Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
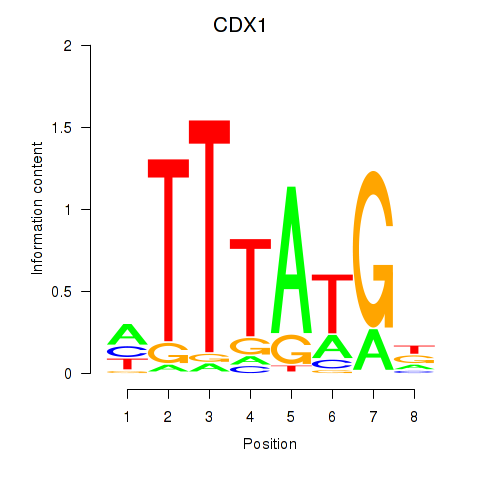
Results for CDX1
Z-value: 0.89
Transcription factors associated with CDX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CDX1
|
ENSG00000113722.12 | caudal type homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CDX1 | hg19_v2_chr5_+_149546334_149546364 | 0.58 | 1.1e-20 | Click! |
Activity profile of CDX1 motif
Sorted Z-values of CDX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_9268707 | 16.86 |
ENST00000318602.7
|
A2M
|
alpha-2-macroglobulin |
| chr12_-_91573249 | 14.20 |
ENST00000550099.1
ENST00000546391.1 ENST00000551354.1 |
DCN
|
decorin |
| chr10_-_69597810 | 13.58 |
ENST00000483798.2
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr10_-_90712520 | 13.43 |
ENST00000224784.6
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chr12_-_91573132 | 13.32 |
ENST00000550563.1
ENST00000546370.1 |
DCN
|
decorin |
| chr15_-_45670924 | 10.68 |
ENST00000396659.3
|
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr16_+_56672571 | 9.91 |
ENST00000290705.8
|
MT1A
|
metallothionein 1A |
| chr11_+_114168085 | 9.87 |
ENST00000541754.1
|
NNMT
|
nicotinamide N-methyltransferase |
| chr12_-_52779433 | 9.83 |
ENST00000257951.3
|
KRT84
|
keratin 84 |
| chr16_+_56685796 | 9.35 |
ENST00000334346.2
ENST00000562399.1 |
MT1B
|
metallothionein 1B |
| chr19_-_51530916 | 8.24 |
ENST00000594768.1
|
KLK11
|
kallikrein-related peptidase 11 |
| chr19_-_51531272 | 8.16 |
ENST00000319720.7
|
KLK11
|
kallikrein-related peptidase 11 |
| chr1_+_89829610 | 8.08 |
ENST00000370456.4
ENST00000535065.1 |
GBP6
|
guanylate binding protein family, member 6 |
| chrX_+_56259316 | 7.92 |
ENST00000468660.1
|
KLF8
|
Kruppel-like factor 8 |
| chr10_-_52645379 | 7.76 |
ENST00000395489.2
|
A1CF
|
APOBEC1 complementation factor |
| chr10_-_69597915 | 7.21 |
ENST00000225171.2
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr10_-_52645416 | 7.00 |
ENST00000374001.2
ENST00000373997.3 ENST00000373995.3 ENST00000282641.2 ENST00000395495.1 ENST00000414883.1 |
A1CF
|
APOBEC1 complementation factor |
| chr5_-_41261540 | 6.75 |
ENST00000263413.3
|
C6
|
complement component 6 |
| chr9_+_124088860 | 6.64 |
ENST00000373806.1
|
GSN
|
gelsolin |
| chr3_-_123512688 | 6.28 |
ENST00000475616.1
|
MYLK
|
myosin light chain kinase |
| chr20_+_43595115 | 6.23 |
ENST00000372806.3
ENST00000396731.4 ENST00000372801.1 ENST00000499879.2 |
STK4
|
serine/threonine kinase 4 |
| chr5_-_147211226 | 6.20 |
ENST00000296695.5
|
SPINK1
|
serine peptidase inhibitor, Kazal type 1 |
| chr20_+_56136136 | 5.89 |
ENST00000319441.4
ENST00000543666.1 |
PCK1
|
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr5_+_140588269 | 5.70 |
ENST00000541609.1
ENST00000239450.2 |
PCDHB12
|
protocadherin beta 12 |
| chr2_-_179672142 | 5.61 |
ENST00000342992.6
ENST00000360870.5 ENST00000460472.2 ENST00000589042.1 ENST00000591111.1 ENST00000342175.6 ENST00000359218.5 |
TTN
|
titin |
| chr12_-_118796910 | 5.48 |
ENST00000541186.1
ENST00000539872.1 |
TAOK3
|
TAO kinase 3 |
| chr2_+_113763031 | 5.48 |
ENST00000259211.6
|
IL36A
|
interleukin 36, alpha |
| chr12_-_11548496 | 5.35 |
ENST00000389362.4
ENST00000565533.1 ENST00000546254.1 |
PRB2
PRB1
|
proline-rich protein BstNI subfamily 2 proline-rich protein BstNI subfamily 1 |
| chr7_-_16840820 | 5.23 |
ENST00000450569.1
|
AGR2
|
anterior gradient 2 |
| chr2_-_152589670 | 5.18 |
ENST00000604864.1
ENST00000603639.1 |
NEB
|
nebulin |
| chr3_-_149095652 | 5.15 |
ENST00000305366.3
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr4_-_73935409 | 5.02 |
ENST00000507544.2
ENST00000295890.4 |
COX18
|
COX18 cytochrome C oxidase assembly factor |
| chr10_-_95360983 | 5.01 |
ENST00000371464.3
|
RBP4
|
retinol binding protein 4, plasma |
| chr6_-_90025011 | 4.97 |
ENST00000402938.3
|
GABRR2
|
gamma-aminobutyric acid (GABA) A receptor, rho 2 |
| chr2_+_32502952 | 4.81 |
ENST00000238831.4
|
YIPF4
|
Yip1 domain family, member 4 |
| chr6_-_33168391 | 4.77 |
ENST00000374685.4
ENST00000413614.2 ENST00000374680.3 |
RXRB
|
retinoid X receptor, beta |
| chr16_+_56691606 | 4.75 |
ENST00000334350.6
|
MT1F
|
metallothionein 1F |
| chr12_+_58013693 | 4.74 |
ENST00000320442.4
ENST00000379218.2 |
SLC26A10
|
solute carrier family 26, member 10 |
| chr9_-_21335240 | 4.74 |
ENST00000537938.1
|
KLHL9
|
kelch-like family member 9 |
| chr2_-_88427568 | 4.65 |
ENST00000393750.3
ENST00000295834.3 |
FABP1
|
fatty acid binding protein 1, liver |
| chr4_-_186732048 | 4.63 |
ENST00000448662.2
ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr16_-_20681177 | 4.56 |
ENST00000524149.1
|
ACSM1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr2_-_163008903 | 4.54 |
ENST00000418842.2
ENST00000375497.3 |
GCG
|
glucagon |
| chr5_+_102201509 | 4.42 |
ENST00000348126.2
ENST00000379787.4 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr1_+_171283331 | 4.40 |
ENST00000367749.3
|
FMO4
|
flavin containing monooxygenase 4 |
| chr19_-_51531210 | 4.33 |
ENST00000391804.3
|
KLK11
|
kallikrein-related peptidase 11 |
| chr17_-_59668550 | 4.28 |
ENST00000521764.1
|
NACA2
|
nascent polypeptide-associated complex alpha subunit 2 |
| chr20_-_17539456 | 4.28 |
ENST00000544874.1
ENST00000377868.2 |
BFSP1
|
beaded filament structural protein 1, filensin |
| chr17_-_73663168 | 4.26 |
ENST00000578201.1
ENST00000423245.2 |
RECQL5
|
RecQ protein-like 5 |
| chr12_-_52715179 | 4.17 |
ENST00000293670.3
|
KRT83
|
keratin 83 |
| chr11_-_2158507 | 4.17 |
ENST00000381392.1
ENST00000381395.1 ENST00000418738.2 |
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr4_-_16085314 | 4.15 |
ENST00000510224.1
|
PROM1
|
prominin 1 |
| chr19_+_8455077 | 4.13 |
ENST00000328024.6
|
RAB11B
|
RAB11B, member RAS oncogene family |
| chr17_+_57232690 | 4.12 |
ENST00000262293.4
|
PRR11
|
proline rich 11 |
| chr5_+_102201430 | 4.10 |
ENST00000438793.3
ENST00000346918.2 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr4_+_74269956 | 4.09 |
ENST00000295897.4
ENST00000415165.2 ENST00000503124.1 ENST00000509063.1 ENST00000401494.3 |
ALB
|
albumin |
| chr2_-_177502254 | 4.05 |
ENST00000339037.3
|
AC017048.3
|
long intergenic non-protein coding RNA 1116 |
| chr16_-_15950868 | 4.04 |
ENST00000396324.3
ENST00000452625.2 ENST00000576790.2 ENST00000300036.5 |
MYH11
|
myosin, heavy chain 11, smooth muscle |
| chr6_-_52774464 | 4.02 |
ENST00000370968.1
ENST00000211122.3 |
GSTA3
|
glutathione S-transferase alpha 3 |
| chr1_-_182921119 | 4.00 |
ENST00000423786.1
|
SHCBP1L
|
SHC SH2-domain binding protein 1-like |
| chr19_+_15852203 | 3.99 |
ENST00000305892.1
|
OR10H3
|
olfactory receptor, family 10, subfamily H, member 3 |
| chr19_-_47164386 | 3.98 |
ENST00000391916.2
ENST00000410105.2 |
DACT3
|
dishevelled-binding antagonist of beta-catenin 3 |
| chr7_-_19748640 | 3.98 |
ENST00000222567.5
|
TWISTNB
|
TWIST neighbor |
| chr12_+_32655048 | 3.97 |
ENST00000427716.2
ENST00000266482.3 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr8_+_22423168 | 3.94 |
ENST00000518912.1
ENST00000428103.1 |
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr5_+_140800638 | 3.87 |
ENST00000398587.2
ENST00000518882.1 |
PCDHGA11
|
protocadherin gamma subfamily A, 11 |
| chr12_-_111358372 | 3.85 |
ENST00000548438.1
ENST00000228841.8 |
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow |
| chr5_-_147211190 | 3.79 |
ENST00000510027.2
|
SPINK1
|
serine peptidase inhibitor, Kazal type 1 |
| chr1_+_196743912 | 3.78 |
ENST00000367425.4
|
CFHR3
|
complement factor H-related 3 |
| chr5_-_55412774 | 3.77 |
ENST00000434982.2
|
ANKRD55
|
ankyrin repeat domain 55 |
| chr6_+_33168597 | 3.76 |
ENST00000374675.3
|
SLC39A7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr12_-_48164812 | 3.68 |
ENST00000549151.1
ENST00000548919.1 |
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr1_+_206223941 | 3.66 |
ENST00000367126.4
|
AVPR1B
|
arginine vasopressin receptor 1B |
| chr11_-_89224508 | 3.63 |
ENST00000525196.1
|
NOX4
|
NADPH oxidase 4 |
| chr15_-_35047166 | 3.63 |
ENST00000290374.4
|
GJD2
|
gap junction protein, delta 2, 36kDa |
| chr7_+_106505912 | 3.58 |
ENST00000359195.3
|
PIK3CG
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr6_+_33168637 | 3.57 |
ENST00000374677.3
|
SLC39A7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr4_-_70826725 | 3.54 |
ENST00000353151.3
|
CSN2
|
casein beta |
| chr4_-_168155169 | 3.52 |
ENST00000534949.1
ENST00000535728.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr1_+_15802594 | 3.50 |
ENST00000375910.3
|
CELA2B
|
chymotrypsin-like elastase family, member 2B |
| chr11_+_28129795 | 3.48 |
ENST00000406787.3
ENST00000342303.5 ENST00000403099.1 ENST00000407364.3 |
METTL15
|
methyltransferase like 15 |
| chr1_+_169079823 | 3.46 |
ENST00000367813.3
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr14_+_74111578 | 3.42 |
ENST00000554113.1
ENST00000555631.2 ENST00000553645.2 ENST00000311089.3 ENST00000555919.3 ENST00000554339.1 ENST00000554871.1 |
DNAL1
|
dynein, axonemal, light chain 1 |
| chr21_-_43735446 | 3.40 |
ENST00000398431.2
|
TFF3
|
trefoil factor 3 (intestinal) |
| chr7_-_143454789 | 3.39 |
ENST00000470691.2
|
CTAGE6
|
CTAGE family, member 6 |
| chr4_-_16085340 | 3.39 |
ENST00000508167.1
|
PROM1
|
prominin 1 |
| chr13_+_23755099 | 3.31 |
ENST00000537476.1
|
SGCG
|
sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein) |
| chr11_-_2182388 | 3.29 |
ENST00000421783.1
ENST00000397262.1 ENST00000250971.3 ENST00000381330.4 ENST00000397270.1 |
INS
INS-IGF2
|
insulin INS-IGF2 readthrough |
| chr21_-_43735628 | 3.28 |
ENST00000291525.10
ENST00000518498.1 |
TFF3
|
trefoil factor 3 (intestinal) |
| chr9_-_21351377 | 3.27 |
ENST00000380210.1
|
IFNA6
|
interferon, alpha 6 |
| chr2_-_79315112 | 3.26 |
ENST00000305089.3
|
REG1B
|
regenerating islet-derived 1 beta |
| chr9_-_95244781 | 3.25 |
ENST00000375544.3
ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN
|
asporin |
| chr2_-_208031943 | 3.15 |
ENST00000421199.1
ENST00000457962.1 |
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr1_-_111970353 | 3.13 |
ENST00000369732.3
|
OVGP1
|
oviductal glycoprotein 1, 120kDa |
| chr1_+_196743943 | 3.10 |
ENST00000471440.2
ENST00000391985.3 |
CFHR3
|
complement factor H-related 3 |
| chr18_+_61575200 | 3.06 |
ENST00000238508.3
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr4_+_69313145 | 3.05 |
ENST00000305363.4
|
TMPRSS11E
|
transmembrane protease, serine 11E |
| chr1_+_104159999 | 3.03 |
ENST00000414303.2
ENST00000423678.1 |
AMY2A
|
amylase, alpha 2A (pancreatic) |
| chr12_-_371994 | 3.01 |
ENST00000343164.4
ENST00000436453.1 ENST00000445055.2 ENST00000546319.1 |
SLC6A13
|
solute carrier family 6 (neurotransmitter transporter), member 13 |
| chr11_+_61015594 | 3.01 |
ENST00000451616.2
|
PGA5
|
pepsinogen 5, group I (pepsinogen A) |
| chr5_+_140718396 | 3.01 |
ENST00000394576.2
|
PCDHGA2
|
protocadherin gamma subfamily A, 2 |
| chr21_+_44313375 | 2.98 |
ENST00000354250.2
ENST00000340344.4 |
NDUFV3
|
NADH dehydrogenase (ubiquinone) flavoprotein 3, 10kDa |
| chr12_+_72080253 | 2.95 |
ENST00000549735.1
|
TMEM19
|
transmembrane protein 19 |
| chr17_-_4544960 | 2.93 |
ENST00000293761.3
|
ALOX15
|
arachidonate 15-lipoxygenase |
| chr2_+_170366203 | 2.92 |
ENST00000284669.1
|
KLHL41
|
kelch-like family member 41 |
| chr5_-_138775177 | 2.92 |
ENST00000302060.5
|
DNAJC18
|
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr8_+_42552533 | 2.91 |
ENST00000289957.2
|
CHRNB3
|
cholinergic receptor, nicotinic, beta 3 (neuronal) |
| chr4_-_47983519 | 2.90 |
ENST00000358519.4
ENST00000544810.1 ENST00000402813.3 |
CNGA1
|
cyclic nucleotide gated channel alpha 1 |
| chr4_-_76957214 | 2.89 |
ENST00000306621.3
|
CXCL11
|
chemokine (C-X-C motif) ligand 11 |
| chr5_+_140739537 | 2.88 |
ENST00000522605.1
|
PCDHGB2
|
protocadherin gamma subfamily B, 2 |
| chr2_+_233527443 | 2.87 |
ENST00000410095.1
|
EFHD1
|
EF-hand domain family, member D1 |
| chr6_+_26087509 | 2.86 |
ENST00000397022.3
ENST00000353147.5 ENST00000352392.4 ENST00000349999.4 ENST00000317896.7 ENST00000357618.5 ENST00000470149.1 ENST00000336625.8 ENST00000461397.1 ENST00000488199.1 |
HFE
|
hemochromatosis |
| chr8_-_120605194 | 2.85 |
ENST00000522167.1
|
ENPP2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr13_+_23755054 | 2.84 |
ENST00000218867.3
|
SGCG
|
sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein) |
| chr10_-_22292613 | 2.79 |
ENST00000376980.3
|
DNAJC1
|
DnaJ (Hsp40) homolog, subfamily C, member 1 |
| chr5_+_64920543 | 2.79 |
ENST00000399438.3
ENST00000510585.2 |
TRAPPC13
CTC-534A2.2
|
trafficking protein particle complex 13 CDNA FLJ26957 fis, clone SLV00486; Uncharacterized protein |
| chr6_+_161123270 | 2.78 |
ENST00000366924.2
ENST00000308192.9 ENST00000418964.1 |
PLG
|
plasminogen |
| chr5_-_160973649 | 2.77 |
ENST00000393959.1
ENST00000517547.1 |
GABRB2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr8_+_12803176 | 2.77 |
ENST00000524591.2
|
KIAA1456
|
KIAA1456 |
| chr17_+_16284104 | 2.76 |
ENST00000577958.1
ENST00000302182.3 ENST00000577640.1 |
UBB
|
ubiquitin B |
| chr9_-_21335356 | 2.75 |
ENST00000359039.4
|
KLHL9
|
kelch-like family member 9 |
| chr7_-_18067478 | 2.70 |
ENST00000506618.2
|
PRPS1L1
|
phosphoribosyl pyrophosphate synthetase 1-like 1 |
| chr16_-_67965756 | 2.66 |
ENST00000571044.1
ENST00000571605.1 |
CTRL
|
chymotrypsin-like |
| chr17_-_56492989 | 2.66 |
ENST00000583753.1
|
RNF43
|
ring finger protein 43 |
| chr15_-_55700457 | 2.61 |
ENST00000442196.3
ENST00000563171.1 ENST00000425574.3 |
CCPG1
|
cell cycle progression 1 |
| chr19_+_9296279 | 2.61 |
ENST00000344248.2
|
OR7D2
|
olfactory receptor, family 7, subfamily D, member 2 |
| chr8_+_22422749 | 2.61 |
ENST00000523900.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr1_+_15986364 | 2.58 |
ENST00000345034.1
|
RSC1A1
|
regulatory solute carrier protein, family 1, member 1 |
| chr10_+_72194585 | 2.57 |
ENST00000420338.2
|
AC022532.1
|
Uncharacterized protein |
| chr14_-_67878917 | 2.57 |
ENST00000216446.4
|
PLEK2
|
pleckstrin 2 |
| chr3_+_135741576 | 2.55 |
ENST00000334546.2
|
PPP2R3A
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr4_+_169418195 | 2.54 |
ENST00000261509.6
ENST00000335742.7 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr5_-_75919253 | 2.54 |
ENST00000296641.4
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2 |
| chrX_+_15767971 | 2.50 |
ENST00000479740.1
ENST00000454127.2 |
CA5B
|
carbonic anhydrase VB, mitochondrial |
| chr8_-_42065187 | 2.50 |
ENST00000270189.6
ENST00000352041.3 ENST00000220809.4 |
PLAT
|
plasminogen activator, tissue |
| chr12_+_72079842 | 2.49 |
ENST00000266673.5
ENST00000550524.1 |
TMEM19
|
transmembrane protein 19 |
| chr6_+_26087646 | 2.47 |
ENST00000309234.6
|
HFE
|
hemochromatosis |
| chr16_-_1275257 | 2.46 |
ENST00000234798.4
|
TPSG1
|
tryptase gamma 1 |
| chrX_+_18725758 | 2.45 |
ENST00000472826.1
ENST00000544635.1 ENST00000496075.2 |
PPEF1
|
protein phosphatase, EF-hand calcium binding domain 1 |
| chr19_+_44598534 | 2.43 |
ENST00000336976.6
|
ZNF224
|
zinc finger protein 224 |
| chr9_-_95186739 | 2.43 |
ENST00000375550.4
|
OMD
|
osteomodulin |
| chr1_+_196857144 | 2.32 |
ENST00000367416.2
ENST00000251424.4 ENST00000367418.2 |
CFHR4
|
complement factor H-related 4 |
| chr14_+_22615942 | 2.29 |
ENST00000390457.2
|
TRAV27
|
T cell receptor alpha variable 27 |
| chr18_+_61445007 | 2.26 |
ENST00000447428.1
ENST00000546027.1 |
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr4_-_175443943 | 2.26 |
ENST00000296522.6
|
HPGD
|
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chr6_+_37225540 | 2.26 |
ENST00000373491.3
|
TBC1D22B
|
TBC1 domain family, member 22B |
| chr11_-_89224638 | 2.24 |
ENST00000535633.1
ENST00000263317.4 |
NOX4
|
NADPH oxidase 4 |
| chr9_-_97356075 | 2.19 |
ENST00000375337.3
|
FBP2
|
fructose-1,6-bisphosphatase 2 |
| chr12_-_101604185 | 2.18 |
ENST00000536262.2
|
SLC5A8
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 8 |
| chr15_-_58571445 | 2.17 |
ENST00000558231.1
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr8_-_42065075 | 2.16 |
ENST00000429089.2
ENST00000519510.1 ENST00000429710.2 ENST00000524009.1 |
PLAT
|
plasminogen activator, tissue |
| chr7_+_120629653 | 2.16 |
ENST00000450913.2
ENST00000340646.5 |
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chrX_+_99899180 | 2.13 |
ENST00000373004.3
|
SRPX2
|
sushi-repeat containing protein, X-linked 2 |
| chr10_-_81320151 | 2.12 |
ENST00000372325.2
ENST00000372327.5 ENST00000417041.1 |
SFTPA2
|
surfactant protein A2 |
| chr2_+_161993465 | 2.10 |
ENST00000457476.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr2_+_166095898 | 2.09 |
ENST00000424833.1
ENST00000375437.2 ENST00000357398.3 |
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chr4_+_156824840 | 2.08 |
ENST00000536354.2
|
TDO2
|
tryptophan 2,3-dioxygenase |
| chr5_+_140535577 | 2.08 |
ENST00000539533.1
|
PCDHB17
|
Protocadherin-psi1; Uncharacterized protein |
| chr17_-_46690839 | 2.08 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chr7_+_119913688 | 2.07 |
ENST00000331113.4
|
KCND2
|
potassium voltage-gated channel, Shal-related subfamily, member 2 |
| chr1_+_241695670 | 2.05 |
ENST00000366557.4
|
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr20_+_16729003 | 2.05 |
ENST00000246081.2
|
OTOR
|
otoraplin |
| chr1_-_152386732 | 2.04 |
ENST00000271835.3
|
CRNN
|
cornulin |
| chr6_-_46048116 | 2.03 |
ENST00000185206.6
|
CLIC5
|
chloride intracellular channel 5 |
| chr14_+_102276192 | 2.01 |
ENST00000557714.1
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr14_+_75179840 | 2.01 |
ENST00000554590.1
ENST00000341162.4 ENST00000534938.2 ENST00000553615.1 |
FCF1
|
FCF1 rRNA-processing protein |
| chr18_-_61329118 | 2.00 |
ENST00000332821.8
ENST00000283752.5 |
SERPINB3
|
serpin peptidase inhibitor, clade B (ovalbumin), member 3 |
| chr11_+_67250490 | 1.98 |
ENST00000528641.2
ENST00000279146.3 |
AIP
|
aryl hydrocarbon receptor interacting protein |
| chr7_-_142176790 | 1.95 |
ENST00000390369.2
|
TRBV7-4
|
T cell receptor beta variable 7-4 (gene/pseudogene) |
| chr1_+_17559776 | 1.94 |
ENST00000537499.1
ENST00000413717.2 ENST00000536552.1 |
PADI1
|
peptidyl arginine deiminase, type I |
| chr5_-_138210977 | 1.94 |
ENST00000274711.6
ENST00000521094.2 |
LRRTM2
|
leucine rich repeat transmembrane neuronal 2 |
| chr12_+_51318513 | 1.91 |
ENST00000332160.4
|
METTL7A
|
methyltransferase like 7A |
| chr2_+_132479948 | 1.91 |
ENST00000355171.4
|
C2orf27A
|
chromosome 2 open reading frame 27A |
| chr15_-_55700522 | 1.90 |
ENST00000564092.1
ENST00000310958.6 |
CCPG1
|
cell cycle progression 1 |
| chr6_-_32374900 | 1.90 |
ENST00000374995.3
ENST00000374993.1 ENST00000414363.1 ENST00000540315.1 ENST00000544175.1 ENST00000429232.2 ENST00000454136.3 ENST00000446536.2 |
BTNL2
|
butyrophilin-like 2 (MHC class II associated) |
| chr1_+_200993071 | 1.89 |
ENST00000446333.1
ENST00000458003.1 |
RP11-168O16.1
|
RP11-168O16.1 |
| chr19_+_52076425 | 1.87 |
ENST00000436511.2
|
ZNF175
|
zinc finger protein 175 |
| chr14_+_65878565 | 1.87 |
ENST00000556518.1
ENST00000557164.1 |
FUT8
|
fucosyltransferase 8 (alpha (1,6) fucosyltransferase) |
| chr4_+_71063641 | 1.83 |
ENST00000514097.1
|
ODAM
|
odontogenic, ameloblast asssociated |
| chr3_-_101039402 | 1.83 |
ENST00000193391.7
|
IMPG2
|
interphotoreceptor matrix proteoglycan 2 |
| chr2_-_224467093 | 1.78 |
ENST00000305409.2
|
SCG2
|
secretogranin II |
| chr7_-_11871815 | 1.78 |
ENST00000423059.4
|
THSD7A
|
thrombospondin, type I, domain containing 7A |
| chrX_-_131623982 | 1.77 |
ENST00000370844.1
|
MBNL3
|
muscleblind-like splicing regulator 3 |
| chr17_-_29624343 | 1.76 |
ENST00000247271.4
|
OMG
|
oligodendrocyte myelin glycoprotein |
| chr6_+_36646435 | 1.76 |
ENST00000244741.5
ENST00000405375.1 ENST00000373711.2 |
CDKN1A
|
cyclin-dependent kinase inhibitor 1A (p21, Cip1) |
| chr3_+_190333097 | 1.75 |
ENST00000412080.1
|
IL1RAP
|
interleukin 1 receptor accessory protein |
| chrY_-_6740649 | 1.72 |
ENST00000383036.1
ENST00000383037.4 |
AMELY
|
amelogenin, Y-linked |
| chr8_-_7320974 | 1.72 |
ENST00000528943.1
ENST00000359758.5 ENST00000361111.2 ENST00000398462.2 ENST00000297498.2 ENST00000317900.5 |
SPAG11B
|
sperm associated antigen 11B |
| chr3_-_108836977 | 1.70 |
ENST00000232603.5
|
MORC1
|
MORC family CW-type zinc finger 1 |
| chr7_-_14942283 | 1.69 |
ENST00000402815.1
|
DGKB
|
diacylglycerol kinase, beta 90kDa |
| chr18_-_3845293 | 1.68 |
ENST00000400145.2
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr11_-_61687739 | 1.68 |
ENST00000531922.1
ENST00000301773.5 |
RAB3IL1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr6_-_43423308 | 1.68 |
ENST00000372485.1
ENST00000372488.3 |
DLK2
|
delta-like 2 homolog (Drosophila) |
| chr6_-_53013620 | 1.67 |
ENST00000259803.7
|
GCM1
|
glial cells missing homolog 1 (Drosophila) |
| chr15_+_42066632 | 1.67 |
ENST00000457542.2
ENST00000221214.6 ENST00000260357.7 ENST00000456763.2 |
MAPKBP1
|
mitogen-activated protein kinase binding protein 1 |
| chr2_+_168675182 | 1.67 |
ENST00000305861.1
|
B3GALT1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr6_-_112575912 | 1.66 |
ENST00000522006.1
ENST00000230538.7 ENST00000519932.1 |
LAMA4
|
laminin, alpha 4 |
| chr9_+_77112244 | 1.65 |
ENST00000376896.3
|
RORB
|
RAR-related orphan receptor B |
| chr19_+_41594377 | 1.64 |
ENST00000330436.3
|
CYP2A13
|
cytochrome P450, family 2, subfamily A, polypeptide 13 |
| chr12_-_28125638 | 1.63 |
ENST00000545234.1
|
PTHLH
|
parathyroid hormone-like hormone |
Network of associatons between targets according to the STRING database.
First level regulatory network of CDX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 9.6 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 2.4 | 14.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 2.3 | 27.5 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 2.2 | 13.4 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 2.1 | 8.5 | GO:0031179 | peptide amidation(GO:0001519) protein amidation(GO:0018032) peptide modification(GO:0031179) |
| 1.9 | 7.5 | GO:2000768 | glomerular parietal epithelial cell differentiation(GO:0072139) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 1.8 | 5.3 | GO:0002590 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) regulation of T cell antigen processing and presentation(GO:0002625) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 1.7 | 6.7 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 1.7 | 10.0 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 1.6 | 7.9 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 1.5 | 4.6 | GO:0018874 | benzoate metabolic process(GO:0018874) butyrate metabolic process(GO:0019605) |
| 1.5 | 4.5 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 1.5 | 5.9 | GO:0006114 | glycerol biosynthetic process(GO:0006114) response to methionine(GO:1904640) |
| 1.4 | 4.3 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 1.4 | 4.1 | GO:0044179 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 1.3 | 6.6 | GO:1903906 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 1.3 | 16.9 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 1.3 | 5.2 | GO:0007525 | somatic muscle development(GO:0007525) |
| 1.2 | 3.5 | GO:1903487 | regulation of lactation(GO:1903487) |
| 1.1 | 3.3 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 1.1 | 3.3 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 1.0 | 3.1 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 1.0 | 12.1 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 1.0 | 5.0 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 1.0 | 2.9 | GO:2001303 | cellular response to interleukin-13(GO:0035963) lipoxin biosynthetic process(GO:2001301) lipoxin A4 metabolic process(GO:2001302) lipoxin A4 biosynthetic process(GO:2001303) |
| 1.0 | 10.7 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.9 | 2.8 | GO:2000048 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.9 | 3.7 | GO:0030822 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.9 | 4.5 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.9 | 5.2 | GO:1903899 | lung goblet cell differentiation(GO:0060480) positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.9 | 3.5 | GO:1903288 | protein transport into plasma membrane raft(GO:0044861) positive regulation of potassium ion import(GO:1903288) |
| 0.9 | 24.0 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.8 | 5.1 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.8 | 6.3 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.8 | 2.3 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.7 | 10.4 | GO:0035878 | nail development(GO:0035878) |
| 0.7 | 3.7 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.7 | 2.1 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.7 | 2.0 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.7 | 4.7 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.7 | 2.7 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.6 | 1.9 | GO:0036071 | N-glycan fucosylation(GO:0036071) |
| 0.5 | 2.2 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 0.5 | 2.7 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.5 | 4.7 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.5 | 4.6 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.5 | 2.0 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.5 | 2.8 | GO:0060613 | fat pad development(GO:0060613) |
| 0.5 | 1.8 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.4 | 1.3 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.4 | 1.8 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.4 | 3.5 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.4 | 3.0 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.4 | 1.3 | GO:0015888 | thiamine transport(GO:0015888) thiamine transmembrane transport(GO:0071934) |
| 0.4 | 1.7 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.4 | 4.1 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.4 | 4.0 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.4 | 1.9 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.4 | 2.3 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.4 | 4.3 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.4 | 1.4 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) negative regulation of skeletal muscle cell proliferation(GO:0014859) regulation of skeletal muscle tissue growth(GO:0048631) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.3 | 2.4 | GO:0035624 | receptor transactivation(GO:0035624) |
| 0.3 | 1.7 | GO:0044026 | DNA hypermethylation(GO:0044026) |
| 0.3 | 2.0 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.3 | 7.3 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.3 | 2.0 | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.3 | 1.6 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.3 | 3.3 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.3 | 3.5 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.3 | 1.6 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.3 | 5.3 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.3 | 2.8 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.3 | 1.2 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.3 | 0.9 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.3 | 1.8 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.3 | 2.9 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.3 | 1.2 | GO:1900827 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.3 | 3.4 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.3 | 0.8 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.3 | 5.2 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.3 | 1.0 | GO:1901162 | primary amino compound biosynthetic process(GO:1901162) |
| 0.3 | 5.0 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.2 | 1.2 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.2 | 3.9 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.2 | 2.1 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.2 | 1.2 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.2 | 1.6 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.2 | 2.0 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.2 | 2.9 | GO:2000291 | regulation of pseudopodium assembly(GO:0031272) regulation of myoblast proliferation(GO:2000291) |
| 0.2 | 8.3 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.2 | 2.8 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.2 | 0.9 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.2 | 1.1 | GO:0010744 | positive regulation of macrophage derived foam cell differentiation(GO:0010744) |
| 0.2 | 1.5 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.2 | 1.5 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.2 | 1.7 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.2 | 2.1 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.2 | 4.7 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.2 | 5.9 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.2 | 0.7 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.2 | 1.9 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.2 | 4.6 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.2 | 1.4 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.2 | 3.3 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.2 | 3.0 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.2 | 5.3 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.1 | 2.5 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 2.1 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.1 | 1.0 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 | 6.5 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 0.6 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.1 | 2.1 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 1.0 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.1 | 2.4 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.1 | 0.4 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 0.1 | 0.8 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 1.8 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.1 | 4.0 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.1 | 3.2 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 | 1.6 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.1 | 1.5 | GO:1902373 | negative regulation of mRNA catabolic process(GO:1902373) |
| 0.1 | 1.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.1 | 1.0 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.1 | 1.7 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.1 | 1.7 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 3.1 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 1.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 2.0 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.1 | 0.9 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 12.2 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 0.6 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 1.3 | GO:0002517 | T cell tolerance induction(GO:0002517) |
| 0.1 | 2.8 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.1 | 5.4 | GO:0070268 | cornification(GO:0070268) |
| 0.1 | 0.4 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.1 | 1.7 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 1.0 | GO:0033622 | integrin activation(GO:0033622) |
| 0.1 | 1.2 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.1 | 2.5 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 0.3 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.1 | 1.5 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.1 | 3.2 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 1.0 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 3.0 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 2.8 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 8.0 | GO:0071346 | cellular response to interferon-gamma(GO:0071346) |
| 0.0 | 0.4 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 2.0 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 1.3 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 4.0 | GO:1990823 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 2.5 | GO:0061053 | somite development(GO:0061053) |
| 0.0 | 2.1 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 2.1 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.0 | 1.2 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 3.4 | GO:0048738 | cardiac muscle tissue development(GO:0048738) |
| 0.0 | 1.8 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.0 | 0.7 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 0.5 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 1.4 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 1.3 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 1.2 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 2.9 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 3.1 | GO:0007411 | axon guidance(GO:0007411) |
| 0.0 | 0.4 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 1.2 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.0 | 0.6 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.7 | GO:0070201 | regulation of establishment of protein localization(GO:0070201) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 14.8 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 3.4 | 13.4 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 2.0 | 27.5 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 1.2 | 3.6 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.9 | 5.3 | GO:1990357 | terminal web(GO:1990357) |
| 0.9 | 7.7 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.8 | 6.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.7 | 11.5 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.6 | 5.0 | GO:0045179 | apical cortex(GO:0045179) |
| 0.6 | 4.3 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.6 | 1.8 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.5 | 2.0 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.5 | 2.8 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.4 | 4.0 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.4 | 6.6 | GO:0030478 | actin cap(GO:0030478) |
| 0.4 | 3.4 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.3 | 14.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.3 | 3.0 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.2 | 1.2 | GO:0033655 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.2 | 3.9 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.2 | 8.3 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.2 | 8.5 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.2 | 4.0 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.2 | 25.1 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.2 | 3.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 1.0 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.2 | 5.0 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.2 | 3.5 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.2 | 2.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 1.5 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.1 | 10.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 7.9 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 12.5 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 1.7 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 0.5 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.1 | 2.9 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 13.2 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 2.1 | GO:0042599 | lamellar body(GO:0042599) |
| 0.1 | 2.8 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 1.8 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 1.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 1.5 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 1.9 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 2.6 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 6.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 3.3 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.1 | 5.7 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 1.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 2.3 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 9.3 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 8.5 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 2.0 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 4.1 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 5.8 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 2.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 86.8 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 2.1 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.0 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 2.4 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 3.0 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.8 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 3.3 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 1.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.5 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 6.1 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.4 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.4 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 1.4 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 1.7 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.6 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.7 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 3.0 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 1.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.8 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 2.0 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 1.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.6 | 16.9 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 2.5 | 9.9 | GO:0030760 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 2.1 | 8.5 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 2.0 | 5.9 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 1.3 | 6.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 1.2 | 4.8 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 1.2 | 3.5 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 1.0 | 2.9 | GO:0047977 | hepoxilin-epoxide hydrolase activity(GO:0047977) |
| 0.8 | 4.0 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.8 | 4.7 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.8 | 3.0 | GO:0016160 | amylase activity(GO:0016160) |
| 0.7 | 10.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.7 | 7.2 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.7 | 2.8 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.7 | 4.3 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.7 | 2.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.7 | 4.7 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.6 | 1.9 | GO:0046921 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.6 | 1.8 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.6 | 2.9 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.6 | 2.3 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.6 | 5.6 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.6 | 1.7 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.6 | 2.8 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.6 | 6.6 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.5 | 2.2 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.5 | 3.6 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.5 | 4.6 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.5 | 3.0 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.5 | 4.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.5 | 5.3 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.5 | 7.2 | GO:0016918 | retinal binding(GO:0016918) |
| 0.5 | 2.7 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.4 | 1.3 | GO:0015403 | thiamine transmembrane transporter activity(GO:0015234) thiamine uptake transmembrane transporter activity(GO:0015403) uptake transmembrane transporter activity(GO:0015563) |
| 0.4 | 10.7 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.4 | 2.5 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.4 | 3.7 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.4 | 2.8 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.4 | 1.9 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.4 | 7.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.4 | 27.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.4 | 5.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.3 | 5.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.3 | 2.8 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.3 | 1.3 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.3 | 3.9 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.3 | 1.2 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.3 | 0.9 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.3 | 1.8 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.3 | 3.1 | GO:0008061 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 0.3 | 6.5 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.3 | 3.7 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.3 | 5.5 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.3 | 0.8 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.3 | 2.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.3 | 4.1 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.3 | 1.5 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.2 | 1.7 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.2 | 3.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 3.6 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.2 | 2.0 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.2 | 2.9 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.2 | 4.0 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.2 | 4.0 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.2 | 3.5 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.2 | 0.6 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.2 | 1.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.2 | 1.1 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.2 | 9.7 | GO:0042805 | actinin binding(GO:0042805) |
| 0.2 | 4.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 7.5 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.2 | 3.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 3.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.2 | 17.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 5.5 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.2 | 4.1 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.2 | 4.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 1.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.2 | 1.2 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.1 | 2.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 3.2 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 3.0 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 2.9 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.1 | 8.9 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 14.4 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 2.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 6.0 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 33.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 1.0 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 1.8 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 2.1 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 1.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.1 | 1.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 1.6 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 2.8 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 1.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 2.4 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 2.7 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 1.0 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.1 | 4.5 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 1.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 2.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.2 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 1.9 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 1.0 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 2.3 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 0.5 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 4.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 2.6 | GO:0008200 | ion channel inhibitor activity(GO:0008200) |
| 0.1 | 3.0 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 2.5 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 0.6 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 1.3 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.1 | 1.0 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 1.0 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 31.2 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.3 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 4.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.4 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.3 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 3.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.4 | GO:0015266 | protein channel activity(GO:0015266) BH3 domain binding(GO:0051434) |
| 0.0 | 1.4 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 1.4 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 1.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 5.8 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.9 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 17.0 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 4.0 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 5.6 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 1.3 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 38.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.4 | 6.4 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.2 | 13.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.2 | 7.1 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.2 | 15.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.2 | 17.2 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.2 | 7.9 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.2 | 9.9 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.2 | 3.7 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.2 | 6.6 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.2 | 8.9 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.2 | 3.6 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.2 | 10.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 11.2 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 3.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 1.1 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 8.5 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 2.9 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 3.0 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 3.0 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 1.5 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 1.2 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.1 | 1.0 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 10.1 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 5.2 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 4.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.8 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.8 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.3 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 6.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 8.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.4 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 5.0 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.0 | 1.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.5 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 6.1 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 26.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.8 | 16.9 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.5 | 30.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.5 | 7.8 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.5 | 7.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.5 | 5.9 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.3 | 16.7 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.3 | 4.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.3 | 3.0 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.2 | 1.6 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.2 | 5.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 6.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.2 | 8.3 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.2 | 3.6 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.2 | 6.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 9.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 19.6 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.2 | 2.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.2 | 2.9 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.2 | 4.1 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.2 | 2.5 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.2 | 5.2 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 1.3 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.1 | 1.8 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 4.7 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.1 | 3.8 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.1 | 6.5 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 3.5 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.1 | 2.5 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.1 | 3.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.6 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 8.1 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 1.8 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 2.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 4.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 3.7 | REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | Genes involved in Integrin alphaIIb beta3 signaling |
| 0.1 | 2.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 3.5 | REACTOME ION CHANNEL TRANSPORT | Genes involved in Ion channel transport |
| 0.1 | 1.2 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 5.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 2.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.2 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 1.0 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 1.0 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 3.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.9 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 2.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 3.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 3.2 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 5.0 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 1.8 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 1.3 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 1.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 2.0 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.0 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.0 | 1.3 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.6 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |