
RALY RNA binding protein-like |
| chr1_-_177133818 | 6.17 |
ENST00000424564.2
ENST00000361833.2 |
ASTN1
|
astrotactin 1 |
| chr17_+_2699697 | 5.76 |
ENST00000254695.8
ENST00000366401.4 ENST00000542807.1 |
RAP1GAP2
|
RAP1 GTPase activating protein 2 |
| chr10_+_80828774 | 5.73 |
ENST00000334512.5
|
ZMIZ1
|
zinc finger, MIZ-type containing 1 |
| chr1_-_21978312 | 5.65 |
ENST00000359708.4
ENST00000290101.4 |
RAP1GAP
|
RAP1 GTPase activating protein |
| chr1_-_161600942 | 5.58 |
ENST00000421702.2
|
FCGR3B
|
Fc fragment of IgG, low affinity IIIb, receptor (CD16b) |
| chr6_-_41909561 | 5.33 |
ENST00000372991.4
|
CCND3
|
cyclin D3 |
| chr1_-_161519682 | 5.32 |
ENST00000367969.3
ENST00000443193.1 |
FCGR3A
|
Fc fragment of IgG, low affinity IIIa, receptor (CD16a) |
| chr7_-_10979750 | 5.32 |
ENST00000339600.5
|
NDUFA4
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4, 9kDa |
| chr16_+_4674814 | 5.27 |
ENST00000415496.1
ENST00000587747.1 ENST00000399577.5 ENST00000588994.1 ENST00000586183.1 |
MGRN1
|
mahogunin ring finger 1, E3 ubiquitin protein ligase |
| chr6_-_41909466 | 5.26 |
ENST00000414200.2
|
CCND3
|
cyclin D3 |
| chr16_+_4674787 | 5.23 |
ENST00000262370.7
|
MGRN1
|
mahogunin ring finger 1, E3 ubiquitin protein ligase |
| chr1_-_161600822 | 5.20 |
ENST00000534776.1
ENST00000540048.1 |
FCGR3B
FCGR3A
|
Fc fragment of IgG, low affinity IIIb, receptor (CD16b) Fc fragment of IgG, low affinity IIIa, receptor (CD16a) |
| chr1_+_233765353 | 5.04 |
ENST00000366620.1
|
KCNK1
|
potassium channel, subfamily K, member 1 |
| chr1_-_161519579 | 4.98 |
ENST00000426740.1
|
FCGR3A
|
Fc fragment of IgG, low affinity IIIa, receptor (CD16a) |
| chr17_+_72462525 | 4.83 |
ENST00000360141.3
|
CD300A
|
CD300a molecule |
| chr8_-_134115118 | 4.78 |
ENST00000395352.3
ENST00000338087.5 |
SLA
|
Src-like-adaptor |
| chr8_+_85095497 | 4.60 |
ENST00000522455.1
ENST00000521695.1 |
RALYL
|
RALY RNA binding protein-like |
| chr16_+_6069664 | 4.34 |
ENST00000422070.4
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr11_-_33774944 | 4.17 |
ENST00000532057.1
ENST00000531080.1 |
FBXO3
|
F-box protein 3 |
| chr8_+_85095553 | 3.65 |
ENST00000521268.1
|
RALYL
|
RALY RNA binding protein-like |
| chr19_+_14491948 | 3.65 |
ENST00000358600.3
|
CD97
|
CD97 molecule |
| chr12_-_54694807 | 3.37 |
ENST00000435572.2
|
NFE2
|
nuclear factor, erythroid 2 |
| chr16_+_16472912 | 3.32 |
ENST00000530217.2
|
NPIPA7
|
nuclear pore complex interacting protein family, member A7 |
| chr19_+_571277 | 3.22 |
ENST00000346916.4
ENST00000545507.2 |
BSG
|
basigin (Ok blood group) |
| chr4_+_74718906 | 3.09 |
ENST00000226524.3
|
PF4V1
|
platelet factor 4 variant 1 |
| chr1_+_226736446 | 3.08 |
ENST00000366788.3
ENST00000366789.4 |
C1orf95
|
chromosome 1 open reading frame 95 |
| chr16_+_56385290 | 3.01 |
ENST00000564727.1
|
GNAO1
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O |
| chr2_-_160473114 | 2.90 |
ENST00000392783.2
|
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B |
| chr3_+_52009110 | 2.83 |
ENST00000491470.1
|
ABHD14A
|
abhydrolase domain containing 14A |
| chr19_+_30863271 | 2.71 |
ENST00000355537.3
|
ZNF536
|
zinc finger protein 536 |
| chr17_-_34207295 | 2.58 |
ENST00000463941.1
ENST00000293272.3 |
CCL5
|
chemokine (C-C motif) ligand 5 |
| chr19_-_3801789 | 2.54 |
ENST00000590849.1
ENST00000395045.2 |
MATK
|
megakaryocyte-associated tyrosine kinase |
| chr8_+_26247878 | 2.49 |
ENST00000518611.1
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr7_-_99063769 | 2.42 |
ENST00000394186.3
ENST00000359832.4 ENST00000449683.1 ENST00000488775.1 ENST00000523680.1 ENST00000292475.3 ENST00000430982.1 ENST00000555673.1 ENST00000413834.1 |
ATP5J2
PTCD1
ATP5J2-PTCD1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F2 pentatricopeptide repeat domain 1 ATP5J2-PTCD1 readthrough |
| chr19_-_17516449 | 2.41 |
ENST00000252593.6
|
BST2
|
bone marrow stromal cell antigen 2 |
| chr3_-_53290016 | 2.18 |
ENST00000423525.2
ENST00000423516.1 ENST00000296289.6 ENST00000462138.1 |
TKT
|
transketolase |
| chr13_-_79980315 | 2.16 |
ENST00000438737.2
|
RBM26
|
RNA binding motif protein 26 |
| chr10_+_18549645 | 2.12 |
ENST00000396576.2
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr3_+_50284321 | 2.10 |
ENST00000451956.1
|
GNAI2
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2 |
| chr8_+_107670064 | 2.08 |
ENST00000312046.6
|
OXR1
|
oxidation resistance 1 |
| chr17_-_56406117 | 2.04 |
ENST00000268893.6
ENST00000355701.3 |
BZRAP1
|
benzodiazepine receptor (peripheral) associated protein 1 |
| chr3_+_130569592 | 2.02 |
ENST00000533801.2
|
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr9_+_103790991 | 1.92 |
ENST00000374874.3
|
LPPR1
|
Lipid phosphate phosphatase-related protein type 1 |
| chr22_-_24384260 | 1.90 |
ENST00000248935.5
|
GSTT1
|
glutathione S-transferase theta 1 |
| chr2_+_237994519 | 1.76 |
ENST00000392008.2
ENST00000409334.1 ENST00000409629.1 |
COPS8
|
COP9 signalosome subunit 8 |
| chr1_+_209929377 | 1.73 |
ENST00000400959.3
ENST00000367025.3 |
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr11_+_61717842 | 1.61 |
ENST00000449131.2
|
BEST1
|
bestrophin 1 |
| chr15_+_78632666 | 1.59 |
ENST00000299529.6
|
CRABP1
|
cellular retinoic acid binding protein 1 |
| chr14_+_53173910 | 1.58 |
ENST00000606149.1
ENST00000555339.1 ENST00000556813.1 |
PSMC6
|
proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
| chr14_+_53173890 | 1.56 |
ENST00000445930.2
|
PSMC6
|
proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
| chr4_-_103682071 | 1.56 |
ENST00000505239.1
|
MANBA
|
mannosidase, beta A, lysosomal |
| chr5_+_176784837 | 1.53 |
ENST00000408923.3
|
RGS14
|
regulator of G-protein signaling 14 |
| chr17_+_63133587 | 1.52 |
ENST00000449996.3
ENST00000262406.9 |
RGS9
|
regulator of G-protein signaling 9 |
| chr19_+_17516624 | 1.51 |
ENST00000596322.1
ENST00000600008.1 ENST00000601885.1 |
CTD-2521M24.9
|
CTD-2521M24.9 |
| chr19_+_17516531 | 1.43 |
ENST00000528911.1
ENST00000528604.1 ENST00000595892.1 ENST00000500836.2 ENST00000598546.1 ENST00000600369.1 ENST00000598356.1 ENST00000594426.1 |
MVB12A
CTD-2521M24.9
|
multivesicular body subunit 12A CTD-2521M24.9 |
| chr16_+_16481306 | 1.41 |
ENST00000422673.2
|
NPIPA7
|
nuclear pore complex interacting protein family, member A7 |
| chr5_+_61602055 | 1.40 |
ENST00000381103.2
|
KIF2A
|
kinesin heavy chain member 2A |
| chr11_-_75062730 | 1.39 |
ENST00000420843.2
ENST00000360025.3 |
ARRB1
|
arrestin, beta 1 |
| chr11_+_111807863 | 1.36 |
ENST00000440460.2
|
DIXDC1
|
DIX domain containing 1 |
| chr22_+_18121356 | 1.34 |
ENST00000317582.5
ENST00000543133.1 ENST00000538149.1 ENST00000337612.5 ENST00000493680.1 |
BCL2L13
|
BCL2-like 13 (apoptosis facilitator) |
| chr1_+_209929494 | 1.33 |
ENST00000367026.3
|
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr12_-_96390108 | 1.29 |
ENST00000538703.1
ENST00000261208.3 |
HAL
|
histidine ammonia-lyase |
| chr19_-_38714847 | 1.27 |
ENST00000420980.2
ENST00000355526.4 |
DPF1
|
D4, zinc and double PHD fingers family 1 |
| chr1_+_231376941 | 1.20 |
ENST00000436239.1
ENST00000366647.4 ENST00000366646.3 ENST00000416000.1 |
GNPAT
|
glyceronephosphate O-acyltransferase |
| chr15_+_63414017 | 1.19 |
ENST00000413507.2
|
LACTB
|
lactamase, beta |
| chr19_+_17516494 | 1.12 |
ENST00000534306.1
|
CTD-2521M24.9
|
CTD-2521M24.9 |
| chr1_+_94883931 | 1.09 |
ENST00000394233.2
ENST00000454898.2 ENST00000536817.1 |
ABCD3
|
ATP-binding cassette, sub-family D (ALD), member 3 |
| chr2_-_101034070 | 1.08 |
ENST00000264249.3
|
CHST10
|
carbohydrate sulfotransferase 10 |
| chr1_+_104615595 | 1.06 |
ENST00000418362.1
|
RP11-364B6.1
|
RP11-364B6.1 |
| chr21_-_46221684 | 1.06 |
ENST00000330942.5
|
UBE2G2
|
ubiquitin-conjugating enzyme E2G 2 |
| chr12_-_96390063 | 1.04 |
ENST00000541929.1
|
HAL
|
histidine ammonia-lyase |
| chr1_+_94883991 | 1.03 |
ENST00000370214.4
|
ABCD3
|
ATP-binding cassette, sub-family D (ALD), member 3 |
| chr4_-_103682145 | 1.02 |
ENST00000226578.4
|
MANBA
|
mannosidase, beta A, lysosomal |
| chr16_+_15737124 | 0.97 |
ENST00000396355.1
ENST00000396353.2 |
NDE1
|
nudE neurodevelopment protein 1 |
| chr19_-_10530784 | 0.95 |
ENST00000593124.1
|
CDC37
|
cell division cycle 37 |
| chr20_-_21494654 | 0.92 |
ENST00000377142.4
|
NKX2-2
|
NK2 homeobox 2 |
| chr12_-_10022735 | 0.92 |
ENST00000228438.2
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr5_-_82969405 | 0.90 |
ENST00000510978.1
|
HAPLN1
|
hyaluronan and proteoglycan link protein 1 |
| chrX_+_1387693 | 0.89 |
ENST00000381529.3
ENST00000432318.2 ENST00000361536.3 ENST00000501036.2 ENST00000381524.3 ENST00000412290.1 |
CSF2RA
|
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
| chr11_-_77791156 | 0.86 |
ENST00000281031.4
|
NDUFC2
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2, 14.5kDa |
| chr11_+_61717535 | 0.83 |
ENST00000534553.1
ENST00000301774.9 |
BEST1
|
bestrophin 1 |
| chr12_+_122242597 | 0.80 |
ENST00000267197.5
|
SETD1B
|
SET domain containing 1B |
| chr1_-_110283138 | 0.70 |
ENST00000256594.3
|
GSTM3
|
glutathione S-transferase mu 3 (brain) |
| chr11_-_4414880 | 0.70 |
ENST00000254436.7
ENST00000543625.1 |
TRIM21
|
tripartite motif containing 21 |
| chr10_-_62332357 | 0.69 |
ENST00000503366.1
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr10_-_101989315 | 0.69 |
ENST00000370397.7
|
CHUK
|
conserved helix-loop-helix ubiquitous kinase |
| chr15_+_28623784 | 0.67 |
ENST00000526619.2
ENST00000337838.7 ENST00000532622.2 |
GOLGA8F
|
golgin A8 family, member F |
| chr2_-_175462934 | 0.63 |
ENST00000392546.2
ENST00000436221.1 |
WIPF1
|
WAS/WASL interacting protein family, member 1 |
| chr11_+_120039685 | 0.62 |
ENST00000530303.1
ENST00000319763.1 |
AP000679.2
|
Uncharacterized protein |
| chr19_+_36426452 | 0.55 |
ENST00000588831.1
|
LRFN3
|
leucine rich repeat and fibronectin type III domain containing 3 |
| chr6_-_34113856 | 0.51 |
ENST00000538487.2
|
GRM4
|
glutamate receptor, metabotropic 4 |
| chr9_+_125137565 | 0.50 |
ENST00000373698.5
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr4_-_152682129 | 0.49 |
ENST00000512306.1
ENST00000508611.1 ENST00000515812.1 ENST00000263985.6 |
PET112
|
PET112 homolog (yeast) |
| chr2_-_99952769 | 0.48 |
ENST00000409434.1
ENST00000434323.1 ENST00000264255.3 |
TXNDC9
|
thioredoxin domain containing 9 |
| chr10_+_133753533 | 0.44 |
ENST00000422256.2
|
PPP2R2D
|
protein phosphatase 2, regulatory subunit B, delta |
| chr6_-_42690312 | 0.42 |
ENST00000230381.5
|
PRPH2
|
peripherin 2 (retinal degeneration, slow) |
| chr3_+_130569429 | 0.40 |
ENST00000505330.1
ENST00000504381.1 ENST00000507488.2 ENST00000393221.4 |
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr1_+_159141397 | 0.38 |
ENST00000368124.4
ENST00000368125.4 ENST00000416746.1 |
CADM3
|
cell adhesion molecule 3 |
| chr20_-_16554078 | 0.35 |
ENST00000354981.2
ENST00000355755.3 ENST00000378003.2 ENST00000408042.1 |
KIF16B
|
kinesin family member 16B |
| chr19_-_40596767 | 0.33 |
ENST00000599972.1
ENST00000450241.2 ENST00000595687.2 |
ZNF780A
|
zinc finger protein 780A |
| chr12_-_48164812 | 0.32 |
ENST00000549151.1
ENST00000548919.1 |
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr18_+_32173276 | 0.31 |
ENST00000591816.1
ENST00000588125.1 ENST00000598334.1 ENST00000588684.1 ENST00000554864.3 ENST00000399121.5 ENST00000595022.1 ENST00000269190.7 ENST00000399097.3 |
DTNA
|
dystrobrevin, alpha |
| chrX_+_1387721 | 0.26 |
ENST00000419094.1
ENST00000381509.3 ENST00000494969.2 ENST00000355805.2 ENST00000355432.3 |
CSF2RA
|
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
| chr19_-_9649253 | 0.26 |
ENST00000593003.1
|
ZNF426
|
zinc finger protein 426 |
| chr7_-_148823387 | 0.23 |
ENST00000483014.1
ENST00000378061.2 |
ZNF425
|
zinc finger protein 425 |
| chr6_-_76203345 | 0.21 |
ENST00000393004.2
|
FILIP1
|
filamin A interacting protein 1 |
| chr6_+_114178512 | 0.21 |
ENST00000368635.4
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate |
| chr15_-_37392703 | 0.20 |
ENST00000382766.2
ENST00000444725.1 |
MEIS2
|
Meis homeobox 2 |
| chr11_+_71938925 | 0.20 |
ENST00000538751.1
|
INPPL1
|
inositol polyphosphate phosphatase-like 1 |
| chr4_-_168155730 | 0.20 |
ENST00000502330.1
ENST00000357154.3 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr19_-_40596828 | 0.20 |
ENST00000414720.2
ENST00000455521.1 ENST00000340963.5 ENST00000595773.1 |
ZNF780A
|
zinc finger protein 780A |
| chr1_-_193075180 | 0.15 |
ENST00000367440.3
|
GLRX2
|
glutaredoxin 2 |
| chr17_-_7080227 | 0.15 |
ENST00000574330.1
|
ASGR1
|
asialoglycoprotein receptor 1 |
| chr17_+_73089382 | 0.14 |
ENST00000538213.2
ENST00000584118.1 |
SLC16A5
|
solute carrier family 16 (monocarboxylate transporter), member 5 |
| chr1_-_8000872 | 0.11 |
ENST00000377507.3
|
TNFRSF9
|
tumor necrosis factor receptor superfamily, member 9 |
| chr4_-_168155700 | 0.10 |
ENST00000357545.4
ENST00000512648.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr3_+_151531810 | 0.09 |
ENST00000232892.7
|
AADAC
|
arylacetamide deacetylase |
| chr17_+_9728828 | 0.08 |
ENST00000262441.5
|
GLP2R
|
glucagon-like peptide 2 receptor |
| chr6_-_31620149 | 0.07 |
ENST00000435080.1
ENST00000375976.4 ENST00000441054.1 |
BAG6
|
BCL2-associated athanogene 6 |
| chr11_-_62783276 | 0.04 |
ENST00000535878.1
ENST00000545207.1 |
SLC22A8
|
solute carrier family 22 (organic anion transporter), member 8 |
| chr19_+_51630287 | 0.03 |
ENST00000599948.1
|
SIGLEC9
|
sialic acid binding Ig-like lectin 9 |
| chr12_+_7033616 | 0.01 |
ENST00000356654.4
|
ATN1
|
atrophin 1 |
| chr17_-_19648916 | 0.00 |
ENST00000444455.1
ENST00000439102.2 |
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZSCAN4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 9.7 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 2.3 | 7.0 | GO:1900368 | regulation of RNA interference(GO:1900368) negative regulation of RNA interference(GO:1900369) |
| 1.9 | 5.7 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 1.6 | 4.8 | GO:1905154 | negative regulation of eosinophil activation(GO:1902567) negative regulation of membrane invagination(GO:1905154) negative regulation of eosinophil migration(GO:2000417) |
| 1.4 | 8.2 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 1.0 | 15.4 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 1.0 | 5.7 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.9 | 10.5 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.8 | 2.4 | GO:1901253 | negative regulation of dendritic cell cytokine production(GO:0002731) negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.8 | 2.3 | GO:0019556 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.7 | 2.2 | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.6 | 2.6 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.6 | 5.0 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.5 | 1.6 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.5 | 2.1 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.4 | 2.5 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.4 | 2.4 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.4 | 2.4 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.4 | 1.2 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.4 | 1.4 | GO:1903772 | regulation of viral budding via host ESCRT complex(GO:1903772) |
| 0.4 | 2.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.3 | 0.7 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.3 | 0.9 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.3 | 1.5 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.3 | 5.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 17.9 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.2 | 2.7 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.2 | 10.6 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.2 | 3.2 | GO:0046689 | response to mercury ion(GO:0046689) |
| 0.2 | 6.2 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.2 | 2.4 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.2 | 0.7 | GO:0072658 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.2 | 2.6 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.2 | 1.1 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 | 2.1 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.1 | 0.7 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.1 | 2.1 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 3.4 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 1.4 | GO:0090240 | positive regulation of histone H4 acetylation(GO:0090240) |
| 0.1 | 3.1 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 1.4 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 | 0.5 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.1 | 0.7 | GO:1902739 | interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 0.1 | 1.8 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 3.1 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.1 | 0.3 | GO:0030822 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.1 | 7.3 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 10.9 | GO:0002433 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.1 | 4.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 4.5 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 1.9 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.1 | 1.0 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 1.0 | GO:0098779 | mitophagy in response to mitochondrial depolarization(GO:0098779) |
| 0.0 | 0.5 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 5.3 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.0 | 0.5 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 2.2 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.4 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.1 | GO:2001179 | regulation of interleukin-10 secretion(GO:2001179) |
| 0.0 | 0.1 | GO:1904327 | maintenance of unfolded protein(GO:0036506) protein localization to cytosolic proteasome complex(GO:1904327) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 1.4 | GO:0090307 | mitotic spindle assembly(GO:0090307) |
| 0.0 | 0.2 | GO:0042262 | DNA protection(GO:0042262) |
| 0.0 | 0.4 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.8 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.2 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.9 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 6.5 | GO:0001558 | regulation of cell growth(GO:0001558) |
| 0.0 | 1.0 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 7.0 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 1.0 | 8.2 | GO:0045179 | apical cortex(GO:0045179) |
| 1.0 | 5.0 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.3 | 4.8 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.3 | 3.1 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.2 | 1.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.2 | 2.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.2 | 10.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 8.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 2.4 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 20.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 3.2 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 15.4 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 1.4 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.1 | 0.7 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 0.8 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 7.6 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 3.3 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 4.8 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.1 | 3.4 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.1 | 0.7 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 0.7 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 1.3 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 21.9 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 13.9 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.8 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 4.9 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 2.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 9.4 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 6.8 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 3.7 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 1.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.1 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.2 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.0 | 1.1 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.7 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 4.7 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 9.7 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 1.8 | 7.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 1.6 | 28.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.7 | 2.2 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.6 | 2.3 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.5 | 5.0 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.5 | 1.4 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.4 | 8.2 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.4 | 4.8 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.4 | 2.6 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.3 | 2.7 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.3 | 2.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.3 | 5.8 | GO:0005537 | mannose binding(GO:0005537) |
| 0.3 | 3.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.3 | 3.0 | GO:0051430 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.3 | 2.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.2 | 3.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.2 | 10.6 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.2 | 0.7 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.2 | 4.8 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 2.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 0.5 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 1.2 | GO:0016413 | palmitoyl-CoA hydrolase activity(GO:0016290) O-acetyltransferase activity(GO:0016413) |
| 0.1 | 1.6 | GO:0016918 | retinal binding(GO:0016918) |
| 0.1 | 0.5 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 1.5 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 2.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 7.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 2.1 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 2.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 3.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 1.3 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.1 | 1.9 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 1.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.7 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 2.4 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.8 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 1.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.6 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 11.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.2 | GO:0030613 | oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) |
| 0.0 | 0.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 7.6 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 1.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.2 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.0 | 0.1 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 1.0 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.9 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 4.3 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.9 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 1.5 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 2.7 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.5 | GO:0008066 | glutamate receptor activity(GO:0008066) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 15.5 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.4 | 9.7 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.3 | 5.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.2 | 8.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.2 | 10.6 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 17.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 1.5 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 1.2 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.1 | 3.2 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 5.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 2.5 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.4 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.7 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 4.8 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.2 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.0 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.6 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 5.0 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.3 | 9.7 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.3 | 8.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.3 | 11.7 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.2 | 7.2 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.2 | 3.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 14.0 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 8.3 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 3.2 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.1 | 2.4 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 2.1 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 7.0 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 6.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.4 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.1 | 0.7 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 1.0 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.1 | 1.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 3.1 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 2.4 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 2.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.2 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 3.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 5.9 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 2.2 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 3.4 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.4 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.9 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.5 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 2.5 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.0 | 0.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.5 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |