Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
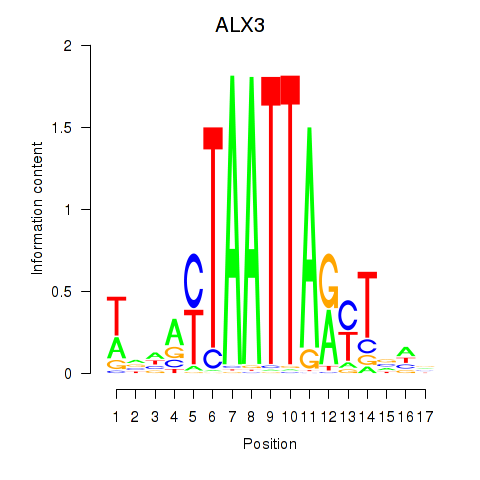
Results for ALX3
Z-value: 0.61
Transcription factors associated with ALX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ALX3
|
ENSG00000156150.6 | ALX homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ALX3 | hg19_v2_chr1_-_110613276_110613322 | 0.01 | 8.9e-01 | Click! |
Activity profile of ALX3 motif
Sorted Z-values of ALX3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_25865159 | 8.36 |
ENST00000502949.1
ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr2_-_89160770 | 6.39 |
ENST00000390240.2
|
IGKJ3
|
immunoglobulin kappa joining 3 |
| chr10_+_5135981 | 6.28 |
ENST00000380554.3
|
AKR1C3
|
aldo-keto reductase family 1, member C3 |
| chr4_+_113568207 | 5.96 |
ENST00000511529.1
|
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr2_+_90077680 | 5.75 |
ENST00000390270.2
|
IGKV3D-20
|
immunoglobulin kappa variable 3D-20 |
| chr16_-_28634874 | 5.64 |
ENST00000395609.1
ENST00000350842.4 |
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr2_-_89385283 | 5.38 |
ENST00000390252.2
|
IGKV3-15
|
immunoglobulin kappa variable 3-15 |
| chr11_-_63376013 | 5.34 |
ENST00000540943.1
|
PLA2G16
|
phospholipase A2, group XVI |
| chr4_-_103749205 | 5.27 |
ENST00000508249.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr4_-_103749179 | 4.43 |
ENST00000502690.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr2_+_90273679 | 4.42 |
ENST00000423080.2
|
IGKV3D-7
|
immunoglobulin kappa variable 3D-7 |
| chr15_-_20193370 | 4.39 |
ENST00000558565.2
|
IGHV3OR15-7
|
immunoglobulin heavy variable 3/OR15-7 (pseudogene) |
| chr14_-_107049312 | 4.33 |
ENST00000390627.2
|
IGHV3-53
|
immunoglobulin heavy variable 3-53 |
| chr16_+_12059091 | 4.12 |
ENST00000562385.1
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr16_-_28608364 | 4.02 |
ENST00000533150.1
|
SULT1A2
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 |
| chr3_-_141747950 | 4.02 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr2_+_90211643 | 3.83 |
ENST00000390277.2
|
IGKV3D-11
|
immunoglobulin kappa variable 3D-11 |
| chr11_-_71823796 | 3.81 |
ENST00000545680.1
ENST00000543587.1 ENST00000538393.1 ENST00000535234.1 ENST00000227618.4 ENST00000535503.1 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chr1_+_28261533 | 3.76 |
ENST00000411604.1
ENST00000373888.4 |
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr11_-_71823715 | 3.73 |
ENST00000545944.1
ENST00000502597.2 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chr11_-_71823266 | 3.61 |
ENST00000538919.1
ENST00000539395.1 ENST00000542531.1 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chr16_+_15489603 | 3.61 |
ENST00000568766.1
ENST00000287594.7 |
RP11-1021N1.1
MPV17L
|
Uncharacterized protein MPV17 mitochondrial membrane protein-like |
| chr14_-_78083112 | 3.56 |
ENST00000216484.2
|
SPTLC2
|
serine palmitoyltransferase, long chain base subunit 2 |
| chr16_-_28621298 | 3.51 |
ENST00000566189.1
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr11_+_71900572 | 3.42 |
ENST00000312293.4
|
FOLR1
|
folate receptor 1 (adult) |
| chr11_+_71900703 | 3.38 |
ENST00000393681.2
|
FOLR1
|
folate receptor 1 (adult) |
| chr2_+_68961934 | 3.30 |
ENST00000409202.3
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr16_-_28621312 | 3.23 |
ENST00000314752.7
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr2_+_68961905 | 3.18 |
ENST00000295381.3
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr2_+_68962014 | 3.06 |
ENST00000467265.1
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr16_+_12059050 | 3.04 |
ENST00000396495.3
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr12_+_56435637 | 3.03 |
ENST00000356464.5
ENST00000552361.1 |
RPS26
|
ribosomal protein S26 |
| chr6_+_34204642 | 2.93 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chr6_+_160542821 | 2.83 |
ENST00000366963.4
|
SLC22A1
|
solute carrier family 22 (organic cation transporter), member 1 |
| chr16_-_28621353 | 2.82 |
ENST00000567512.1
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr1_-_24126023 | 2.81 |
ENST00000429356.1
|
GALE
|
UDP-galactose-4-epimerase |
| chr2_-_220264703 | 2.78 |
ENST00000519905.1
ENST00000523282.1 ENST00000434339.1 ENST00000457935.1 |
DNPEP
|
aspartyl aminopeptidase |
| chr2_-_136678123 | 2.67 |
ENST00000422708.1
|
DARS
|
aspartyl-tRNA synthetase |
| chr2_+_187371440 | 2.57 |
ENST00000445547.1
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr3_+_139063372 | 2.53 |
ENST00000478464.1
|
MRPS22
|
mitochondrial ribosomal protein S22 |
| chr17_-_64225508 | 2.51 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr7_-_87856280 | 2.47 |
ENST00000490437.1
ENST00000431660.1 |
SRI
|
sorcin |
| chr6_+_160542870 | 2.45 |
ENST00000324965.4
ENST00000457470.2 |
SLC22A1
|
solute carrier family 22 (organic cation transporter), member 1 |
| chr16_-_28608424 | 2.36 |
ENST00000335715.4
|
SULT1A2
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 |
| chr5_-_98262240 | 2.34 |
ENST00000284049.3
|
CHD1
|
chromodomain helicase DNA binding protein 1 |
| chr1_-_24306835 | 2.31 |
ENST00000484146.2
|
SRSF10
|
serine/arginine-rich splicing factor 10 |
| chr15_+_64680003 | 2.25 |
ENST00000261884.3
|
TRIP4
|
thyroid hormone receptor interactor 4 |
| chr10_-_95242044 | 2.25 |
ENST00000371501.4
ENST00000371502.4 ENST00000371489.1 |
MYOF
|
myoferlin |
| chr7_-_87856303 | 2.23 |
ENST00000394641.3
|
SRI
|
sorcin |
| chr6_-_36515177 | 2.20 |
ENST00000229812.7
|
STK38
|
serine/threonine kinase 38 |
| chr10_-_95241951 | 2.11 |
ENST00000358334.5
ENST00000359263.4 ENST00000371488.3 |
MYOF
|
myoferlin |
| chr16_-_55866997 | 2.09 |
ENST00000360526.3
ENST00000361503.4 |
CES1
|
carboxylesterase 1 |
| chr19_+_50016411 | 2.09 |
ENST00000426395.3
ENST00000600273.1 ENST00000599988.1 |
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr19_+_50016610 | 2.07 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr2_-_89340242 | 2.02 |
ENST00000480492.1
|
IGKV1-12
|
immunoglobulin kappa variable 1-12 |
| chr17_-_7167279 | 2.01 |
ENST00000571932.2
|
CLDN7
|
claudin 7 |
| chr14_-_106552755 | 1.90 |
ENST00000390600.2
|
IGHV3-9
|
immunoglobulin heavy variable 3-9 |
| chr16_+_72088376 | 1.88 |
ENST00000570083.1
ENST00000355906.5 ENST00000398131.2 ENST00000569639.1 ENST00000564499.1 ENST00000357763.4 ENST00000562526.1 ENST00000565574.1 ENST00000568417.2 ENST00000356967.5 |
HP
HPR
|
haptoglobin haptoglobin-related protein |
| chr19_+_36139125 | 1.87 |
ENST00000246554.3
|
COX6B1
|
cytochrome c oxidase subunit VIb polypeptide 1 (ubiquitous) |
| chr1_+_220267429 | 1.80 |
ENST00000366922.1
ENST00000302637.5 |
IARS2
|
isoleucyl-tRNA synthetase 2, mitochondrial |
| chr5_+_115177178 | 1.69 |
ENST00000316788.7
|
AP3S1
|
adaptor-related protein complex 3, sigma 1 subunit |
| chrX_-_77225135 | 1.69 |
ENST00000458128.1
|
PGAM4
|
phosphoglycerate mutase family member 4 |
| chr20_-_33735070 | 1.68 |
ENST00000374491.3
ENST00000542871.1 ENST00000374492.3 |
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chr11_-_62323702 | 1.67 |
ENST00000530285.1
|
AHNAK
|
AHNAK nucleoprotein |
| chr7_-_144435985 | 1.66 |
ENST00000549981.1
|
TPK1
|
thiamin pyrophosphokinase 1 |
| chr17_+_35851570 | 1.64 |
ENST00000394386.1
|
DUSP14
|
dual specificity phosphatase 14 |
| chr9_-_115095123 | 1.61 |
ENST00000458258.1
|
PTBP3
|
polypyrimidine tract binding protein 3 |
| chr9_+_90112741 | 1.58 |
ENST00000469640.2
|
DAPK1
|
death-associated protein kinase 1 |
| chr1_+_28261621 | 1.57 |
ENST00000549094.1
|
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr1_+_28261492 | 1.53 |
ENST00000373894.3
|
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr15_+_58702742 | 1.53 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chr3_-_141719195 | 1.53 |
ENST00000397991.4
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr20_-_43133491 | 1.52 |
ENST00000411544.1
|
SERINC3
|
serine incorporator 3 |
| chr1_-_150738261 | 1.51 |
ENST00000448301.2
ENST00000368985.3 |
CTSS
|
cathepsin S |
| chr11_+_57480046 | 1.47 |
ENST00000378312.4
ENST00000278422.4 |
TMX2
|
thioredoxin-related transmembrane protein 2 |
| chrX_+_77154935 | 1.46 |
ENST00000481445.1
|
COX7B
|
cytochrome c oxidase subunit VIIb |
| chr9_+_90112590 | 1.43 |
ENST00000472284.1
|
DAPK1
|
death-associated protein kinase 1 |
| chr11_-_327537 | 1.43 |
ENST00000602735.1
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr9_+_90112767 | 1.39 |
ENST00000408954.3
|
DAPK1
|
death-associated protein kinase 1 |
| chr1_-_54411255 | 1.36 |
ENST00000371377.3
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr12_+_104337515 | 1.33 |
ENST00000550595.1
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1 |
| chr9_+_90112117 | 1.32 |
ENST00000358077.5
|
DAPK1
|
death-associated protein kinase 1 |
| chr1_-_54411240 | 1.32 |
ENST00000371378.2
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr11_-_107729887 | 1.31 |
ENST00000525815.1
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr1_+_62439037 | 1.30 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr3_+_138340049 | 1.25 |
ENST00000464668.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr1_-_150669500 | 1.23 |
ENST00000271732.3
|
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr4_+_71859156 | 1.20 |
ENST00000286648.5
ENST00000504730.1 ENST00000504952.1 |
DCK
|
deoxycytidine kinase |
| chr14_-_67878917 | 1.18 |
ENST00000216446.4
|
PLEK2
|
pleckstrin 2 |
| chr14_-_106668095 | 1.17 |
ENST00000390606.2
|
IGHV3-20
|
immunoglobulin heavy variable 3-20 |
| chr14_-_54423529 | 1.16 |
ENST00000245451.4
ENST00000559087.1 |
BMP4
|
bone morphogenetic protein 4 |
| chr1_+_186265399 | 1.16 |
ENST00000367486.3
ENST00000367484.3 ENST00000533951.1 ENST00000367482.4 ENST00000367483.4 ENST00000367485.4 ENST00000445192.2 |
PRG4
|
proteoglycan 4 |
| chr3_+_121774202 | 1.14 |
ENST00000469710.1
ENST00000493101.1 ENST00000330540.2 ENST00000264468.5 |
CD86
|
CD86 molecule |
| chr10_-_105845674 | 1.14 |
ENST00000353479.5
ENST00000369733.3 |
COL17A1
|
collagen, type XVII, alpha 1 |
| chr3_+_186353756 | 1.13 |
ENST00000431018.1
ENST00000450521.1 ENST00000539949.1 |
FETUB
|
fetuin B |
| chr19_+_42817527 | 1.10 |
ENST00000598766.1
|
TMEM145
|
transmembrane protein 145 |
| chr4_-_103748880 | 1.03 |
ENST00000453744.2
ENST00000349311.8 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr4_-_120243545 | 1.01 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr14_+_39736582 | 0.99 |
ENST00000556148.1
ENST00000348007.3 |
CTAGE5
|
CTAGE family, member 5 |
| chr12_+_7014126 | 0.98 |
ENST00000415834.1
ENST00000436789.1 |
LRRC23
|
leucine rich repeat containing 23 |
| chr2_+_102615416 | 0.97 |
ENST00000393414.2
|
IL1R2
|
interleukin 1 receptor, type II |
| chr12_-_10282836 | 0.95 |
ENST00000304084.8
ENST00000353231.5 ENST00000525605.1 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr11_+_35201826 | 0.93 |
ENST00000531873.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr8_-_91095099 | 0.92 |
ENST00000265431.3
|
CALB1
|
calbindin 1, 28kDa |
| chr12_+_28410128 | 0.92 |
ENST00000381259.1
ENST00000381256.1 |
CCDC91
|
coiled-coil domain containing 91 |
| chr12_-_122985067 | 0.89 |
ENST00000540586.1
ENST00000543897.1 |
ZCCHC8
|
zinc finger, CCHC domain containing 8 |
| chr15_+_58724184 | 0.88 |
ENST00000433326.2
|
LIPC
|
lipase, hepatic |
| chr2_+_217524323 | 0.87 |
ENST00000456764.1
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa |
| chr6_-_7313381 | 0.87 |
ENST00000489567.1
ENST00000479365.1 ENST00000462112.1 ENST00000397511.2 ENST00000534851.1 ENST00000474597.1 ENST00000244763.4 |
SSR1
|
signal sequence receptor, alpha |
| chr3_+_138340067 | 0.86 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr6_-_87804815 | 0.86 |
ENST00000369582.2
|
CGA
|
glycoprotein hormones, alpha polypeptide |
| chr16_+_31271274 | 0.80 |
ENST00000287497.8
ENST00000544665.3 |
ITGAM
|
integrin, alpha M (complement component 3 receptor 3 subunit) |
| chr4_-_103749313 | 0.77 |
ENST00000394803.5
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr1_+_115572415 | 0.77 |
ENST00000256592.1
|
TSHB
|
thyroid stimulating hormone, beta |
| chr12_-_86650077 | 0.76 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chrX_+_135618258 | 0.72 |
ENST00000440515.1
ENST00000456412.1 |
VGLL1
|
vestigial like 1 (Drosophila) |
| chr4_-_103749105 | 0.71 |
ENST00000394801.4
ENST00000394804.2 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr8_+_107738240 | 0.70 |
ENST00000449762.2
ENST00000297447.6 |
OXR1
|
oxidation resistance 1 |
| chr19_+_48949030 | 0.69 |
ENST00000253237.5
|
GRWD1
|
glutamate-rich WD repeat containing 1 |
| chr11_+_114168085 | 0.68 |
ENST00000541754.1
|
NNMT
|
nicotinamide N-methyltransferase |
| chr7_+_120628731 | 0.68 |
ENST00000310396.5
|
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chr14_+_22977587 | 0.67 |
ENST00000390504.1
|
TRAJ33
|
T cell receptor alpha joining 33 |
| chr17_-_39191107 | 0.64 |
ENST00000344363.5
|
KRTAP1-3
|
keratin associated protein 1-3 |
| chr16_-_30122717 | 0.64 |
ENST00000566613.1
|
GDPD3
|
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr9_+_34652164 | 0.63 |
ENST00000441545.2
ENST00000553620.1 |
IL11RA
|
interleukin 11 receptor, alpha |
| chr14_+_32798462 | 0.61 |
ENST00000280979.4
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr5_+_176811431 | 0.61 |
ENST00000512593.1
ENST00000324417.5 |
SLC34A1
|
solute carrier family 34 (type II sodium/phosphate contransporter), member 1 |
| chr4_+_80584903 | 0.59 |
ENST00000506460.1
|
RP11-452C8.1
|
RP11-452C8.1 |
| chr1_+_157963063 | 0.58 |
ENST00000360089.4
ENST00000368173.3 ENST00000392272.2 |
KIRREL
|
kin of IRRE like (Drosophila) |
| chr3_-_151034734 | 0.55 |
ENST00000260843.4
|
GPR87
|
G protein-coupled receptor 87 |
| chr2_+_166095898 | 0.53 |
ENST00000424833.1
ENST00000375437.2 ENST00000357398.3 |
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chr7_-_87342564 | 0.51 |
ENST00000265724.3
ENST00000416177.1 |
ABCB1
|
ATP-binding cassette, sub-family B (MDR/TAP), member 1 |
| chr12_+_7013897 | 0.51 |
ENST00000007969.8
ENST00000323702.5 |
LRRC23
|
leucine rich repeat containing 23 |
| chr11_+_5710919 | 0.50 |
ENST00000379965.3
ENST00000425490.1 |
TRIM22
|
tripartite motif containing 22 |
| chr19_-_36304201 | 0.49 |
ENST00000301175.3
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr14_+_32798547 | 0.49 |
ENST00000557354.1
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr4_+_37455536 | 0.48 |
ENST00000381980.4
ENST00000508175.1 |
C4orf19
|
chromosome 4 open reading frame 19 |
| chr12_-_86650045 | 0.47 |
ENST00000604798.1
|
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr12_+_7014064 | 0.47 |
ENST00000443597.2
|
LRRC23
|
leucine rich repeat containing 23 |
| chr10_+_70661014 | 0.45 |
ENST00000373585.3
|
DDX50
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 50 |
| chr20_-_50722183 | 0.45 |
ENST00000371523.4
|
ZFP64
|
ZFP64 zinc finger protein |
| chr17_+_1674982 | 0.45 |
ENST00000572048.1
ENST00000573763.1 |
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr1_-_92952433 | 0.45 |
ENST00000294702.5
|
GFI1
|
growth factor independent 1 transcription repressor |
| chr4_+_110736659 | 0.45 |
ENST00000394631.3
ENST00000226796.6 |
GAR1
|
GAR1 ribonucleoprotein |
| chr12_-_22063787 | 0.44 |
ENST00000544039.1
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr12_+_15699286 | 0.44 |
ENST00000442921.2
ENST00000542557.1 ENST00000445537.2 ENST00000544244.1 |
PTPRO
|
protein tyrosine phosphatase, receptor type, O |
| chr4_+_142142035 | 0.44 |
ENST00000262990.4
ENST00000512809.1 ENST00000503649.1 ENST00000512738.1 ENST00000421169.2 |
ZNF330
|
zinc finger protein 330 |
| chr20_+_1099233 | 0.43 |
ENST00000246015.4
ENST00000335877.6 ENST00000438768.2 |
PSMF1
|
proteasome (prosome, macropain) inhibitor subunit 1 (PI31) |
| chr14_+_74034310 | 0.43 |
ENST00000538782.1
|
ACOT2
|
acyl-CoA thioesterase 2 |
| chr6_+_158733692 | 0.37 |
ENST00000367094.2
ENST00000367097.3 |
TULP4
|
tubby like protein 4 |
| chr6_+_161123270 | 0.37 |
ENST00000366924.2
ENST00000308192.9 ENST00000418964.1 |
PLG
|
plasminogen |
| chr4_-_66536057 | 0.35 |
ENST00000273854.3
|
EPHA5
|
EPH receptor A5 |
| chr2_+_234826016 | 0.33 |
ENST00000324695.4
ENST00000433712.2 |
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr2_+_102953608 | 0.30 |
ENST00000311734.2
ENST00000409584.1 |
IL1RL1
|
interleukin 1 receptor-like 1 |
| chr9_+_105757590 | 0.27 |
ENST00000374798.3
ENST00000487798.1 |
CYLC2
|
cylicin, basic protein of sperm head cytoskeleton 2 |
| chr4_-_164534657 | 0.27 |
ENST00000339875.5
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr19_+_49199209 | 0.26 |
ENST00000522966.1
ENST00000425340.2 ENST00000391876.4 |
FUT2
|
fucosyltransferase 2 (secretor status included) |
| chr4_-_66536196 | 0.26 |
ENST00000511294.1
|
EPHA5
|
EPH receptor A5 |
| chr8_+_30244580 | 0.25 |
ENST00000523115.1
ENST00000519647.1 |
RBPMS
|
RNA binding protein with multiple splicing |
| chr11_+_7618413 | 0.23 |
ENST00000528883.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr6_+_26183958 | 0.22 |
ENST00000356530.3
|
HIST1H2BE
|
histone cluster 1, H2be |
| chrX_+_153533275 | 0.19 |
ENST00000426989.1
ENST00000426203.1 ENST00000369912.2 |
TKTL1
|
transketolase-like 1 |
| chr6_+_154360357 | 0.18 |
ENST00000330432.7
ENST00000360422.4 |
OPRM1
|
opioid receptor, mu 1 |
| chr12_+_122688090 | 0.15 |
ENST00000324189.4
ENST00000546192.1 |
B3GNT4
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 4 |
| chr8_+_50824233 | 0.14 |
ENST00000522124.1
|
SNTG1
|
syntrophin, gamma 1 |
| chr7_+_107224364 | 0.13 |
ENST00000491150.1
|
BCAP29
|
B-cell receptor-associated protein 29 |
| chr1_+_81771806 | 0.12 |
ENST00000370721.1
ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2
|
latrophilin 2 |
| chr3_+_45927994 | 0.11 |
ENST00000357632.2
ENST00000395963.2 |
CCR9
|
chemokine (C-C motif) receptor 9 |
| chr6_+_26402517 | 0.11 |
ENST00000414912.2
|
BTN3A1
|
butyrophilin, subfamily 3, member A1 |
| chr5_+_140201183 | 0.09 |
ENST00000529619.1
ENST00000529859.1 ENST00000378126.3 |
PCDHA5
|
protocadherin alpha 5 |
| chr1_-_24306798 | 0.09 |
ENST00000374452.5
ENST00000492112.2 ENST00000343255.5 ENST00000344989.6 |
SRSF10
|
serine/arginine-rich splicing factor 10 |
| chr18_+_29027696 | 0.09 |
ENST00000257189.4
|
DSG3
|
desmoglein 3 |
| chr3_+_183853052 | 0.09 |
ENST00000273783.3
ENST00000432569.1 ENST00000444495.1 |
EIF2B5
|
eukaryotic translation initiation factor 2B, subunit 5 epsilon, 82kDa |
| chr1_-_151431647 | 0.07 |
ENST00000368863.2
ENST00000409503.1 ENST00000491586.1 ENST00000533351.1 ENST00000540984.1 |
POGZ
|
pogo transposable element with ZNF domain |
| chr1_+_225600404 | 0.04 |
ENST00000366845.2
|
AC092811.1
|
AC092811.1 |
| chr12_-_112123524 | 0.03 |
ENST00000327551.6
|
BRAP
|
BRCA1 associated protein |
| chr3_+_148709128 | 0.03 |
ENST00000345003.4
ENST00000296048.6 ENST00000483267.1 |
GYG1
|
glycogenin 1 |
| chr21_-_15918618 | 0.03 |
ENST00000400564.1
ENST00000400566.1 |
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr18_-_67624160 | 0.02 |
ENST00000581982.1
ENST00000280200.4 |
CD226
|
CD226 molecule |
| chr6_+_26402465 | 0.01 |
ENST00000476549.2
ENST00000289361.6 ENST00000450085.2 ENST00000425234.2 ENST00000427334.1 ENST00000506698.1 |
BTN3A1
|
butyrophilin, subfamily 3, member A1 |
| chr10_+_35484793 | 0.00 |
ENST00000488741.1
ENST00000474931.1 ENST00000468236.1 ENST00000344351.5 ENST00000490511.1 |
CREM
|
cAMP responsive element modulator |
| chrX_+_108779004 | 0.00 |
ENST00000218004.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ALX3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 6.8 | GO:0003147 | neural crest cell migration involved in heart formation(GO:0003147) cell migration involved in heart formation(GO:0060974) anterior neural tube closure(GO:0061713) |
| 2.1 | 6.3 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.9 | 3.6 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.9 | 3.6 | GO:1904502 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.9 | 2.7 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.9 | 21.6 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.8 | 5.3 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.8 | 5.3 | GO:0048241 | epinephrine transport(GO:0048241) |
| 0.7 | 2.9 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.7 | 6.9 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.7 | 4.7 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.6 | 1.9 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.6 | 1.8 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.6 | 4.2 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.6 | 11.2 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.6 | 1.7 | GO:0036509 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.4 | 1.7 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.4 | 2.5 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.4 | 1.7 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.4 | 1.2 | GO:2000005 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm development(GO:0048389) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) pattern specification involved in mesonephros development(GO:0061227) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in kidney development(GO:0072098) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.4 | 1.1 | GO:0032641 | negative regulation of tolerance induction(GO:0002644) lymphotoxin A production(GO:0032641) interleukin-4 biosynthetic process(GO:0042097) lymphotoxin A biosynthetic process(GO:0042109) regulation of interleukin-4 biosynthetic process(GO:0045402) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.4 | 6.4 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.4 | 4.4 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.3 | 1.1 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.3 | 11.9 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.2 | 1.0 | GO:0032690 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.2 | 1.5 | GO:0009597 | detection of virus(GO:0009597) |
| 0.2 | 2.8 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.2 | 2.1 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.2 | 2.7 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.2 | 0.6 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.2 | 2.4 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.2 | 24.9 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.2 | 7.0 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.2 | 3.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.4 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.1 | 0.4 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 5.6 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 0.4 | GO:2000048 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.1 | 0.9 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 | 1.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 2.3 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.1 | 1.7 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 0.9 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.1 | 0.3 | GO:0002826 | negative regulation of T-helper 1 type immune response(GO:0002826) |
| 0.1 | 1.3 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.5 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 6.4 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.1 | 0.9 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.1 | 0.5 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.1 | 2.0 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 0.3 | GO:0050955 | thermoception(GO:0050955) |
| 0.1 | 0.4 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.1 | 0.2 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.1 | 2.0 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.1 | 0.8 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.1 | 0.7 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.1 | 1.4 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.1 | 8.3 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 1.2 | GO:0043101 | purine-containing compound salvage(GO:0043101) |
| 0.0 | 0.1 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.9 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.5 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.0 | 2.3 | GO:0045661 | regulation of myoblast differentiation(GO:0045661) |
| 0.0 | 1.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 1.0 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.0 | 1.6 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 1.6 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.7 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 2.5 | GO:0070126 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 0.0 | 0.6 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.6 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 1.0 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 1.1 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.3 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.3 | GO:0042354 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 1.4 | GO:0043407 | negative regulation of MAP kinase activity(GO:0043407) |
| 0.0 | 0.0 | GO:0060369 | positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.0 | 1.5 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.8 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.2 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.1 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.0 | 0.6 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 5.3 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 0.5 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.0 | 0.4 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.6 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.7 | 4.7 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.5 | 1.9 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.4 | 6.8 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.4 | 11.2 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.3 | 2.9 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.2 | 1.5 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.2 | 0.8 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.2 | 1.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 0.4 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 1.6 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.1 | 2.5 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 0.9 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.1 | 2.7 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 2.7 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 5.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 9.0 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 2.5 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 0.3 | GO:0033150 | perinuclear theca(GO:0033011) cytoskeletal calyx(GO:0033150) |
| 0.1 | 1.5 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.1 | 0.4 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.1 | 1.7 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.1 | 1.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.4 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.1 | 2.0 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 2.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 1.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 1.7 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 8.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.1 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 6.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.7 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 3.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 4.4 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.4 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 1.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 1.0 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.9 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 2.3 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.6 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 1.9 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 4.3 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 3.8 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 9.3 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 2.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 2.4 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 2.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 2.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.4 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.9 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 17.1 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.4 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.1 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 21.6 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 1.7 | 6.8 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 1.6 | 6.3 | GO:0047023 | enone reductase activity(GO:0035671) androsterone dehydrogenase activity(GO:0047023) indanol dehydrogenase activity(GO:0047718) |
| 1.3 | 5.3 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 1.2 | 3.6 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 1.1 | 5.3 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 1.0 | 4.2 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.9 | 2.8 | GO:0003978 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.7 | 6.9 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.6 | 1.8 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.5 | 2.7 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.5 | 2.5 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.4 | 1.7 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.4 | 2.1 | GO:0004771 | sterol esterase activity(GO:0004771) methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.3 | 2.4 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.3 | 1.9 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.3 | 11.9 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.2 | 0.7 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) |
| 0.2 | 2.9 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.2 | 1.7 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.2 | 1.0 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.2 | 5.7 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.2 | 1.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.2 | 0.7 | GO:0008112 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.2 | 1.3 | GO:0046790 | virion binding(GO:0046790) |
| 0.2 | 0.9 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.2 | 1.7 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 31.6 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 1.2 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 0.5 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.1 | 1.5 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 0.5 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 2.4 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.1 | 2.8 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 4.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 3.3 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 2.2 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 0.3 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.1 | 1.7 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 1.2 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 0.8 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.1 | 0.9 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.6 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.1 | 0.4 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 1.5 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 1.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 1.9 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 0.2 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.1 | 0.6 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 2.3 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 1.1 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.6 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 1.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.4 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.4 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 0.2 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.6 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 5.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.1 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.5 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.4 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.6 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 0.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 1.0 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.3 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 6.1 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.7 | GO:0004896 | cytokine receptor activity(GO:0004896) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 12.2 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 5.7 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 2.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 5.6 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 2.8 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 3.6 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 1.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.9 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.6 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.1 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.2 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 1.3 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.1 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.6 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.8 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.4 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.2 | PID BMP PATHWAY | BMP receptor signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 21.6 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.4 | 5.8 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.4 | 5.7 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.3 | 5.3 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.3 | 12.2 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.3 | 2.9 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.2 | 2.8 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 1.6 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 2.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 3.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.7 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 3.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 4.5 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.1 | 1.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 1.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.9 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 1.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 8.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 3.0 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 3.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.7 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.7 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 1.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.4 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 1.1 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |