Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
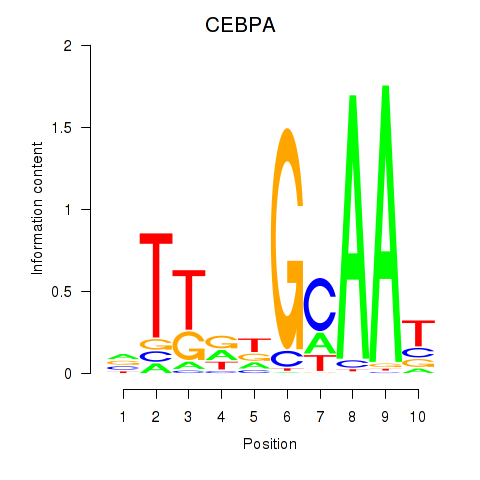
Results for CEBPA
Z-value: 1.42
Transcription factors associated with CEBPA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CEBPA
|
ENSG00000245848.2 | CCAAT enhancer binding protein alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CEBPA | hg19_v2_chr19_-_33793430_33793470 | 0.61 | 3.1e-23 | Click! |
Activity profile of CEBPA motif
Sorted Z-values of CEBPA motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_69742121 | 41.01 |
ENST00000261267.2
ENST00000549690.1 ENST00000548839.1 |
LYZ
|
lysozyme |
| chr20_-_43883197 | 39.77 |
ENST00000338380.2
|
SLPI
|
secretory leukocyte peptidase inhibitor |
| chr6_-_32557610 | 32.18 |
ENST00000360004.5
|
HLA-DRB1
|
major histocompatibility complex, class II, DR beta 1 |
| chr9_+_130911770 | 27.71 |
ENST00000372998.1
|
LCN2
|
lipocalin 2 |
| chr9_+_130911723 | 26.64 |
ENST00000277480.2
ENST00000373013.2 ENST00000540948.1 |
LCN2
|
lipocalin 2 |
| chr17_+_1665345 | 21.00 |
ENST00000576406.1
ENST00000571149.1 |
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr12_-_10007448 | 20.36 |
ENST00000538152.1
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr10_-_21786179 | 19.11 |
ENST00000377113.5
|
CASC10
|
cancer susceptibility candidate 10 |
| chr19_-_6720686 | 17.79 |
ENST00000245907.6
|
C3
|
complement component 3 |
| chr8_-_6837602 | 17.17 |
ENST00000382692.2
|
DEFA1
|
defensin, alpha 1 |
| chr11_-_5271122 | 17.06 |
ENST00000330597.3
|
HBG1
|
hemoglobin, gamma A |
| chr1_+_196621156 | 16.37 |
ENST00000359637.2
|
CFH
|
complement factor H |
| chr1_+_196621002 | 16.12 |
ENST00000367429.4
ENST00000439155.2 |
CFH
|
complement factor H |
| chr16_+_72088376 | 15.80 |
ENST00000570083.1
ENST00000355906.5 ENST00000398131.2 ENST00000569639.1 ENST00000564499.1 ENST00000357763.4 ENST00000562526.1 ENST00000565574.1 ENST00000568417.2 ENST00000356967.5 |
HP
HPR
|
haptoglobin haptoglobin-related protein |
| chr1_-_153348067 | 15.46 |
ENST00000368737.3
|
S100A12
|
S100 calcium binding protein A12 |
| chr21_-_46348694 | 14.92 |
ENST00000355153.4
ENST00000397850.2 |
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr1_+_22963158 | 14.63 |
ENST00000438241.1
|
C1QA
|
complement component 1, q subcomponent, A chain |
| chr14_+_95078714 | 14.55 |
ENST00000393078.3
ENST00000393080.4 ENST00000467132.1 |
SERPINA3
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 |
| chr12_-_123201337 | 14.39 |
ENST00000528880.2
|
HCAR3
|
hydroxycarboxylic acid receptor 3 |
| chr12_-_91573249 | 14.39 |
ENST00000550099.1
ENST00000546391.1 ENST00000551354.1 |
DCN
|
decorin |
| chr8_-_6875778 | 13.78 |
ENST00000535841.1
ENST00000327857.2 |
DEFA1B
DEFA3
|
defensin, alpha 1B defensin, alpha 3, neutrophil-specific |
| chr12_-_7656357 | 13.03 |
ENST00000396620.3
ENST00000432237.2 ENST00000359156.4 |
CD163
|
CD163 molecule |
| chr17_-_7017968 | 12.86 |
ENST00000355035.5
|
ASGR2
|
asialoglycoprotein receptor 2 |
| chr17_+_20059302 | 12.46 |
ENST00000395530.2
|
SPECC1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr4_-_185747188 | 12.39 |
ENST00000507295.1
ENST00000504900.1 ENST00000281455.2 ENST00000454703.2 |
ACSL1
|
acyl-CoA synthetase long-chain family member 1 |
| chr12_+_7167980 | 12.36 |
ENST00000360817.5
ENST00000402681.3 |
C1S
|
complement component 1, s subcomponent |
| chr8_+_12808834 | 12.27 |
ENST00000400069.3
|
KIAA1456
|
KIAA1456 |
| chr10_+_5005598 | 12.07 |
ENST00000442997.1
|
AKR1C1
|
aldo-keto reductase family 1, member C1 |
| chr12_-_123187890 | 12.02 |
ENST00000328880.5
|
HCAR2
|
hydroxycarboxylic acid receptor 2 |
| chr11_-_10590118 | 11.97 |
ENST00000529598.1
|
LYVE1
|
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr2_+_90060377 | 11.95 |
ENST00000436451.2
|
IGKV6D-21
|
immunoglobulin kappa variable 6D-21 (non-functional) |
| chr17_+_1665253 | 11.94 |
ENST00000254722.4
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr17_-_7018128 | 11.84 |
ENST00000380952.2
ENST00000254850.7 |
ASGR2
|
asialoglycoprotein receptor 2 |
| chr11_-_33913708 | 11.55 |
ENST00000257818.2
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr19_-_37019562 | 11.52 |
ENST00000523638.1
|
ZNF260
|
zinc finger protein 260 |
| chr14_-_23285011 | 11.49 |
ENST00000397532.3
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr12_-_91572278 | 11.17 |
ENST00000425043.1
ENST00000420120.2 ENST00000441303.2 ENST00000456569.2 |
DCN
|
decorin |
| chr6_+_30457244 | 10.70 |
ENST00000376630.4
|
HLA-E
|
major histocompatibility complex, class I, E |
| chr4_-_100242549 | 10.68 |
ENST00000305046.8
ENST00000394887.3 |
ADH1B
|
alcohol dehydrogenase 1B (class I), beta polypeptide |
| chrX_-_47489244 | 10.43 |
ENST00000469388.1
ENST00000396992.3 ENST00000377005.2 |
CFP
|
complement factor properdin |
| chr6_+_31895467 | 10.39 |
ENST00000556679.1
ENST00000456570.1 |
CFB
CFB
|
complement factor B Complement factor B; Uncharacterized protein; cDNA FLJ55673, highly similar to Complement factor B |
| chr10_+_5005445 | 10.15 |
ENST00000380872.4
|
AKR1C1
|
aldo-keto reductase family 1, member C1 |
| chr6_-_32920794 | 10.13 |
ENST00000395305.3
ENST00000395303.3 ENST00000374843.4 ENST00000429234.1 |
HLA-DMA
XXbac-BPG181M17.5
|
major histocompatibility complex, class II, DM alpha Uncharacterized protein |
| chr3_-_46249878 | 9.85 |
ENST00000296140.3
|
CCR1
|
chemokine (C-C motif) receptor 1 |
| chr10_-_70092671 | 9.72 |
ENST00000358769.2
ENST00000432941.1 ENST00000495025.2 |
PBLD
|
phenazine biosynthesis-like protein domain containing |
| chr16_-_55866997 | 9.64 |
ENST00000360526.3
ENST00000361503.4 |
CES1
|
carboxylesterase 1 |
| chr19_-_40971667 | 9.60 |
ENST00000263368.4
|
BLVRB
|
biliverdin reductase B (flavin reductase (NADPH)) |
| chr14_-_23285069 | 9.55 |
ENST00000554758.1
ENST00000397528.4 |
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr1_+_111415757 | 9.54 |
ENST00000429072.2
ENST00000271324.5 |
CD53
|
CD53 molecule |
| chr2_-_43453734 | 9.34 |
ENST00000282388.3
|
ZFP36L2
|
ZFP36 ring finger protein-like 2 |
| chr4_-_155533787 | 9.28 |
ENST00000407946.1
ENST00000405164.1 ENST00000336098.3 ENST00000393846.2 ENST00000404648.3 ENST00000443553.1 |
FGG
|
fibrinogen gamma chain |
| chr11_-_59633951 | 9.17 |
ENST00000257264.3
|
TCN1
|
transcobalamin I (vitamin B12 binding protein, R binder family) |
| chr2_+_102721023 | 8.94 |
ENST00000409589.1
ENST00000409329.1 |
IL1R1
|
interleukin 1 receptor, type I |
| chr17_+_56315936 | 8.87 |
ENST00000543544.1
|
LPO
|
lactoperoxidase |
| chr11_+_60050026 | 8.82 |
ENST00000395016.3
|
MS4A4A
|
membrane-spanning 4-domains, subfamily A, member 4A |
| chr12_-_12491608 | 8.80 |
ENST00000545735.1
|
MANSC1
|
MANSC domain containing 1 |
| chr6_-_133055815 | 8.44 |
ENST00000509351.1
ENST00000417437.2 ENST00000414302.2 ENST00000423615.2 ENST00000427187.2 ENST00000275223.3 ENST00000519686.2 |
VNN3
|
vanin 3 |
| chr19_-_40971643 | 8.43 |
ENST00000595483.1
|
BLVRB
|
biliverdin reductase B (flavin reductase (NADPH)) |
| chr10_-_45474237 | 8.32 |
ENST00000448778.1
ENST00000298295.3 |
C10orf10
|
chromosome 10 open reading frame 10 |
| chr10_+_97515409 | 8.27 |
ENST00000371207.3
ENST00000543964.1 |
ENTPD1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr17_-_34625719 | 8.21 |
ENST00000422211.2
ENST00000542124.1 |
CCL3L1
|
chemokine (C-C motif) ligand 3-like 1 |
| chr3_+_46395219 | 8.19 |
ENST00000445132.2
ENST00000292301.4 |
CCR2
|
chemokine (C-C motif) receptor 2 |
| chr12_-_96390063 | 7.97 |
ENST00000541929.1
|
HAL
|
histidine ammonia-lyase |
| chr11_-_10590238 | 7.84 |
ENST00000256178.3
|
LYVE1
|
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr19_+_56652556 | 7.76 |
ENST00000337080.3
|
ZNF444
|
zinc finger protein 444 |
| chr6_-_33037019 | 7.66 |
ENST00000437811.1
|
HLA-DPA1
|
major histocompatibility complex, class II, DP alpha 1 |
| chr5_-_180229833 | 7.62 |
ENST00000307826.4
|
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr11_-_104972158 | 7.60 |
ENST00000598974.1
ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1
CARD16
CARD17
|
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr15_-_64648273 | 7.57 |
ENST00000607537.1
ENST00000303052.7 ENST00000303032.6 |
CSNK1G1
|
casein kinase 1, gamma 1 |
| chr11_-_64510409 | 7.46 |
ENST00000394429.1
ENST00000394428.1 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr17_-_6983550 | 7.45 |
ENST00000576617.1
ENST00000416562.2 |
CLEC10A
|
C-type lectin domain family 10, member A |
| chr20_+_56136136 | 7.39 |
ENST00000319441.4
ENST00000543666.1 |
PCK1
|
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr8_-_6795823 | 7.31 |
ENST00000297435.2
|
DEFA4
|
defensin, alpha 4, corticostatin |
| chr1_+_171154347 | 7.20 |
ENST00000209929.7
ENST00000441535.1 |
FMO2
|
flavin containing monooxygenase 2 (non-functional) |
| chr3_+_186330712 | 7.13 |
ENST00000411641.2
ENST00000273784.5 |
AHSG
|
alpha-2-HS-glycoprotein |
| chr1_+_111770232 | 7.10 |
ENST00000369744.2
|
CHI3L2
|
chitinase 3-like 2 |
| chr3_+_186435137 | 7.09 |
ENST00000447445.1
|
KNG1
|
kininogen 1 |
| chr1_+_111770278 | 7.06 |
ENST00000369748.4
|
CHI3L2
|
chitinase 3-like 2 |
| chr5_+_49963239 | 6.92 |
ENST00000505554.1
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr4_+_74606223 | 6.89 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr1_-_153029980 | 6.77 |
ENST00000392653.2
|
SPRR2A
|
small proline-rich protein 2A |
| chr14_-_92414055 | 6.75 |
ENST00000342058.4
|
FBLN5
|
fibulin 5 |
| chr5_-_39219705 | 6.74 |
ENST00000351578.6
|
FYB
|
FYN binding protein |
| chr3_+_186435065 | 6.74 |
ENST00000287611.2
ENST00000265023.4 |
KNG1
|
kininogen 1 |
| chr16_+_56965960 | 6.70 |
ENST00000439977.2
ENST00000344114.4 ENST00000300302.5 ENST00000379792.2 |
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr6_+_31895480 | 6.68 |
ENST00000418949.2
ENST00000383177.3 ENST00000477310.1 |
C2
CFB
|
complement component 2 Complement factor B; Uncharacterized protein; cDNA FLJ55673, highly similar to Complement factor B |
| chr17_-_66951474 | 6.68 |
ENST00000269080.2
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr17_-_6983594 | 6.65 |
ENST00000571664.1
ENST00000254868.4 |
CLEC10A
|
C-type lectin domain family 10, member A |
| chr5_+_170736243 | 6.62 |
ENST00000296921.5
|
TLX3
|
T-cell leukemia homeobox 3 |
| chrX_+_12924732 | 6.55 |
ENST00000218032.6
ENST00000311912.5 |
TLR8
|
toll-like receptor 8 |
| chr9_+_125137565 | 6.49 |
ENST00000373698.5
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr16_+_2587965 | 6.48 |
ENST00000342085.4
ENST00000566659.1 |
PDPK1
|
3-phosphoinositide dependent protein kinase-1 |
| chr2_-_21266935 | 6.48 |
ENST00000233242.1
|
APOB
|
apolipoprotein B |
| chr20_+_46130671 | 6.45 |
ENST00000371998.3
ENST00000371997.3 |
NCOA3
|
nuclear receptor coactivator 3 |
| chr10_+_114133773 | 6.41 |
ENST00000354655.4
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5 |
| chr17_+_56315787 | 6.33 |
ENST00000262290.4
ENST00000421678.2 |
LPO
|
lactoperoxidase |
| chr6_-_133055896 | 6.23 |
ENST00000367927.5
ENST00000425515.2 ENST00000207771.3 ENST00000392393.3 ENST00000450865.2 ENST00000392394.2 |
VNN3
|
vanin 3 |
| chr6_+_150920999 | 6.23 |
ENST00000367328.1
ENST00000367326.1 |
PLEKHG1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr19_+_840963 | 6.20 |
ENST00000234347.5
|
PRTN3
|
proteinase 3 |
| chr2_-_175462456 | 6.13 |
ENST00000409891.1
ENST00000410117.1 |
WIPF1
|
WAS/WASL interacting protein family, member 1 |
| chr17_-_2415169 | 6.12 |
ENST00000263092.6
ENST00000538844.1 ENST00000576976.1 |
METTL16
|
methyltransferase like 16 |
| chr3_+_157154578 | 6.11 |
ENST00000295927.3
|
PTX3
|
pentraxin 3, long |
| chr19_-_48389651 | 6.07 |
ENST00000222002.3
|
SULT2A1
|
sulfotransferase family, cytosolic, 2A, dehydroepiandrosterone (DHEA)-preferring, member 1 |
| chr1_+_245133278 | 6.05 |
ENST00000366522.2
|
EFCAB2
|
EF-hand calcium binding domain 2 |
| chr12_-_10251576 | 6.03 |
ENST00000315330.4
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr3_-_151047327 | 5.97 |
ENST00000325602.5
|
P2RY13
|
purinergic receptor P2Y, G-protein coupled, 13 |
| chr19_-_54876558 | 5.96 |
ENST00000391742.2
ENST00000434277.2 |
LAIR1
|
leukocyte-associated immunoglobulin-like receptor 1 |
| chr17_-_4167142 | 5.95 |
ENST00000570535.1
ENST00000574367.1 ENST00000341657.4 ENST00000433651.1 |
ANKFY1
|
ankyrin repeat and FYVE domain containing 1 |
| chr6_-_49712147 | 5.93 |
ENST00000433368.2
ENST00000354620.4 |
CRISP3
|
cysteine-rich secretory protein 3 |
| chr14_+_21423611 | 5.91 |
ENST00000304625.2
|
RNASE2
|
ribonuclease, RNase A family, 2 (liver, eosinophil-derived neurotoxin) |
| chr8_+_27348649 | 5.83 |
ENST00000521780.1
ENST00000380476.3 ENST00000518379.1 ENST00000521684.1 |
EPHX2
|
epoxide hydrolase 2, cytoplasmic |
| chr1_+_117297007 | 5.83 |
ENST00000369478.3
ENST00000369477.1 |
CD2
|
CD2 molecule |
| chr19_+_852291 | 5.79 |
ENST00000263621.1
|
ELANE
|
elastase, neutrophil expressed |
| chr12_-_10251603 | 5.79 |
ENST00000457018.2
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr5_-_39219641 | 5.78 |
ENST00000509072.1
ENST00000504542.1 ENST00000505428.1 ENST00000506557.1 |
FYB
|
FYN binding protein |
| chr18_+_61554932 | 5.77 |
ENST00000299502.4
ENST00000457692.1 ENST00000413956.1 |
SERPINB2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
| chr6_+_106534192 | 5.75 |
ENST00000369091.2
ENST00000369096.4 |
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr3_+_40498783 | 5.72 |
ENST00000338970.6
ENST00000396203.2 ENST00000416518.1 |
RPL14
|
ribosomal protein L14 |
| chrX_+_16668278 | 5.70 |
ENST00000380200.3
|
S100G
|
S100 calcium binding protein G |
| chr2_-_175462934 | 5.65 |
ENST00000392546.2
ENST00000436221.1 |
WIPF1
|
WAS/WASL interacting protein family, member 1 |
| chr20_+_46130619 | 5.63 |
ENST00000372004.3
|
NCOA3
|
nuclear receptor coactivator 3 |
| chr20_+_46130601 | 5.61 |
ENST00000341724.6
|
NCOA3
|
nuclear receptor coactivator 3 |
| chr19_+_49258775 | 5.57 |
ENST00000593756.1
|
FGF21
|
fibroblast growth factor 21 |
| chr6_+_31895254 | 5.52 |
ENST00000299367.5
ENST00000442278.2 |
C2
|
complement component 2 |
| chr12_-_96390108 | 5.51 |
ENST00000538703.1
ENST00000261208.3 |
HAL
|
histidine ammonia-lyase |
| chr12_+_54674482 | 5.50 |
ENST00000547708.1
ENST00000340913.6 ENST00000551702.1 ENST00000330752.8 ENST00000547276.1 |
HNRNPA1
|
heterogeneous nuclear ribonucleoprotein A1 |
| chr2_+_163200598 | 5.50 |
ENST00000437150.2
ENST00000453113.2 |
GCA
|
grancalcin, EF-hand calcium binding protein |
| chr11_+_60048053 | 5.50 |
ENST00000337908.4
|
MS4A4A
|
membrane-spanning 4-domains, subfamily A, member 4A |
| chr4_+_71296204 | 5.44 |
ENST00000413702.1
|
MUC7
|
mucin 7, secreted |
| chr5_-_64920115 | 5.44 |
ENST00000381018.3
ENST00000274327.7 |
TRIM23
|
tripartite motif containing 23 |
| chr11_-_18258342 | 5.42 |
ENST00000278222.4
|
SAA4
|
serum amyloid A4, constitutive |
| chr5_-_130970723 | 5.38 |
ENST00000308008.6
ENST00000296859.6 ENST00000507093.1 ENST00000510071.1 ENST00000509018.1 ENST00000307984.5 |
RAPGEF6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr16_+_2588012 | 5.38 |
ENST00000354836.5
ENST00000389224.3 |
PDPK1
|
3-phosphoinositide dependent protein kinase-1 |
| chr3_+_48264816 | 5.35 |
ENST00000296435.2
ENST00000576243.1 |
CAMP
|
cathelicidin antimicrobial peptide |
| chr11_+_116700614 | 5.33 |
ENST00000375345.1
|
APOC3
|
apolipoprotein C-III |
| chr1_-_186649543 | 5.31 |
ENST00000367468.5
|
PTGS2
|
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) |
| chrX_+_15767971 | 5.24 |
ENST00000479740.1
ENST00000454127.2 |
CA5B
|
carbonic anhydrase VB, mitochondrial |
| chr19_+_55014085 | 5.22 |
ENST00000351841.2
|
LAIR2
|
leukocyte-associated immunoglobulin-like receptor 2 |
| chr1_+_101185290 | 5.21 |
ENST00000370119.4
ENST00000347652.2 ENST00000294728.2 ENST00000370115.1 |
VCAM1
|
vascular cell adhesion molecule 1 |
| chr1_-_200992827 | 5.21 |
ENST00000332129.2
ENST00000422435.2 |
KIF21B
|
kinesin family member 21B |
| chr4_+_159593418 | 5.18 |
ENST00000507475.1
ENST00000307738.5 |
ETFDH
|
electron-transferring-flavoprotein dehydrogenase |
| chr12_-_114841703 | 5.17 |
ENST00000526441.1
|
TBX5
|
T-box 5 |
| chr2_-_211179883 | 5.16 |
ENST00000352451.3
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast |
| chr11_+_116700600 | 5.12 |
ENST00000227667.3
|
APOC3
|
apolipoprotein C-III |
| chr11_+_67806467 | 5.12 |
ENST00000265686.3
ENST00000524598.1 ENST00000529657.1 |
TCIRG1
|
T-cell, immune regulator 1, ATPase, H+ transporting, lysosomal V0 subunit A3 |
| chr15_+_58430368 | 5.08 |
ENST00000558772.1
ENST00000219919.4 |
AQP9
|
aquaporin 9 |
| chr17_-_34524157 | 4.99 |
ENST00000378354.4
ENST00000394484.1 |
CCL3L3
|
chemokine (C-C motif) ligand 3-like 3 |
| chr20_-_1600642 | 4.98 |
ENST00000381603.3
ENST00000381605.4 ENST00000279477.7 ENST00000568365.1 ENST00000564763.1 |
SIRPB1
RP4-576H24.4
|
signal-regulatory protein beta 1 Uncharacterized protein |
| chr10_+_26727125 | 4.95 |
ENST00000376236.4
|
APBB1IP
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr6_-_49712123 | 4.94 |
ENST00000263045.4
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr3_+_10857885 | 4.94 |
ENST00000254488.2
ENST00000454147.1 |
SLC6A11
|
solute carrier family 6 (neurotransmitter transporter), member 11 |
| chr1_-_92952433 | 4.91 |
ENST00000294702.5
|
GFI1
|
growth factor independent 1 transcription repressor |
| chr22_+_35776828 | 4.86 |
ENST00000216117.8
|
HMOX1
|
heme oxygenase (decycling) 1 |
| chr1_+_196743912 | 4.83 |
ENST00000367425.4
|
CFHR3
|
complement factor H-related 3 |
| chr12_-_71551868 | 4.80 |
ENST00000247829.3
|
TSPAN8
|
tetraspanin 8 |
| chr11_+_59824060 | 4.76 |
ENST00000395032.2
ENST00000358152.2 |
MS4A3
|
membrane-spanning 4-domains, subfamily A, member 3 (hematopoietic cell-specific) |
| chr19_+_55014013 | 4.73 |
ENST00000301202.2
|
LAIR2
|
leukocyte-associated immunoglobulin-like receptor 2 |
| chr22_-_30901637 | 4.73 |
ENST00000381982.3
ENST00000255858.7 ENST00000540456.1 ENST00000392772.2 |
SEC14L4
|
SEC14-like 4 (S. cerevisiae) |
| chr10_+_17270214 | 4.69 |
ENST00000544301.1
|
VIM
|
vimentin |
| chr11_+_60048129 | 4.63 |
ENST00000355131.3
|
MS4A4A
|
membrane-spanning 4-domains, subfamily A, member 4A |
| chr6_-_27782548 | 4.59 |
ENST00000333151.3
|
HIST1H2AJ
|
histone cluster 1, H2aj |
| chr19_+_6887571 | 4.59 |
ENST00000250572.8
ENST00000381407.5 ENST00000312053.4 ENST00000450315.3 ENST00000381404.4 |
EMR1
|
egf-like module containing, mucin-like, hormone receptor-like 1 |
| chr6_+_27782788 | 4.54 |
ENST00000359465.4
|
HIST1H2BM
|
histone cluster 1, H2bm |
| chrX_+_15518923 | 4.50 |
ENST00000348343.6
|
BMX
|
BMX non-receptor tyrosine kinase |
| chr13_-_72441315 | 4.43 |
ENST00000305425.4
ENST00000313174.7 ENST00000354591.4 |
DACH1
|
dachshund homolog 1 (Drosophila) |
| chr3_-_49726486 | 4.43 |
ENST00000449682.2
|
MST1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr13_-_46679185 | 4.41 |
ENST00000439329.3
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr3_-_194072019 | 4.41 |
ENST00000429275.1
ENST00000323830.3 |
CPN2
|
carboxypeptidase N, polypeptide 2 |
| chr5_-_159739532 | 4.36 |
ENST00000520748.1
ENST00000393977.3 ENST00000257536.7 |
CCNJL
|
cyclin J-like |
| chr1_+_207277590 | 4.30 |
ENST00000367070.3
|
C4BPA
|
complement component 4 binding protein, alpha |
| chr13_-_46679144 | 4.30 |
ENST00000181383.4
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr11_+_59824127 | 4.29 |
ENST00000278865.3
|
MS4A3
|
membrane-spanning 4-domains, subfamily A, member 3 (hematopoietic cell-specific) |
| chr6_+_117002339 | 4.25 |
ENST00000413340.1
ENST00000368564.1 ENST00000356348.1 |
KPNA5
|
karyopherin alpha 5 (importin alpha 6) |
| chr4_-_186877502 | 4.24 |
ENST00000431902.1
ENST00000284776.7 ENST00000415274.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr11_-_104905840 | 4.24 |
ENST00000526568.1
ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1
|
caspase 1, apoptosis-related cysteine peptidase |
| chr1_-_153321301 | 4.23 |
ENST00000368739.3
|
PGLYRP4
|
peptidoglycan recognition protein 4 |
| chr1_-_163172625 | 4.21 |
ENST00000527988.1
ENST00000531476.1 ENST00000530507.1 |
RGS5
|
regulator of G-protein signaling 5 |
| chr12_-_110434096 | 4.20 |
ENST00000320063.9
ENST00000457474.2 ENST00000547815.1 ENST00000361006.5 |
GIT2
|
G protein-coupled receptor kinase interacting ArfGAP 2 |
| chr4_-_119274121 | 4.16 |
ENST00000296498.3
|
PRSS12
|
protease, serine, 12 (neurotrypsin, motopsin) |
| chr5_+_49962772 | 4.13 |
ENST00000281631.5
ENST00000513738.1 ENST00000503665.1 ENST00000514067.2 ENST00000503046.1 |
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr9_+_33290491 | 4.11 |
ENST00000379540.3
ENST00000379521.4 ENST00000318524.6 |
NFX1
|
nuclear transcription factor, X-box binding 1 |
| chr13_-_33112956 | 4.11 |
ENST00000505213.1
|
N4BP2L2
|
NEDD4 binding protein 2-like 2 |
| chr6_+_29910301 | 4.11 |
ENST00000376809.5
ENST00000376802.2 |
HLA-A
|
major histocompatibility complex, class I, A |
| chr10_+_114710516 | 4.00 |
ENST00000542695.1
ENST00000346198.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr2_+_89999259 | 3.98 |
ENST00000558026.1
|
IGKV2D-28
|
immunoglobulin kappa variable 2D-28 |
| chr2_+_71693812 | 3.96 |
ENST00000409651.1
ENST00000394120.2 ENST00000409744.1 ENST00000409366.1 ENST00000410020.3 ENST00000410041.1 |
DYSF
|
dysferlin |
| chr13_-_99667960 | 3.95 |
ENST00000448493.2
|
DOCK9
|
dedicator of cytokinesis 9 |
| chr16_+_82090028 | 3.87 |
ENST00000568090.1
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr11_-_130786400 | 3.83 |
ENST00000265909.4
|
SNX19
|
sorting nexin 19 |
| chr12_-_110434021 | 3.80 |
ENST00000355312.3
ENST00000551209.1 ENST00000550186.1 |
GIT2
|
G protein-coupled receptor kinase interacting ArfGAP 2 |
| chr1_+_209941827 | 3.80 |
ENST00000367023.1
|
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr14_+_22446680 | 3.79 |
ENST00000390443.3
|
TRAV8-6
|
T cell receptor alpha variable 8-6 |
| chr3_-_158450231 | 3.77 |
ENST00000479756.1
|
RARRES1
|
retinoic acid receptor responder (tazarotene induced) 1 |
| chr18_-_28622774 | 3.75 |
ENST00000434452.1
|
DSC3
|
desmocollin 3 |
| chr22_-_22307199 | 3.73 |
ENST00000397495.4
ENST00000263212.5 |
PPM1F
|
protein phosphatase, Mg2+/Mn2+ dependent, 1F |
| chr18_-_28622699 | 3.71 |
ENST00000360428.4
|
DSC3
|
desmocollin 3 |
| chr1_+_119957554 | 3.71 |
ENST00000543831.1
ENST00000433745.1 ENST00000369416.3 |
HSD3B2
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 |
| chr1_+_209941942 | 3.71 |
ENST00000487271.1
ENST00000477431.1 |
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr11_-_18270182 | 3.69 |
ENST00000528349.1
ENST00000526900.1 ENST00000529528.1 ENST00000414546.2 ENST00000256733.4 |
SAA2
|
serum amyloid A2 |
| chr10_-_70092635 | 3.68 |
ENST00000309049.4
|
PBLD
|
phenazine biosynthesis-like protein domain containing |
Network of associatons between targets according to the STRING database.
First level regulatory network of CEBPA
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.9 | 54.3 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 8.5 | 42.3 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 8.4 | 42.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 6.6 | 32.9 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 5.9 | 17.8 | GO:0001798 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 5.7 | 17.2 | GO:0035915 | pore formation in membrane of other organism(GO:0035915) |
| 5.3 | 15.8 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 4.5 | 13.5 | GO:0019556 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 3.6 | 10.7 | GO:0032759 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) TRAIL production(GO:0032639) regulation of TRAIL production(GO:0032679) positive regulation of TRAIL production(GO:0032759) positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 3.5 | 14.1 | GO:1904640 | response to methionine(GO:1904640) |
| 3.5 | 21.0 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) basic amino acid transmembrane transport(GO:1990822) |
| 3.5 | 10.4 | GO:2000910 | regulation of cholesterol import(GO:0060620) negative regulation of cholesterol import(GO:0060621) regulation of sterol import(GO:2000909) negative regulation of sterol import(GO:2000910) |
| 3.3 | 13.4 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 3.2 | 22.2 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 3.0 | 8.9 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 3.0 | 11.8 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 2.7 | 8.2 | GO:2000439 | negative regulation of eosinophil activation(GO:1902567) positive regulation of monocyte extravasation(GO:2000439) |
| 2.7 | 62.8 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 2.5 | 17.7 | GO:0035624 | receptor transactivation(GO:0035624) |
| 2.4 | 12.0 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 2.2 | 8.7 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 2.1 | 12.4 | GO:0034201 | response to oleic acid(GO:0034201) |
| 2.0 | 24.0 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 1.9 | 7.6 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 1.8 | 5.3 | GO:0031394 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 1.7 | 5.2 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 1.7 | 5.1 | GO:0030264 | nuclear fragmentation involved in apoptotic nuclear change(GO:0030264) positive regulation of cardiac vascular smooth muscle cell differentiation(GO:2000724) |
| 1.7 | 25.4 | GO:0042167 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 1.7 | 77.9 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 1.6 | 6.6 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 1.6 | 6.6 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 1.6 | 4.9 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 1.6 | 17.7 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 1.6 | 6.3 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 1.6 | 9.3 | GO:1904628 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 1.5 | 15.3 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 1.5 | 6.1 | GO:0044793 | negative regulation by host of viral process(GO:0044793) |
| 1.5 | 5.8 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 1.4 | 4.2 | GO:0051710 | regulation of cytolysis in other organism(GO:0051710) |
| 1.4 | 5.5 | GO:1903936 | response to sodium arsenite(GO:1903935) cellular response to sodium arsenite(GO:1903936) |
| 1.4 | 12.3 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 1.3 | 3.8 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 1.2 | 3.7 | GO:0002590 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) regulation of T cell antigen processing and presentation(GO:0002625) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 1.2 | 6.1 | GO:0080009 | mRNA methylation(GO:0080009) |
| 1.2 | 3.6 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 1.2 | 5.8 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 1.2 | 5.8 | GO:0033078 | extrathymic T cell differentiation(GO:0033078) |
| 1.1 | 3.4 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) negative regulation of DNA catabolic process(GO:1903625) regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 1.1 | 17.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 1.1 | 3.4 | GO:0055099 | detection of endogenous stimulus(GO:0009726) response to high density lipoprotein particle(GO:0055099) |
| 1.1 | 3.3 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 1.1 | 2.2 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 1.1 | 3.2 | GO:0018874 | benzoate metabolic process(GO:0018874) butyrate metabolic process(GO:0019605) |
| 1.0 | 13.5 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 1.0 | 6.2 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 1.0 | 5.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) pyrimidine nucleobase transport(GO:0015855) urea transmembrane transport(GO:0071918) purine nucleobase transmembrane transport(GO:1904823) |
| 1.0 | 10.1 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 1.0 | 8.0 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 1.0 | 3.0 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 1.0 | 6.0 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 1.0 | 6.9 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 1.0 | 9.6 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 1.0 | 2.9 | GO:0046136 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.9 | 3.8 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.9 | 3.8 | GO:1902159 | regulation of cyclic nucleotide-gated ion channel activity(GO:1902159) |
| 0.9 | 6.5 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.9 | 2.8 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.9 | 9.2 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.9 | 2.7 | GO:2001248 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.9 | 2.7 | GO:0019474 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.9 | 5.2 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.9 | 14.5 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.9 | 3.4 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.8 | 20.0 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.8 | 14.9 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.8 | 2.5 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.8 | 1.6 | GO:1900736 | regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900736) |
| 0.8 | 23.1 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.8 | 2.3 | GO:1901877 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.8 | 2.3 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.7 | 6.7 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.7 | 4.3 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.7 | 3.5 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.7 | 6.3 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.7 | 2.7 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.7 | 19.8 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.7 | 7.4 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.7 | 3.4 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.7 | 4.0 | GO:0044334 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.6 | 15.5 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.6 | 2.5 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.6 | 5.5 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.6 | 4.5 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.6 | 1.7 | GO:0097052 | tryptophan catabolic process to acetyl-CoA(GO:0019442) L-kynurenine metabolic process(GO:0097052) |
| 0.6 | 12.8 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.6 | 4.4 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.6 | 14.9 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.5 | 3.8 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.5 | 1.6 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.5 | 1.6 | GO:0097460 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.5 | 3.1 | GO:0071321 | positive regulation of oocyte development(GO:0060282) cellular response to cGMP(GO:0071321) |
| 0.5 | 8.3 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.5 | 4.1 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.5 | 1.5 | GO:0015888 | thiamine transport(GO:0015888) thiamine transmembrane transport(GO:0071934) |
| 0.5 | 11.6 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.5 | 4.9 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.5 | 9.4 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.5 | 2.3 | GO:1900920 | regulation of amino acid import(GO:0010958) regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 0.5 | 0.5 | GO:0046668 | regulation of retinal cell programmed cell death(GO:0046668) |
| 0.5 | 23.8 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.5 | 16.8 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.4 | 7.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.4 | 3.5 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.4 | 6.6 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.4 | 4.3 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.4 | 24.7 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.4 | 2.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.4 | 3.3 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.4 | 7.5 | GO:0006068 | ethanol metabolic process(GO:0006067) ethanol catabolic process(GO:0006068) |
| 0.4 | 3.7 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.4 | 3.0 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.4 | 5.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.4 | 5.8 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.4 | 2.1 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.4 | 14.7 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.3 | 2.1 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.3 | 0.7 | GO:0032730 | positive regulation of interleukin-1 alpha production(GO:0032730) |
| 0.3 | 5.4 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.3 | 2.7 | GO:0000075 | cell cycle checkpoint(GO:0000075) |
| 0.3 | 1.3 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.3 | 4.0 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.3 | 2.2 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.3 | 4.4 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.3 | 3.7 | GO:0006702 | androgen biosynthetic process(GO:0006702) |
| 0.3 | 7.5 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.3 | 3.6 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.3 | 1.8 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.3 | 6.0 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.3 | 10.4 | GO:0003341 | cilium movement(GO:0003341) |
| 0.3 | 11.4 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.3 | 6.8 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.3 | 2.8 | GO:0002475 | antigen processing and presentation via MHC class Ib(GO:0002475) |
| 0.3 | 7.0 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.3 | 8.1 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.3 | 3.3 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.3 | 5.4 | GO:0031640 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) |
| 0.3 | 2.7 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.3 | 3.6 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.3 | 3.1 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.3 | 3.9 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.2 | 2.0 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.2 | 1.6 | GO:0072501 | cellular divalent inorganic anion homeostasis(GO:0072501) |
| 0.2 | 0.9 | GO:0050705 | regulation of interleukin-1 alpha secretion(GO:0050705) |
| 0.2 | 0.2 | GO:0043379 | memory T cell differentiation(GO:0043379) |
| 0.2 | 1.3 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.2 | 0.4 | GO:0050955 | thermoception(GO:0050955) |
| 0.2 | 1.9 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.2 | 1.6 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.2 | 14.7 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.2 | 0.8 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.2 | 1.2 | GO:0010615 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.2 | 2.3 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.2 | 1.8 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.2 | 1.8 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.2 | 1.9 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.2 | 4.7 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.2 | 1.6 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.2 | 0.5 | GO:0014859 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.2 | 0.5 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.2 | 2.0 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.2 | 1.0 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.2 | 0.7 | GO:0030177 | positive regulation of Wnt signaling pathway(GO:0030177) positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.2 | 0.8 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.2 | 3.0 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.1 | 14.0 | GO:2000257 | regulation of complement activation(GO:0030449) regulation of protein activation cascade(GO:2000257) |
| 0.1 | 1.0 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.1 | 0.7 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.1 | 0.8 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 2.1 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 0.9 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 5.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 2.5 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.1 | 3.0 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.1 | 2.3 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.1 | 0.7 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.1 | 1.7 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 0.4 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.1 | 0.2 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.1 | 0.3 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.1 | 7.5 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.1 | 5.0 | GO:0032729 | positive regulation of interferon-gamma production(GO:0032729) |
| 0.1 | 11.3 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 1.3 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.1 | 5.2 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.1 | 0.2 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 | 0.3 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 5.7 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.1 | 0.9 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 0.7 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.1 | 0.5 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.1 | 7.3 | GO:0070268 | cornification(GO:0070268) |
| 0.1 | 3.2 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.1 | 2.3 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.1 | 2.0 | GO:0060119 | inner ear receptor cell development(GO:0060119) |
| 0.1 | 1.2 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 4.5 | GO:0007588 | excretion(GO:0007588) |
| 0.1 | 2.1 | GO:0048477 | oogenesis(GO:0048477) |
| 0.1 | 3.5 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.8 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 1.4 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 1.8 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 4.5 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 0.6 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 13.8 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.0 | 2.7 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.7 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 4.3 | GO:0002433 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.0 | 0.4 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 5.5 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.8 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.5 | GO:0050961 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 | 0.5 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.0 | 0.7 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 2.0 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 5.3 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 3.1 | GO:0046718 | viral entry into host cell(GO:0046718) |
| 0.0 | 0.5 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.5 | GO:0007184 | SMAD protein import into nucleus(GO:0007184) |
| 0.0 | 0.6 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.0 | 1.0 | GO:0002763 | positive regulation of myeloid leukocyte differentiation(GO:0002763) |
| 0.0 | 1.5 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.6 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 15.6 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 1.2 | GO:0055001 | muscle cell development(GO:0055001) |
| 0.0 | 0.6 | GO:0060512 | prostate gland morphogenesis(GO:0060512) |
| 0.0 | 1.3 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.7 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 0.2 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.2 | GO:0000733 | DNA strand renaturation(GO:0000733) t-circle formation(GO:0090656) |
| 0.0 | 3.5 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.4 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.1 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 1.1 | GO:0042552 | myelination(GO:0042552) |
| 0.0 | 0.6 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 1.6 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 0.2 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 1.5 | GO:0061025 | membrane fusion(GO:0061025) |
| 0.0 | 0.5 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.4 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 15.8 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 3.0 | 14.9 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 2.9 | 14.6 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 2.6 | 31.0 | GO:0043203 | axon hillock(GO:0043203) |
| 2.4 | 50.0 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 2.4 | 11.8 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 2.1 | 12.9 | GO:1990037 | Lewy body core(GO:1990037) |
| 2.1 | 16.9 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 1.9 | 177.2 | GO:0035580 | specific granule lumen(GO:0035580) |
| 1.9 | 18.5 | GO:0042611 | MHC protein complex(GO:0042611) |
| 1.8 | 25.6 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 1.8 | 5.4 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 1.7 | 5.2 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 1.4 | 17.1 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 1.2 | 6.0 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.9 | 6.6 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.9 | 84.3 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.7 | 6.7 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.7 | 9.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.7 | 4.0 | GO:0071664 | beta-catenin-TCF7L2 complex(GO:0070369) catenin-TCF7L2 complex(GO:0071664) |
| 0.7 | 3.3 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.6 | 14.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.6 | 2.3 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.5 | 111.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.4 | 4.9 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.4 | 5.1 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.4 | 3.7 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.4 | 1.6 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.4 | 1.6 | GO:0033655 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.3 | 2.4 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.3 | 6.6 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.3 | 3.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.3 | 4.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.3 | 3.5 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.3 | 1.8 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.3 | 2.3 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.2 | 7.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.2 | 28.1 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.2 | 1.6 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.2 | 0.9 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.2 | 12.3 | GO:0000786 | nucleosome(GO:0000786) |
| 0.2 | 5.9 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.2 | 5.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.2 | 5.2 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.2 | 4.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.2 | 2.8 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.2 | 5.2 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.2 | 0.5 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.2 | 1.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 13.0 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 11.5 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 2.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 14.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.7 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 9.3 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.1 | 3.0 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 8.5 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 2.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 1.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 31.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 2.0 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 10.9 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 14.8 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 1.9 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 10.4 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 13.0 | GO:0030666 | endocytic vesicle membrane(GO:0030666) |
| 0.1 | 1.0 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 15.7 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 7.3 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 4.6 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 7.5 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 4.5 | GO:0030286 | dynein complex(GO:0030286) |
| 0.1 | 5.3 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 2.0 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.1 | 4.2 | GO:0044216 | host(GO:0018995) host cell(GO:0043657) other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.1 | 1.6 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 19.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 2.0 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 3.5 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 3.0 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.1 | 5.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 5.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 4.7 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 1.0 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 5.7 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 3.0 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 1.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.7 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 59.3 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 17.4 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 4.0 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 2.7 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.1 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.0 | 19.7 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.6 | GO:0048786 | presynaptic membrane(GO:0042734) presynaptic active zone(GO:0048786) |
| 0.0 | 38.2 | GO:0016021 | integral component of membrane(GO:0016021) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.0 | 42.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 6.5 | 26.1 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 4.5 | 18.0 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 4.0 | 11.9 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 3.4 | 13.5 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 3.1 | 24.7 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 3.0 | 11.8 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 2.9 | 14.7 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 2.7 | 8.2 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 2.6 | 15.8 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 2.6 | 10.4 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 2.5 | 7.4 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 2.5 | 12.3 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 2.2 | 8.9 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 2.1 | 14.9 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 2.0 | 6.1 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 1.9 | 9.6 | GO:0004771 | sterol esterase activity(GO:0004771) methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 1.9 | 21.0 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 1.9 | 7.6 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 1.8 | 5.5 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 1.8 | 17.7 | GO:0004568 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 1.7 | 5.1 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 1.7 | 10.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 1.6 | 4.9 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 1.6 | 6.5 | GO:0035473 | lipase binding(GO:0035473) |
| 1.6 | 58.4 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 1.6 | 17.1 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 1.5 | 3.1 | GO:0019862 | IgA binding(GO:0019862) |
| 1.4 | 4.2 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 1.4 | 9.7 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 1.3 | 32.5 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 1.3 | 5.1 | GO:0015254 | glycerol channel activity(GO:0015254) urea channel activity(GO:0015265) |
| 1.2 | 3.7 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) steroid delta-isomerase activity(GO:0004769) |
| 1.1 | 3.4 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 1.1 | 4.5 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 1.1 | 6.3 | GO:1990254 | keratin filament binding(GO:1990254) |
| 1.0 | 3.1 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 1.0 | 5.9 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 1.0 | 5.8 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 1.0 | 5.7 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.9 | 2.7 | GO:1902122 | bile acid receptor activity(GO:0038181) chenodeoxycholic acid binding(GO:1902122) |
| 0.9 | 8.0 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.9 | 95.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.8 | 11.8 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.8 | 6.7 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.8 | 9.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.8 | 14.7 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.8 | 1.6 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.8 | 3.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.8 | 15.5 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.8 | 2.3 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.8 | 9.9 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.7 | 23.1 | GO:0001848 | complement binding(GO:0001848) |
| 0.7 | 4.5 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.7 | 5.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.6 | 17.7 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.6 | 3.8 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.6 | 18.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.6 | 3.0 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.6 | 3.6 | GO:0015185 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.6 | 8.3 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.6 | 1.2 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.5 | 1.6 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.5 | 5.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.5 | 1.5 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) thiamine uptake transmembrane transporter activity(GO:0015403) uptake transmembrane transporter activity(GO:0015563) azole transmembrane transporter activity(GO:1901474) |
| 0.5 | 8.7 | GO:0005522 | profilin binding(GO:0005522) |
| 0.5 | 15.1 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.5 | 2.7 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.4 | 2.7 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.4 | 1.3 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.4 | 2.2 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.4 | 27.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.4 | 9.7 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.4 | 1.7 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.4 | 7.5 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.4 | 59.7 | GO:0002020 | protease binding(GO:0002020) |
| 0.4 | 1.6 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.4 | 3.9 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.4 | 3.7 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.4 | 20.0 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.4 | 10.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.4 | 11.0 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.4 | 3.2 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.3 | 1.0 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.3 | 6.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.3 | 4.4 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.3 | 1.6 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.3 | 5.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.3 | 3.4 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.3 | 27.7 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.3 | 6.0 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.3 | 9.5 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.3 | 2.7 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.3 | 2.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.3 | 10.6 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.3 | 1.5 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.3 | 0.9 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.3 | 2.8 | GO:0030883 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.3 | 15.2 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.3 | 4.5 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.3 | 5.9 | GO:0016722 | oxidoreductase activity, oxidizing metal ions(GO:0016722) |
| 0.3 | 0.8 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.3 | 2.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.2 | 3.5 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.2 | 56.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.2 | 0.7 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.2 | 2.9 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.2 | 5.9 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.2 | 11.6 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.2 | 7.0 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.2 | 0.4 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.2 | 2.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.2 | 0.9 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.2 | 2.6 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.2 | 0.5 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.2 | 15.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.2 | 2.8 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.2 | 0.7 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.2 | 3.4 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.2 | 5.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 1.4 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.2 | 0.5 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.2 | 5.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.2 | 1.8 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.1 | 3.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 2.9 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 0.8 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.1 | 1.4 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 4.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 30.5 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 3.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 8.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 2.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 4.0 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 1.7 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 14.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 2.3 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.1 | 0.7 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.1 | 1.3 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.1 | 2.0 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 2.8 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 2.3 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 0.9 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 0.4 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.1 | 13.3 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 6.1 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 4.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 6.9 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 1.0 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 2.5 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 8.0 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 5.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 4.6 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 0.4 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 0.7 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.1 | 15.3 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 0.2 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.1 | 22.7 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.1 | 7.7 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 0.9 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.8 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.9 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.5 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.2 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0032145 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.0 | 1.4 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.2 | GO:0036310 | annealing helicase activity(GO:0036310) annealing activity(GO:0097617) |
| 0.0 | 0.2 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 2.4 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 2.3 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 0.5 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 35.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.9 | 46.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.7 | 22.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.7 | 33.7 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.6 | 15.4 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.6 | 39.0 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.5 | 9.6 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.4 | 54.9 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.4 | 11.9 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.4 | 95.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.4 | 64.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.3 | 11.8 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.3 | 12.3 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.3 | 10.9 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.3 | 2.3 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.3 | 9.6 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 4.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 9.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 8.5 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.1 | 3.5 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 2.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 4.6 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 7.0 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 8.7 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 7.6 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 39.1 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.1 | 1.8 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.1 | 2.7 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 1.4 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 7.2 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 3.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 3.3 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 4.0 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 6.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 1.7 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 4.5 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 5.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 3.0 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 0.8 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.1 | 0.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.9 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.0 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.7 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.0 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.9 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.1 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.5 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.2 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.5 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.8 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 64.5 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 2.6 | 46.4 | REACTOME DEFENSINS | Genes involved in Defensins |
| 2.2 | 43.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 1.8 | 39.8 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 1.3 | 10.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 1.2 | 25.6 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 1.0 | 19.8 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.9 | 18.8 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.8 | 12.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.8 | 11.8 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.8 | 16.9 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.8 | 13.8 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.7 | 18.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.7 | 59.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.6 | 13.6 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.6 | 17.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.6 | 15.5 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.6 | 7.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.6 | 35.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.5 | 6.6 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.5 | 4.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.4 | 3.7 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.4 | 22.9 | REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | Genes involved in Integrin alphaIIb beta3 signaling |
| 0.4 | 5.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.3 | 5.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.3 | 9.7 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.3 | 6.7 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.3 | 9.5 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.3 | 6.0 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.3 | 2.8 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.3 | 23.8 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.3 | 5.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.3 | 3.4 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.3 | 20.1 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.3 | 5.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.2 | 7.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 3.9 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.2 | 3.0 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.2 | 8.4 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.2 | 0.8 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.2 | 7.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 2.2 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.2 | 4.5 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.2 | 8.6 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.2 | 3.6 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.2 | 5.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 25.4 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 2.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 7.4 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.1 | 0.7 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.1 | 1.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 1.7 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 2.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 2.7 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 2.3 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 6.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 9.0 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 2.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 3.0 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 3.0 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 3.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 0.7 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 9.2 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 0.9 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.1 | 1.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 4.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 4.2 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 1.4 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.7 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 1.7 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 2.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.6 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.8 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.8 | REACTOME ENERGY DEPENDENT REGULATION OF MTOR BY LKB1 AMPK | Genes involved in Energy dependent regulation of mTOR by LKB1-AMPK |
| 0.0 | 0.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 1.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.7 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 1.1 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.5 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |