Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
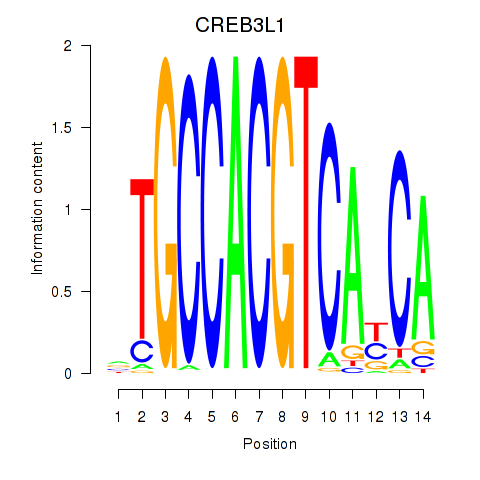
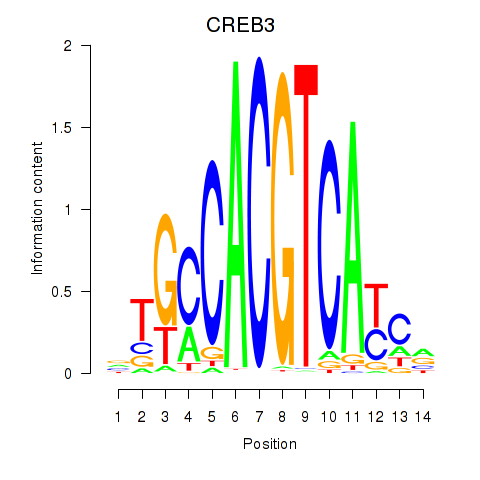
Results for CREB3L1_CREB3
Z-value: 1.22
Transcription factors associated with CREB3L1_CREB3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CREB3L1
|
ENSG00000157613.6 | cAMP responsive element binding protein 3 like 1 |
|
CREB3
|
ENSG00000107175.6 | cAMP responsive element binding protein 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CREB3 | hg19_v2_chr9_+_35732312_35732332 | 0.35 | 8.8e-08 | Click! |
| CREB3L1 | hg19_v2_chr11_+_46316677_46316719 | -0.08 | 2.5e-01 | Click! |
Activity profile of CREB3L1_CREB3 motif
Sorted Z-values of CREB3L1_CREB3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_106641728 | 27.31 |
ENST00000378026.4
|
CKAP4
|
cytoskeleton-associated protein 4 |
| chr19_-_48894104 | 24.49 |
ENST00000597017.1
|
KDELR1
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 |
| chr7_+_128379346 | 23.72 |
ENST00000535011.2
ENST00000542996.2 ENST00000535623.1 ENST00000538546.1 ENST00000249364.4 ENST00000449187.2 |
CALU
|
calumenin |
| chr6_+_116692102 | 23.62 |
ENST00000359564.2
|
DSE
|
dermatan sulfate epimerase |
| chr7_+_128379449 | 22.74 |
ENST00000479257.1
|
CALU
|
calumenin |
| chr3_-_57583052 | 22.45 |
ENST00000496292.1
ENST00000489843.1 |
ARF4
|
ADP-ribosylation factor 4 |
| chr3_-_57583185 | 22.33 |
ENST00000463880.1
|
ARF4
|
ADP-ribosylation factor 4 |
| chr3_+_100211412 | 21.88 |
ENST00000323523.4
ENST00000403410.1 ENST00000449609.1 |
TMEM45A
|
transmembrane protein 45A |
| chr2_-_69614373 | 20.42 |
ENST00000361060.5
ENST00000357308.4 |
GFPT1
|
glutamine--fructose-6-phosphate transaminase 1 |
| chr3_-_57583130 | 20.11 |
ENST00000303436.6
|
ARF4
|
ADP-ribosylation factor 4 |
| chr9_+_100818976 | 17.48 |
ENST00000210444.5
|
NANS
|
N-acetylneuraminic acid synthase |
| chr22_+_38864041 | 17.47 |
ENST00000216014.4
ENST00000409006.3 |
KDELR3
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
| chr5_-_131562935 | 16.64 |
ENST00000379104.2
ENST00000379100.2 ENST00000428369.1 |
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II |
| chrX_+_47441712 | 16.52 |
ENST00000218388.4
ENST00000377018.2 ENST00000456754.2 ENST00000377017.1 ENST00000441738.1 |
TIMP1
|
TIMP metallopeptidase inhibitor 1 |
| chr11_+_32112431 | 15.74 |
ENST00000054950.3
|
RCN1
|
reticulocalbin 1, EF-hand calcium binding domain |
| chr9_+_101984577 | 15.54 |
ENST00000223641.4
|
SEC61B
|
Sec61 beta subunit |
| chr9_+_114393634 | 14.07 |
ENST00000556107.1
ENST00000374294.3 |
DNAJC25
DNAJC25-GNG10
|
DnaJ (Hsp40) homolog, subfamily C , member 25 DNAJC25-GNG10 readthrough |
| chr8_-_71519889 | 14.04 |
ENST00000521425.1
|
TRAM1
|
translocation associated membrane protein 1 |
| chr12_-_56123444 | 13.97 |
ENST00000546457.1
ENST00000549117.1 |
CD63
|
CD63 molecule |
| chr1_+_162531294 | 13.73 |
ENST00000367926.4
ENST00000271469.3 |
UAP1
|
UDP-N-acteylglucosamine pyrophosphorylase 1 |
| chr5_-_131563501 | 13.62 |
ENST00000401867.1
ENST00000379086.1 ENST00000418055.1 ENST00000453286.1 ENST00000166534.4 |
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II |
| chr5_+_122110691 | 12.92 |
ENST00000379516.2
ENST00000505934.1 ENST00000514949.1 |
SNX2
|
sorting nexin 2 |
| chr18_-_47017956 | 12.15 |
ENST00000584895.1
ENST00000332968.6 ENST00000580210.1 ENST00000579408.1 |
RPL17-C18orf32
RPL17
|
RPL17-C18orf32 readthrough ribosomal protein L17 |
| chr8_+_64081118 | 11.82 |
ENST00000539294.1
|
YTHDF3
|
YTH domain family, member 3 |
| chr20_-_5100591 | 11.68 |
ENST00000379143.5
|
PCNA
|
proliferating cell nuclear antigen |
| chr8_+_64081214 | 11.13 |
ENST00000542911.2
|
YTHDF3
|
YTH domain family, member 3 |
| chr5_-_57756087 | 10.76 |
ENST00000274289.3
|
PLK2
|
polo-like kinase 2 |
| chr11_-_14521379 | 10.75 |
ENST00000249923.3
ENST00000529866.1 ENST00000439561.2 ENST00000534771.1 |
COPB1
|
coatomer protein complex, subunit beta 1 |
| chr7_+_39663061 | 10.72 |
ENST00000005257.2
|
RALA
|
v-ral simian leukemia viral oncogene homolog A (ras related) |
| chr12_-_50616382 | 10.62 |
ENST00000552783.1
|
LIMA1
|
LIM domain and actin binding 1 |
| chr19_-_4670345 | 10.33 |
ENST00000599630.1
ENST00000262947.3 |
C19orf10
|
chromosome 19 open reading frame 10 |
| chr6_+_24775153 | 10.17 |
ENST00000356509.3
ENST00000230056.3 |
GMNN
|
geminin, DNA replication inhibitor |
| chr3_-_107941230 | 10.16 |
ENST00000264538.3
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr12_-_50616122 | 9.93 |
ENST00000552823.1
ENST00000552909.1 |
LIMA1
|
LIM domain and actin binding 1 |
| chr7_-_30066233 | 9.90 |
ENST00000222803.5
|
FKBP14
|
FK506 binding protein 14, 22 kDa |
| chr7_+_94023873 | 9.86 |
ENST00000297268.6
|
COL1A2
|
collagen, type I, alpha 2 |
| chr9_+_112542572 | 9.64 |
ENST00000374530.3
|
PALM2-AKAP2
|
PALM2-AKAP2 readthrough |
| chr3_+_133292574 | 9.50 |
ENST00000264993.3
|
CDV3
|
CDV3 homolog (mouse) |
| chr1_+_45205498 | 9.34 |
ENST00000372218.4
|
KIF2C
|
kinesin family member 2C |
| chr14_-_35344093 | 9.21 |
ENST00000382422.2
|
BAZ1A
|
bromodomain adjacent to zinc finger domain, 1A |
| chr12_-_56709674 | 9.17 |
ENST00000551286.1
ENST00000549318.1 |
CNPY2
RP11-977G19.10
|
canopy FGF signaling regulator 2 Uncharacterized protein |
| chr19_+_50180317 | 9.16 |
ENST00000534465.1
|
PRMT1
|
protein arginine methyltransferase 1 |
| chr10_-_98346801 | 9.14 |
ENST00000371142.4
|
TM9SF3
|
transmembrane 9 superfamily member 3 |
| chr11_+_69455855 | 9.04 |
ENST00000227507.2
ENST00000536559.1 |
CCND1
|
cyclin D1 |
| chr12_-_120638902 | 9.00 |
ENST00000551150.1
ENST00000313104.5 ENST00000547191.1 ENST00000546989.1 ENST00000392514.4 ENST00000228306.4 ENST00000550856.1 |
RPLP0
|
ribosomal protein, large, P0 |
| chr1_+_45205478 | 8.84 |
ENST00000452259.1
ENST00000372224.4 |
KIF2C
|
kinesin family member 2C |
| chr19_+_16187816 | 8.80 |
ENST00000588410.1
|
TPM4
|
tropomyosin 4 |
| chr7_-_93633658 | 8.73 |
ENST00000433727.1
|
BET1
|
Bet1 golgi vesicular membrane trafficking protein |
| chr19_+_5681153 | 8.60 |
ENST00000579559.1
ENST00000577222.1 |
HSD11B1L
RPL36
|
hydroxysteroid (11-beta) dehydrogenase 1-like ribosomal protein L36 |
| chr7_-_93633684 | 8.55 |
ENST00000222547.3
ENST00000425626.1 |
BET1
|
Bet1 golgi vesicular membrane trafficking protein |
| chr5_+_65440032 | 8.49 |
ENST00000334121.6
|
SREK1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chr5_-_10761206 | 8.49 |
ENST00000432074.2
ENST00000230895.6 |
DAP
|
death-associated protein |
| chr2_+_183580954 | 8.21 |
ENST00000264065.7
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10 |
| chr2_+_27255806 | 8.21 |
ENST00000238788.9
ENST00000404032.3 |
TMEM214
|
transmembrane protein 214 |
| chr11_-_62341445 | 8.08 |
ENST00000329251.4
|
EEF1G
|
eukaryotic translation elongation factor 1 gamma |
| chr2_-_33824336 | 7.94 |
ENST00000431950.1
ENST00000403368.1 ENST00000441530.2 |
FAM98A
|
family with sequence similarity 98, member A |
| chrX_+_153060090 | 7.72 |
ENST00000370086.3
ENST00000370085.3 |
SSR4
|
signal sequence receptor, delta |
| chr8_+_64081156 | 7.62 |
ENST00000517371.1
|
YTHDF3
|
YTH domain family, member 3 |
| chr4_-_122744998 | 7.47 |
ENST00000274026.5
|
CCNA2
|
cyclin A2 |
| chr10_+_121410882 | 7.41 |
ENST00000369085.3
|
BAG3
|
BCL2-associated athanogene 3 |
| chr5_-_39425222 | 7.16 |
ENST00000320816.6
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr2_-_33824382 | 6.97 |
ENST00000238823.8
|
FAM98A
|
family with sequence similarity 98, member A |
| chr10_-_120840309 | 6.95 |
ENST00000369144.3
|
EIF3A
|
eukaryotic translation initiation factor 3, subunit A |
| chr6_-_7313381 | 6.94 |
ENST00000489567.1
ENST00000479365.1 ENST00000462112.1 ENST00000397511.2 ENST00000534851.1 ENST00000474597.1 ENST00000244763.4 |
SSR1
|
signal sequence receptor, alpha |
| chr5_-_39425290 | 6.87 |
ENST00000545653.1
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr12_-_49318715 | 6.80 |
ENST00000444214.2
|
FKBP11
|
FK506 binding protein 11, 19 kDa |
| chr1_+_228270361 | 6.80 |
ENST00000272102.5
ENST00000540651.1 |
ARF1
|
ADP-ribosylation factor 1 |
| chr13_-_31040060 | 6.69 |
ENST00000326004.4
ENST00000341423.5 |
HMGB1
|
high mobility group box 1 |
| chr7_+_16793160 | 6.67 |
ENST00000262067.4
|
TSPAN13
|
tetraspanin 13 |
| chr19_+_50180409 | 6.64 |
ENST00000391851.4
|
PRMT1
|
protein arginine methyltransferase 1 |
| chr3_-_150264272 | 6.57 |
ENST00000491660.1
ENST00000487153.1 ENST00000239944.2 |
SERP1
|
stress-associated endoplasmic reticulum protein 1 |
| chr17_-_15466742 | 6.42 |
ENST00000584811.1
ENST00000419890.2 |
TVP23C
|
trans-golgi network vesicle protein 23 homolog C (S. cerevisiae) |
| chr2_+_171785824 | 6.40 |
ENST00000452526.2
|
GORASP2
|
golgi reassembly stacking protein 2, 55kDa |
| chr1_+_228270784 | 6.29 |
ENST00000541182.1
|
ARF1
|
ADP-ribosylation factor 1 |
| chr3_-_49395705 | 6.23 |
ENST00000419349.1
|
GPX1
|
glutathione peroxidase 1 |
| chr3_+_122785895 | 6.12 |
ENST00000316218.7
|
PDIA5
|
protein disulfide isomerase family A, member 5 |
| chr3_-_49395892 | 6.07 |
ENST00000419783.1
|
GPX1
|
glutathione peroxidase 1 |
| chrX_+_153059608 | 6.05 |
ENST00000370087.1
|
SSR4
|
signal sequence receptor, delta |
| chr3_-_113465065 | 5.98 |
ENST00000497255.1
ENST00000478020.1 ENST00000240922.3 ENST00000493900.1 |
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr3_-_139108475 | 5.96 |
ENST00000515006.1
ENST00000513274.1 ENST00000514508.1 ENST00000507777.1 ENST00000512153.1 ENST00000333188.5 |
COPB2
|
coatomer protein complex, subunit beta 2 (beta prime) |
| chr5_-_39425068 | 5.88 |
ENST00000515700.1
ENST00000339788.6 |
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr4_-_119757239 | 5.86 |
ENST00000280551.6
|
SEC24D
|
SEC24 family member D |
| chr5_+_149340282 | 5.86 |
ENST00000286298.4
|
SLC26A2
|
solute carrier family 26 (anion exchanger), member 2 |
| chr5_-_94890648 | 5.81 |
ENST00000513823.1
ENST00000514952.1 ENST00000358746.2 |
TTC37
|
tetratricopeptide repeat domain 37 |
| chr6_+_31916733 | 5.81 |
ENST00000483004.1
|
CFB
|
complement factor B |
| chr4_-_47465666 | 5.79 |
ENST00000381571.4
|
COMMD8
|
COMM domain containing 8 |
| chrX_-_102941596 | 5.76 |
ENST00000441076.2
ENST00000422355.1 ENST00000442614.1 ENST00000422154.2 ENST00000451301.1 |
MORF4L2
|
mortality factor 4 like 2 |
| chrX_-_10544942 | 5.69 |
ENST00000380779.1
|
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr11_+_118443098 | 5.57 |
ENST00000392859.3
ENST00000359415.4 ENST00000534182.2 ENST00000264028.4 |
ARCN1
|
archain 1 |
| chr5_-_114961858 | 5.56 |
ENST00000282382.4
ENST00000456936.3 ENST00000408996.4 |
TMED7-TICAM2
TMED7
TICAM2
|
TMED7-TICAM2 readthrough transmembrane emp24 protein transport domain containing 7 toll-like receptor adaptor molecule 2 |
| chr17_+_57642886 | 5.40 |
ENST00000251241.4
ENST00000451169.2 ENST00000425628.3 ENST00000584385.1 ENST00000580030.1 |
DHX40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
| chr21_+_45138941 | 5.32 |
ENST00000398081.1
ENST00000468090.1 ENST00000291565.4 |
PDXK
|
pyridoxal (pyridoxine, vitamin B6) kinase |
| chr17_+_66511540 | 5.28 |
ENST00000588188.2
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr17_-_15466850 | 5.20 |
ENST00000438826.3
ENST00000225576.3 ENST00000519970.1 ENST00000518321.1 ENST00000428082.2 ENST00000522212.2 |
TVP23C
TVP23C-CDRT4
|
trans-golgi network vesicle protein 23 homolog C (S. cerevisiae) TVP23C-CDRT4 readthrough |
| chr1_+_151372010 | 5.03 |
ENST00000290541.6
|
PSMB4
|
proteasome (prosome, macropain) subunit, beta type, 4 |
| chr21_-_38445011 | 4.88 |
ENST00000464265.1
ENST00000399102.1 |
PIGP
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr8_+_109455845 | 4.86 |
ENST00000220853.3
|
EMC2
|
ER membrane protein complex subunit 2 |
| chr19_+_50180507 | 4.84 |
ENST00000454376.2
ENST00000524771.1 |
PRMT1
|
protein arginine methyltransferase 1 |
| chr6_-_101329191 | 4.82 |
ENST00000324723.6
ENST00000369162.2 ENST00000522650.1 |
ASCC3
|
activating signal cointegrator 1 complex subunit 3 |
| chr8_-_29208183 | 4.82 |
ENST00000240100.2
|
DUSP4
|
dual specificity phosphatase 4 |
| chr4_-_83350580 | 4.76 |
ENST00000349655.4
ENST00000602300.1 |
HNRNPDL
|
heterogeneous nuclear ribonucleoprotein D-like |
| chr11_+_65686802 | 4.74 |
ENST00000376991.2
|
DRAP1
|
DR1-associated protein 1 (negative cofactor 2 alpha) |
| chr14_-_50087312 | 4.73 |
ENST00000298289.6
|
RPL36AL
|
ribosomal protein L36a-like |
| chr1_+_42921761 | 4.72 |
ENST00000372562.1
|
PPCS
|
phosphopantothenoylcysteine synthetase |
| chr10_-_71930222 | 4.61 |
ENST00000458634.2
ENST00000373239.2 ENST00000373242.2 ENST00000373241.4 |
SAR1A
|
SAR1 homolog A (S. cerevisiae) |
| chr1_-_153940097 | 4.56 |
ENST00000413622.1
ENST00000310483.6 |
SLC39A1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr8_+_133787586 | 4.54 |
ENST00000395379.1
ENST00000395386.2 ENST00000337920.4 |
PHF20L1
|
PHD finger protein 20-like 1 |
| chr6_-_109702885 | 4.40 |
ENST00000504373.1
|
CD164
|
CD164 molecule, sialomucin |
| chr8_-_57906362 | 4.36 |
ENST00000262644.4
|
IMPAD1
|
inositol monophosphatase domain containing 1 |
| chr10_-_97416400 | 4.33 |
ENST00000371224.2
ENST00000371221.3 |
ALDH18A1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr6_-_74231444 | 4.32 |
ENST00000331523.2
ENST00000356303.2 |
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1 |
| chr19_-_42759300 | 4.29 |
ENST00000222329.4
|
ERF
|
Ets2 repressor factor |
| chr3_-_118959716 | 4.23 |
ENST00000467604.1
ENST00000491906.1 ENST00000475803.1 ENST00000479150.1 ENST00000470111.1 ENST00000459820.1 |
B4GALT4
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4 |
| chr5_-_54603368 | 4.19 |
ENST00000508346.1
ENST00000251636.5 |
DHX29
|
DEAH (Asp-Glu-Ala-His) box polypeptide 29 |
| chr7_+_100271355 | 4.17 |
ENST00000436220.1
ENST00000424361.1 |
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr2_-_130886795 | 4.14 |
ENST00000409914.2
|
POTEF
|
POTE ankyrin domain family, member F |
| chr11_+_65686728 | 4.12 |
ENST00000312515.2
ENST00000525501.1 |
DRAP1
|
DR1-associated protein 1 (negative cofactor 2 alpha) |
| chr3_+_37903432 | 4.12 |
ENST00000443503.2
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
| chr21_-_34960930 | 4.09 |
ENST00000437395.1
|
DONSON
|
downstream neighbor of SON |
| chr16_+_81040794 | 4.06 |
ENST00000439957.3
ENST00000393335.3 ENST00000428963.2 ENST00000564669.1 |
CENPN
|
centromere protein N |
| chr18_-_47018869 | 4.05 |
ENST00000583036.1
ENST00000580261.1 |
RPL17
|
ribosomal protein L17 |
| chr4_+_75311019 | 4.00 |
ENST00000502307.1
|
AREG
|
amphiregulin |
| chr21_-_38445297 | 3.88 |
ENST00000430792.1
ENST00000399103.1 |
PIGP
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr7_+_100271446 | 3.87 |
ENST00000419828.1
ENST00000427895.1 |
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr1_-_43833628 | 3.86 |
ENST00000413844.2
ENST00000372458.3 |
ELOVL1
|
ELOVL fatty acid elongase 1 |
| chr2_-_120980939 | 3.82 |
ENST00000426077.2
|
TMEM185B
|
transmembrane protein 185B |
| chr1_-_119682812 | 3.79 |
ENST00000537870.1
|
WARS2
|
tryptophanyl tRNA synthetase 2, mitochondrial |
| chr15_-_72668805 | 3.78 |
ENST00000268097.5
|
HEXA
|
hexosaminidase A (alpha polypeptide) |
| chr18_-_47018897 | 3.78 |
ENST00000418495.1
|
RPL17
|
ribosomal protein L17 |
| chr12_-_76953573 | 3.74 |
ENST00000549646.1
ENST00000550628.1 ENST00000553139.1 ENST00000261183.3 ENST00000393250.4 |
OSBPL8
|
oxysterol binding protein-like 8 |
| chr11_-_3818932 | 3.73 |
ENST00000324932.7
ENST00000359171.4 |
NUP98
|
nucleoporin 98kDa |
| chr3_-_121468513 | 3.73 |
ENST00000494517.1
ENST00000393667.3 |
GOLGB1
|
golgin B1 |
| chr14_-_50053081 | 3.71 |
ENST00000396020.3
ENST00000245458.6 |
RPS29
|
ribosomal protein S29 |
| chr16_+_56642489 | 3.67 |
ENST00000561491.1
|
MT2A
|
metallothionein 2A |
| chr11_-_67276100 | 3.65 |
ENST00000301488.3
|
CDK2AP2
|
cyclin-dependent kinase 2 associated protein 2 |
| chr7_-_73184588 | 3.65 |
ENST00000395145.2
|
CLDN3
|
claudin 3 |
| chr5_+_54603566 | 3.64 |
ENST00000230640.5
|
SKIV2L2
|
superkiller viralicidic activity 2-like 2 (S. cerevisiae) |
| chr5_+_54603807 | 3.61 |
ENST00000545714.1
|
SKIV2L2
|
superkiller viralicidic activity 2-like 2 (S. cerevisiae) |
| chr11_+_125495862 | 3.60 |
ENST00000428830.2
ENST00000544373.1 ENST00000527013.1 ENST00000526937.1 ENST00000534685.1 |
CHEK1
|
checkpoint kinase 1 |
| chr6_+_63921351 | 3.58 |
ENST00000370659.1
|
FKBP1C
|
FK506 binding protein 1C |
| chr18_-_47018769 | 3.58 |
ENST00000583637.1
ENST00000578528.1 ENST00000578532.1 ENST00000580387.1 ENST00000579248.1 ENST00000581373.1 |
RPL17
|
ribosomal protein L17 |
| chr1_+_32645269 | 3.51 |
ENST00000373610.3
|
TXLNA
|
taxilin alpha |
| chr6_-_79944336 | 3.48 |
ENST00000344726.5
ENST00000275036.7 |
HMGN3
|
high mobility group nucleosomal binding domain 3 |
| chr16_-_47007545 | 3.46 |
ENST00000317089.5
|
DNAJA2
|
DnaJ (Hsp40) homolog, subfamily A, member 2 |
| chr20_+_23331373 | 3.46 |
ENST00000254998.2
|
NXT1
|
NTF2-like export factor 1 |
| chr16_+_56642041 | 3.46 |
ENST00000245185.5
|
MT2A
|
metallothionein 2A |
| chr11_+_125496124 | 3.41 |
ENST00000533778.2
ENST00000534070.1 |
CHEK1
|
checkpoint kinase 1 |
| chr3_-_118959733 | 3.40 |
ENST00000459778.1
ENST00000359213.3 |
B4GALT4
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4 |
| chr6_+_151561085 | 3.39 |
ENST00000402676.2
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr12_-_10766184 | 3.37 |
ENST00000539554.1
ENST00000381881.2 ENST00000320756.2 |
MAGOHB
|
mago-nashi homolog B (Drosophila) |
| chr12_-_101801505 | 3.35 |
ENST00000539055.1
ENST00000551688.1 ENST00000551671.1 ENST00000261636.8 |
ARL1
|
ADP-ribosylation factor-like 1 |
| chr11_-_118927816 | 3.30 |
ENST00000534233.1
ENST00000532752.1 ENST00000525859.1 ENST00000404233.3 ENST00000532421.1 ENST00000543287.1 ENST00000527310.2 ENST00000529972.1 |
HYOU1
|
hypoxia up-regulated 1 |
| chr6_-_74231303 | 3.29 |
ENST00000309268.6
|
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1 |
| chr4_-_54930790 | 3.28 |
ENST00000263921.3
|
CHIC2
|
cysteine-rich hydrophobic domain 2 |
| chr14_-_78227476 | 3.20 |
ENST00000554775.1
ENST00000555761.1 ENST00000554324.1 ENST00000261531.7 |
SNW1
|
SNW domain containing 1 |
| chr11_-_118972575 | 3.20 |
ENST00000432443.2
|
DPAGT1
|
dolichyl-phosphate (UDP-N-acetylglucosamine) N-acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase) |
| chr16_+_53164833 | 3.16 |
ENST00000564845.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr9_+_117350009 | 3.05 |
ENST00000374050.3
|
ATP6V1G1
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G1 |
| chr14_+_55518349 | 3.02 |
ENST00000395468.4
|
MAPK1IP1L
|
mitogen-activated protein kinase 1 interacting protein 1-like |
| chr4_+_75310851 | 3.02 |
ENST00000395748.3
ENST00000264487.2 |
AREG
|
amphiregulin |
| chr1_-_38156153 | 3.01 |
ENST00000464085.1
ENST00000486637.1 ENST00000358011.4 ENST00000461359.1 |
C1orf109
|
chromosome 1 open reading frame 109 |
| chr17_+_7155343 | 2.99 |
ENST00000573513.1
ENST00000354429.2 ENST00000574255.1 ENST00000396627.2 ENST00000356683.2 |
ELP5
|
elongator acetyltransferase complex subunit 5 |
| chr19_+_797443 | 2.94 |
ENST00000394601.4
ENST00000589575.1 |
PTBP1
|
polypyrimidine tract binding protein 1 |
| chr19_+_54619125 | 2.91 |
ENST00000445811.1
ENST00000419967.1 ENST00000445124.1 ENST00000447810.1 |
PRPF31
|
pre-mRNA processing factor 31 |
| chr12_+_56552128 | 2.87 |
ENST00000548580.1
ENST00000293422.5 ENST00000348108.4 ENST00000549017.1 ENST00000549566.1 ENST00000536128.1 ENST00000547649.1 ENST00000547408.1 ENST00000551589.1 ENST00000549392.1 ENST00000548400.1 ENST00000548293.1 |
MYL6
|
myosin, light chain 6, alkali, smooth muscle and non-muscle |
| chr21_-_38445443 | 2.85 |
ENST00000360525.4
|
PIGP
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr3_+_169684553 | 2.84 |
ENST00000337002.4
ENST00000480708.1 |
SEC62
|
SEC62 homolog (S. cerevisiae) |
| chr2_-_198299726 | 2.82 |
ENST00000409915.4
ENST00000487698.1 ENST00000414963.2 ENST00000335508.6 |
SF3B1
|
splicing factor 3b, subunit 1, 155kDa |
| chr2_+_28615669 | 2.76 |
ENST00000379619.1
ENST00000264716.4 |
FOSL2
|
FOS-like antigen 2 |
| chr8_+_22422749 | 2.76 |
ENST00000523900.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr5_-_32174369 | 2.75 |
ENST00000265070.6
|
GOLPH3
|
golgi phosphoprotein 3 (coat-protein) |
| chr7_+_100860949 | 2.75 |
ENST00000305105.2
|
ZNHIT1
|
zinc finger, HIT-type containing 1 |
| chr1_-_65432171 | 2.74 |
ENST00000342505.4
|
JAK1
|
Janus kinase 1 |
| chr1_-_42801540 | 2.70 |
ENST00000372573.1
|
FOXJ3
|
forkhead box J3 |
| chr9_-_74980113 | 2.67 |
ENST00000376962.5
ENST00000376960.4 ENST00000237937.3 |
ZFAND5
|
zinc finger, AN1-type domain 5 |
| chr4_+_110736659 | 2.64 |
ENST00000394631.3
ENST00000226796.6 |
GAR1
|
GAR1 ribonucleoprotein |
| chr10_-_111683183 | 2.56 |
ENST00000403138.2
ENST00000369683.1 |
XPNPEP1
|
X-prolyl aminopeptidase (aminopeptidase P) 1, soluble |
| chr3_-_134204815 | 2.56 |
ENST00000514612.1
ENST00000510994.1 ENST00000354910.5 |
ANAPC13
|
anaphase promoting complex subunit 13 |
| chr8_+_140943416 | 2.50 |
ENST00000507535.3
|
C8orf17
|
chromosome 8 open reading frame 17 |
| chr7_+_150929550 | 2.46 |
ENST00000482173.1
ENST00000495645.1 ENST00000035307.2 |
CHPF2
|
chondroitin polymerizing factor 2 |
| chr12_+_70636765 | 2.43 |
ENST00000552231.1
ENST00000229195.3 ENST00000547780.1 ENST00000418359.3 |
CNOT2
|
CCR4-NOT transcription complex, subunit 2 |
| chr3_-_121468602 | 2.42 |
ENST00000340645.5
|
GOLGB1
|
golgin B1 |
| chr20_+_55926583 | 2.38 |
ENST00000395840.2
|
RAE1
|
ribonucleic acid export 1 |
| chr3_-_25824872 | 2.34 |
ENST00000308710.5
|
NGLY1
|
N-glycanase 1 |
| chr7_+_23145366 | 2.31 |
ENST00000339077.5
ENST00000322275.5 ENST00000539124.1 ENST00000542558.1 |
KLHL7
|
kelch-like family member 7 |
| chr1_-_160313025 | 2.29 |
ENST00000368069.3
ENST00000241704.7 |
COPA
|
coatomer protein complex, subunit alpha |
| chr10_+_89622870 | 2.28 |
ENST00000371953.3
|
PTEN
|
phosphatase and tensin homolog |
| chr10_-_111683308 | 2.25 |
ENST00000502935.1
ENST00000322238.8 ENST00000369680.4 |
XPNPEP1
|
X-prolyl aminopeptidase (aminopeptidase P) 1, soluble |
| chrX_+_101854426 | 2.25 |
ENST00000536530.1
ENST00000473968.1 ENST00000604957.1 ENST00000477663.1 ENST00000479502.1 |
ARMCX5
|
armadillo repeat containing, X-linked 5 |
| chr1_-_119683251 | 2.23 |
ENST00000369426.5
ENST00000235521.4 |
WARS2
|
tryptophanyl tRNA synthetase 2, mitochondrial |
| chr17_-_7218403 | 2.23 |
ENST00000570780.1
|
GPS2
|
G protein pathway suppressor 2 |
| chr14_+_31091511 | 2.21 |
ENST00000544052.2
ENST00000421551.3 ENST00000541123.1 ENST00000557076.1 ENST00000553693.1 ENST00000396629.2 |
SCFD1
|
sec1 family domain containing 1 |
| chr10_+_75504105 | 2.06 |
ENST00000535742.1
ENST00000546025.1 ENST00000345254.4 ENST00000540668.1 ENST00000339365.2 ENST00000411652.2 |
SEC24C
|
SEC24 family member C |
| chr20_-_62738524 | 2.06 |
ENST00000369768.1
|
NPBWR2
|
neuropeptides B/W receptor 2 |
| chr6_+_32936353 | 2.04 |
ENST00000374825.4
|
BRD2
|
bromodomain containing 2 |
| chr16_-_73082274 | 2.00 |
ENST00000268489.5
|
ZFHX3
|
zinc finger homeobox 3 |
| chr11_-_3818688 | 2.00 |
ENST00000355260.3
ENST00000397004.4 ENST00000397007.4 ENST00000532475.1 |
NUP98
|
nucleoporin 98kDa |
| chr8_+_110552337 | 2.00 |
ENST00000337573.5
|
EBAG9
|
estrogen receptor binding site associated, antigen, 9 |
| chr3_-_119182523 | 1.99 |
ENST00000319172.5
|
TMEM39A
|
transmembrane protein 39A |
| chr1_-_113498943 | 1.95 |
ENST00000369626.3
|
SLC16A1
|
solute carrier family 16 (monocarboxylate transporter), member 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of CREB3L1_CREB3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.2 | 68.2 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 5.0 | 19.9 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 4.4 | 13.1 | GO:0036090 | cleavage furrow ingression(GO:0036090) lysosomal membrane organization(GO:0097212) regulation of late endosome to lysosome transport(GO:1902822) |
| 3.8 | 42.0 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 3.7 | 18.4 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 3.4 | 20.7 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 2.9 | 11.7 | GO:1902990 | leading strand elongation(GO:0006272) mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 2.6 | 34.2 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 2.5 | 12.3 | GO:0009608 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 2.3 | 30.3 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 2.3 | 6.9 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) translation reinitiation(GO:0002188) |
| 2.2 | 15.7 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 2.1 | 16.5 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 2.0 | 11.7 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 1.9 | 7.5 | GO:0071373 | cellular response to cocaine(GO:0071314) cellular response to luteinizing hormone stimulus(GO:0071373) |
| 1.8 | 18.2 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 1.8 | 5.3 | GO:0042823 | pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 1.7 | 6.7 | GO:0002840 | plasmacytoid dendritic cell activation(GO:0002270) T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) regulation of restriction endodeoxyribonuclease activity(GO:0032072) negative regulation of apoptotic cell clearance(GO:2000426) |
| 1.4 | 7.1 | GO:1990169 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 1.4 | 5.6 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 1.2 | 6.0 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 1.2 | 13.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 1.2 | 5.9 | GO:0034036 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 1.2 | 7.0 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 1.1 | 5.7 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 1.1 | 28.6 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 1.1 | 17.5 | GO:0009226 | nucleotide-sugar biosynthetic process(GO:0009226) |
| 1.1 | 14.0 | GO:2000680 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 1.0 | 9.2 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 1.0 | 12.9 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.9 | 23.6 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.9 | 13.0 | GO:0061000 | negative regulation of dendritic spine development(GO:0061000) |
| 0.9 | 5.6 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.9 | 9.0 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.9 | 10.7 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.9 | 7.0 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.8 | 3.2 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.8 | 3.0 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.7 | 6.0 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.7 | 3.7 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.7 | 4.4 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.7 | 4.3 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.7 | 4.8 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.7 | 2.8 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) asymmetric Golgi ribbon formation(GO:0090164) |
| 0.7 | 9.9 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.6 | 3.9 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.6 | 3.2 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.6 | 3.5 | GO:0046649 | B cell activation(GO:0042113) lymphocyte activation(GO:0046649) |
| 0.6 | 11.6 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.6 | 5.6 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.5 | 3.7 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.5 | 6.6 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.5 | 2.0 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.5 | 2.9 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.5 | 9.6 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.5 | 1.4 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.5 | 2.4 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.5 | 1.4 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.4 | 2.2 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.4 | 5.7 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.4 | 4.8 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.4 | 7.4 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.4 | 20.3 | GO:0018208 | peptidyl-proline modification(GO:0018208) |
| 0.4 | 67.6 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.4 | 4.8 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.4 | 7.6 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.4 | 2.7 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.4 | 1.5 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.4 | 4.2 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.4 | 2.6 | GO:0031118 | snoRNA metabolic process(GO:0016074) rRNA pseudouridine synthesis(GO:0031118) |
| 0.4 | 3.3 | GO:1903298 | regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) |
| 0.4 | 0.4 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.4 | 20.0 | GO:0030835 | negative regulation of actin filament depolymerization(GO:0030835) |
| 0.4 | 18.1 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.3 | 2.4 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.3 | 1.4 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.3 | 7.3 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.3 | 5.3 | GO:2000480 | negative regulation of meiotic nuclear division(GO:0045835) negative regulation of activated T cell proliferation(GO:0046007) negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.3 | 1.7 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.3 | 0.3 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.3 | 1.3 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.2 | 9.5 | GO:0008283 | cell proliferation(GO:0008283) |
| 0.2 | 5.8 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 6.4 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.2 | 7.3 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.2 | 3.4 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.2 | 4.7 | GO:0034030 | coenzyme A biosynthetic process(GO:0015937) nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.2 | 0.9 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.2 | 1.7 | GO:0015846 | polyamine transport(GO:0015846) |
| 0.2 | 2.3 | GO:0002775 | antimicrobial peptide production(GO:0002775) antibacterial peptide production(GO:0002778) |
| 0.2 | 8.0 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.2 | 5.8 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.2 | 7.7 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 0.2 | 1.4 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.2 | 2.5 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.2 | 1.3 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.2 | 2.9 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.2 | 2.1 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.1 | 2.5 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.1 | 0.6 | GO:0071027 | nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.1 | 4.1 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 | 8.5 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 2.7 | GO:0043486 | histone exchange(GO:0043486) |
| 0.1 | 11.0 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.1 | 3.6 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 2.8 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 8.5 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.1 | 2.3 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.1 | 58.4 | GO:0043687 | post-translational protein modification(GO:0043687) |
| 0.1 | 3.2 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.1 | 3.5 | GO:0042026 | protein refolding(GO:0042026) |
| 0.1 | 1.7 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.1 | 0.7 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.1 | 6.1 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 1.3 | GO:0006505 | GPI anchor metabolic process(GO:0006505) GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 2.7 | GO:0003016 | respiratory system process(GO:0003016) |
| 0.1 | 8.1 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 4.1 | GO:0033260 | nuclear DNA replication(GO:0033260) |
| 0.1 | 4.4 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 3.3 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.1 | 3.6 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 0.2 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 | 0.2 | GO:1903061 | regulation of protein lipidation(GO:1903059) positive regulation of protein lipidation(GO:1903061) |
| 0.1 | 2.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 1.8 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 3.4 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 4.1 | GO:0001895 | retina homeostasis(GO:0001895) |
| 0.1 | 6.5 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.1 | 3.0 | GO:0030335 | positive regulation of cell migration(GO:0030335) |
| 0.1 | 4.2 | GO:0048701 | embryonic cranial skeleton morphogenesis(GO:0048701) |
| 0.1 | 0.9 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.1 | 0.9 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.7 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.0 | 0.6 | GO:0033182 | regulation of histone ubiquitination(GO:0033182) |
| 0.0 | 2.9 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 9.6 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.4 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.1 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 2.8 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.7 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.0 | 0.9 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 4.8 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.3 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 2.3 | GO:1903169 | regulation of calcium ion transmembrane transport(GO:1903169) |
| 0.0 | 1.8 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 1.6 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 0.3 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.6 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.9 | GO:0006413 | translational initiation(GO:0006413) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 29.3 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 4.2 | 46.5 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 3.9 | 54.6 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 3.9 | 11.7 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 2.5 | 7.5 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 2.2 | 19.9 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 2.2 | 12.9 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 2.1 | 14.9 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 1.5 | 5.8 | GO:0055087 | Ski complex(GO:0055087) |
| 1.4 | 30.3 | GO:0042599 | lamellar body(GO:0042599) |
| 1.3 | 13.1 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 1.2 | 14.0 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 1.2 | 11.6 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 1.1 | 20.7 | GO:0034709 | methylosome(GO:0034709) |
| 1.0 | 5.7 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.9 | 9.0 | GO:0097413 | Lewy body(GO:0097413) |
| 0.9 | 6.0 | GO:0031415 | NatA complex(GO:0031415) |
| 0.8 | 10.2 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.8 | 9.2 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.8 | 9.9 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.8 | 6.9 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.8 | 7.6 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.6 | 11.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.6 | 13.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.6 | 18.2 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.6 | 1.7 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.5 | 4.9 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.5 | 4.9 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.5 | 8.8 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.5 | 1.4 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) CNTFR-CLCF1 complex(GO:0097059) |
| 0.4 | 2.6 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.4 | 46.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.4 | 2.9 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.4 | 31.3 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.4 | 21.3 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.4 | 2.7 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.4 | 2.9 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.4 | 47.2 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.3 | 3.0 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.3 | 5.0 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.3 | 11.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.3 | 13.5 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.3 | 7.3 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.3 | 5.3 | GO:0031588 | cAMP-dependent protein kinase complex(GO:0005952) nucleotide-activated protein kinase complex(GO:0031588) |
| 0.2 | 2.4 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.2 | 2.5 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.2 | 2.4 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.2 | 0.6 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.2 | 1.5 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 2.3 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.2 | 9.0 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.2 | 5.8 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.2 | 12.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.2 | 0.9 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 15.1 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 46.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 1.8 | GO:0001741 | XY body(GO:0001741) |
| 0.1 | 0.9 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 3.4 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 3.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 5.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 1.3 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 10.1 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 1.6 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 2.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 39.9 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.1 | 3.3 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.1 | 1.4 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 8.3 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 5.3 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.1 | 5.2 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.1 | 2.8 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 4.1 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.1 | 3.7 | GO:0005874 | microtubule(GO:0005874) |
| 0.1 | 4.4 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 10.5 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.1 | 9.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 3.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 7.4 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.7 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 4.2 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 12.5 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.4 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 2.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.7 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 1.3 | GO:0005643 | nuclear pore(GO:0005643) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.9 | 23.6 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 6.1 | 24.5 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 6.1 | 30.3 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 3.5 | 17.5 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 3.4 | 20.7 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 3.1 | 80.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 2.9 | 11.7 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 2.6 | 12.9 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 2.4 | 30.6 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 1.8 | 18.2 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 1.3 | 7.7 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 1.2 | 13.7 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 1.2 | 6.0 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 1.2 | 7.0 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 1.2 | 17.5 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 1.2 | 11.6 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 1.1 | 6.7 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 1.1 | 4.3 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 1.1 | 3.2 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 1.0 | 5.9 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 1.0 | 2.9 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.9 | 20.4 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.9 | 19.9 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.9 | 10.8 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.9 | 4.4 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.8 | 2.5 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.8 | 2.3 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.7 | 2.9 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.7 | 9.9 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.7 | 20.3 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.7 | 4.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.7 | 13.1 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.7 | 8.2 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.7 | 16.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.6 | 3.9 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.6 | 7.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.6 | 4.8 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.6 | 14.1 | GO:0004871 | signal transducer activity(GO:0004871) |
| 0.5 | 7.1 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.5 | 8.8 | GO:0070513 | death domain binding(GO:0070513) |
| 0.5 | 13.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.5 | 20.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.5 | 1.4 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) procollagen galactosyltransferase activity(GO:0050211) |
| 0.5 | 6.0 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.4 | 2.6 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.4 | 1.7 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.4 | 12.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.3 | 2.7 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.3 | 6.1 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.3 | 2.4 | GO:0001224 | RNA polymerase II transcription cofactor binding(GO:0001224) |
| 0.3 | 4.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.3 | 16.5 | GO:0097472 | cyclin-dependent protein kinase activity(GO:0097472) |
| 0.3 | 1.4 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.3 | 5.3 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.3 | 5.3 | GO:0008603 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.2 | 23.0 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.2 | 50.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.2 | 3.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.2 | 3.4 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.2 | 4.8 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.2 | 4.7 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.2 | 0.6 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.2 | 6.4 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.2 | 10.0 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.2 | 4.8 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.2 | 4.9 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.2 | 1.5 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.2 | 5.0 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.2 | 5.8 | GO:0001848 | complement binding(GO:0001848) |
| 0.2 | 2.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.2 | 0.9 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.2 | 1.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 12.9 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.1 | 10.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 3.1 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.1 | 3.5 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 11.7 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 8.4 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 0.7 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 2.9 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 2.3 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 3.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 2.9 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 6.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 8.8 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 4.6 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 3.7 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 1.9 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 0.7 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.1 | 3.2 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 0.9 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 42.3 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 0.3 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.0 | 2.7 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 2.3 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 2.1 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.1 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.6 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 10.6 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 3.2 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 1.0 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 3.0 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.2 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 0.3 | GO:0015386 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) potassium ion antiporter activity(GO:0022821) |
| 0.0 | 2.0 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 2.5 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.6 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 13.2 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 5.1 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 1.2 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 5.6 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 1.3 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 2.0 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 1.0 | GO:0004620 | phospholipase activity(GO:0004620) |
| 0.0 | 0.5 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 39.9 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.7 | 16.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.5 | 7.0 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.4 | 9.9 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.4 | 71.1 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.3 | 5.8 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.3 | 18.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.3 | 6.9 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.3 | 2.7 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.3 | 15.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.3 | 7.0 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.3 | 26.2 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.3 | 19.9 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.2 | 4.1 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.2 | 2.9 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.2 | 6.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 45.0 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.2 | 11.9 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.2 | 4.8 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 4.2 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 5.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 4.5 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 25.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 4.1 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.0 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.9 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.9 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 37.7 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 1.5 | 9.2 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 1.1 | 12.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 1.1 | 19.9 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 1.0 | 11.7 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.8 | 58.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.7 | 18.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.6 | 11.7 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.6 | 12.2 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.5 | 11.6 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.5 | 76.4 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.5 | 91.0 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.5 | 5.6 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.4 | 25.8 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.4 | 7.5 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.4 | 6.4 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.4 | 10.7 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.4 | 7.0 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.4 | 6.7 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.3 | 8.0 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.3 | 4.7 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.3 | 6.9 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.3 | 7.7 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.3 | 8.9 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.3 | 5.8 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.3 | 4.8 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.3 | 7.9 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.3 | 6.0 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.3 | 14.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.3 | 2.3 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.2 | 8.1 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.2 | 13.6 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.2 | 3.9 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.2 | 2.7 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.2 | 5.3 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.2 | 4.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 2.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 3.0 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 3.0 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 3.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 2.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 0.6 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 5.0 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.1 | 0.9 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.1 | 1.6 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.1 | 6.2 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 8.0 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 3.9 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 1.9 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 1.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.7 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 2.9 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.2 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |