Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for DUXA
Z-value: 1.01
Transcription factors associated with DUXA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DUXA
|
ENSG00000258873.2 | double homeobox A |
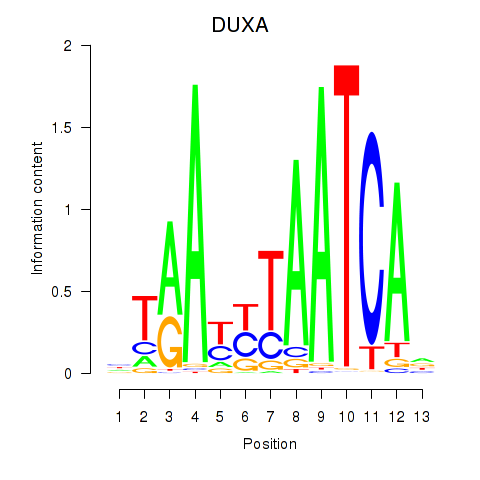
Activity profile of DUXA motif
Sorted Z-values of DUXA motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_64708615 | 47.66 |
ENST00000338957.4
ENST00000423889.3 |
ZC3H12B
|
zinc finger CCCH-type containing 12B |
| chr1_-_13390765 | 22.39 |
ENST00000357367.2
|
PRAMEF8
|
PRAME family member 8 |
| chr3_+_153839149 | 18.16 |
ENST00000465093.1
ENST00000465817.1 |
ARHGEF26
|
Rho guanine nucleotide exchange factor (GEF) 26 |
| chr1_+_12976450 | 17.45 |
ENST00000361079.2
|
PRAMEF7
|
PRAME family member 7 |
| chr11_+_49050504 | 16.37 |
ENST00000332682.7
|
TRIM49B
|
tripartite motif containing 49B |
| chr1_-_13452656 | 15.63 |
ENST00000376132.3
|
PRAMEF13
|
PRAME family member 13 |
| chr2_+_149632783 | 12.99 |
ENST00000435030.1
|
KIF5C
|
kinesin family member 5C |
| chr16_+_6533729 | 12.91 |
ENST00000551752.1
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr11_+_55029628 | 11.99 |
ENST00000417545.2
|
TRIM48
|
tripartite motif containing 48 |
| chr16_+_6533380 | 11.84 |
ENST00000552089.1
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr9_-_99417562 | 11.33 |
ENST00000375234.3
ENST00000446045.1 |
AAED1
|
AhpC/TSA antioxidant enzyme domain containing 1 |
| chr2_-_86333244 | 11.13 |
ENST00000263857.6
ENST00000409681.1 |
POLR1A
|
polymerase (RNA) I polypeptide A, 194kDa |
| chr11_-_89540388 | 11.07 |
ENST00000532501.2
|
TRIM49
|
tripartite motif containing 49 |
| chr14_-_90798418 | 10.73 |
ENST00000354366.3
|
NRDE2
|
NRDE-2, necessary for RNA interference, domain containing |
| chr1_-_13005246 | 9.88 |
ENST00000415464.2
|
PRAMEF6
|
PRAME family member 6 |
| chr2_-_224467093 | 8.93 |
ENST00000305409.2
|
SCG2
|
secretogranin II |
| chr1_+_92632542 | 8.90 |
ENST00000409154.4
ENST00000370378.4 |
KIAA1107
|
KIAA1107 |
| chr12_-_91546926 | 8.70 |
ENST00000550758.1
|
DCN
|
decorin |
| chr1_+_12916941 | 8.11 |
ENST00000240189.2
|
PRAMEF2
|
PRAME family member 2 |
| chr5_+_173472607 | 7.95 |
ENST00000303177.3
ENST00000519867.1 |
NSG2
|
Neuron-specific protein family member 2 |
| chr1_-_153113927 | 7.89 |
ENST00000368752.4
|
SPRR2B
|
small proline-rich protein 2B |
| chr15_+_80733570 | 7.70 |
ENST00000533983.1
ENST00000527771.1 ENST00000525103.1 |
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr6_+_150920999 | 7.66 |
ENST00000367328.1
ENST00000367326.1 |
PLEKHG1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr2_-_175629164 | 7.61 |
ENST00000409323.1
ENST00000261007.5 ENST00000348749.5 |
CHRNA1
|
cholinergic receptor, nicotinic, alpha 1 (muscle) |
| chr17_+_74733744 | 7.17 |
ENST00000586689.1
ENST00000587661.1 ENST00000593181.1 ENST00000336509.4 ENST00000355954.3 |
MFSD11
|
major facilitator superfamily domain containing 11 |
| chr1_-_13673511 | 7.02 |
ENST00000344998.3
ENST00000334600.6 |
PRAMEF14
|
PRAME family member 14 |
| chr2_+_54342574 | 7.01 |
ENST00000303536.4
ENST00000394666.3 |
ACYP2
|
acylphosphatase 2, muscle type |
| chr4_-_87028478 | 6.95 |
ENST00000515400.1
ENST00000395157.3 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr19_-_19302931 | 6.85 |
ENST00000444486.3
ENST00000514819.3 ENST00000585679.1 ENST00000162023.5 |
MEF2BNB-MEF2B
MEF2BNB
MEF2B
|
MEF2BNB-MEF2B readthrough MEF2B neighbor myocyte enhancer factor 2B |
| chr17_-_4167142 | 6.78 |
ENST00000570535.1
ENST00000574367.1 ENST00000341657.4 ENST00000433651.1 |
ANKFY1
|
ankyrin repeat and FYVE domain containing 1 |
| chr1_-_12946025 | 6.71 |
ENST00000235349.5
|
PRAMEF4
|
PRAME family member 4 |
| chr1_-_13115578 | 6.57 |
ENST00000414205.2
|
PRAMEF6
|
PRAME family member 6 |
| chr16_-_21314360 | 6.53 |
ENST00000219599.3
ENST00000576703.1 |
CRYM
|
crystallin, mu |
| chr7_+_77325738 | 6.26 |
ENST00000334955.8
|
RSBN1L
|
round spermatid basic protein 1-like |
| chr14_-_31926701 | 6.24 |
ENST00000310850.4
|
DTD2
|
D-tyrosyl-tRNA deacylase 2 (putative) |
| chr11_-_89653576 | 6.13 |
ENST00000420869.1
|
TRIM49D1
|
tripartite motif containing 49D1 |
| chr5_-_115872142 | 6.11 |
ENST00000510263.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr1_+_50459990 | 6.02 |
ENST00000448346.1
|
AL645730.2
|
AL645730.2 |
| chr12_+_14572070 | 5.93 |
ENST00000545769.1
ENST00000428217.2 ENST00000396279.2 ENST00000542514.1 ENST00000536279.1 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr11_-_123525289 | 5.89 |
ENST00000392770.2
ENST00000299333.3 ENST00000530277.1 |
SCN3B
|
sodium channel, voltage-gated, type III, beta subunit |
| chr7_-_137028534 | 5.86 |
ENST00000348225.2
|
PTN
|
pleiotrophin |
| chr1_+_12851545 | 5.53 |
ENST00000332296.7
|
PRAMEF1
|
PRAME family member 1 |
| chr12_-_114211444 | 5.53 |
ENST00000510694.2
ENST00000550223.1 |
RP11-438N16.1
|
RP11-438N16.1 |
| chr5_+_150404904 | 5.51 |
ENST00000521632.1
|
GPX3
|
glutathione peroxidase 3 (plasma) |
| chr18_+_39766626 | 5.42 |
ENST00000593234.1
ENST00000585627.1 ENST00000591199.1 ENST00000586990.1 ENST00000593051.1 ENST00000593316.1 ENST00000591381.1 ENST00000585639.1 ENST00000589068.1 |
LINC00907
|
long intergenic non-protein coding RNA 907 |
| chr11_-_5526834 | 5.41 |
ENST00000380237.1
ENST00000396895.1 ENST00000380252.1 |
HBE1
HBG2
|
hemoglobin, epsilon 1 hemoglobin, gamma G |
| chr7_-_137028498 | 5.37 |
ENST00000393083.2
|
PTN
|
pleiotrophin |
| chr1_+_152975488 | 5.35 |
ENST00000542696.1
|
SPRR3
|
small proline-rich protein 3 |
| chr11_+_28129795 | 5.29 |
ENST00000406787.3
ENST00000342303.5 ENST00000403099.1 ENST00000407364.3 |
METTL15
|
methyltransferase like 15 |
| chr11_-_133826852 | 5.22 |
ENST00000533871.2
ENST00000321016.8 |
IGSF9B
|
immunoglobulin superfamily, member 9B |
| chr5_+_149877334 | 5.21 |
ENST00000523767.1
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr3_-_138725110 | 5.07 |
ENST00000383163.2
|
PRR23A
|
proline rich 23A |
| chr8_+_19796381 | 5.02 |
ENST00000524029.1
ENST00000522701.1 ENST00000311322.8 |
LPL
|
lipoprotein lipase |
| chr6_-_133055815 | 5.01 |
ENST00000509351.1
ENST00000417437.2 ENST00000414302.2 ENST00000423615.2 ENST00000427187.2 ENST00000275223.3 ENST00000519686.2 |
VNN3
|
vanin 3 |
| chr1_+_13359819 | 4.94 |
ENST00000376168.1
|
PRAMEF5
|
PRAME family member 5 |
| chrX_+_77166172 | 4.93 |
ENST00000343533.5
ENST00000350425.4 ENST00000341514.6 |
ATP7A
|
ATPase, Cu++ transporting, alpha polypeptide |
| chr1_+_13516066 | 4.82 |
ENST00000332192.6
|
PRAMEF21
|
PRAME family member 21 |
| chr16_-_57219966 | 4.81 |
ENST00000565760.1
ENST00000309137.8 ENST00000570184.1 ENST00000562324.1 |
FAM192A
|
family with sequence similarity 192, member A |
| chr12_-_110434096 | 4.55 |
ENST00000320063.9
ENST00000457474.2 ENST00000547815.1 ENST00000361006.5 |
GIT2
|
G protein-coupled receptor kinase interacting ArfGAP 2 |
| chr19_-_17488143 | 4.52 |
ENST00000599426.1
ENST00000252590.4 |
PLVAP
|
plasmalemma vesicle associated protein |
| chr3_-_33759699 | 4.26 |
ENST00000399362.4
ENST00000359576.5 ENST00000307312.7 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr2_+_166428839 | 4.25 |
ENST00000342316.4
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3 |
| chr11_-_47736896 | 4.22 |
ENST00000525123.1
ENST00000528244.1 ENST00000532595.1 ENST00000529154.1 ENST00000530969.1 |
AGBL2
|
ATP/GTP binding protein-like 2 |
| chr4_-_100212132 | 4.22 |
ENST00000209668.2
|
ADH1A
|
alcohol dehydrogenase 1A (class I), alpha polypeptide |
| chr9_-_13279563 | 4.21 |
ENST00000541718.1
|
MPDZ
|
multiple PDZ domain protein |
| chr20_-_44600810 | 4.11 |
ENST00000322927.2
ENST00000426788.1 |
ZNF335
|
zinc finger protein 335 |
| chr2_+_54342533 | 4.10 |
ENST00000406041.1
|
ACYP2
|
acylphosphatase 2, muscle type |
| chr3_-_39321512 | 4.01 |
ENST00000399220.2
|
CX3CR1
|
chemokine (C-X3-C motif) receptor 1 |
| chr3_-_93781750 | 3.94 |
ENST00000314636.2
|
DHFRL1
|
dihydrofolate reductase-like 1 |
| chr2_-_175629135 | 3.88 |
ENST00000409542.1
ENST00000409219.1 |
CHRNA1
|
cholinergic receptor, nicotinic, alpha 1 (muscle) |
| chr1_-_13007420 | 3.84 |
ENST00000376189.1
|
PRAMEF6
|
PRAME family member 6 |
| chr20_-_17539456 | 3.80 |
ENST00000544874.1
ENST00000377868.2 |
BFSP1
|
beaded filament structural protein 1, filensin |
| chr12_+_120740119 | 3.79 |
ENST00000536460.1
ENST00000202967.4 |
SIRT4
|
sirtuin 4 |
| chr17_+_56315936 | 3.70 |
ENST00000543544.1
|
LPO
|
lactoperoxidase |
| chr1_+_180601139 | 3.70 |
ENST00000367590.4
ENST00000367589.3 |
XPR1
|
xenotropic and polytropic retrovirus receptor 1 |
| chr2_+_196313239 | 3.70 |
ENST00000413290.1
|
AC064834.1
|
AC064834.1 |
| chr9_-_95298314 | 3.68 |
ENST00000344604.5
ENST00000375540.1 |
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chr7_+_120702819 | 3.66 |
ENST00000423795.1
|
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chr8_-_18744528 | 3.65 |
ENST00000523619.1
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr1_+_13421176 | 3.63 |
ENST00000376152.1
|
PRAMEF9
|
PRAME family member 9 |
| chr10_-_61900762 | 3.63 |
ENST00000355288.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr22_+_45725524 | 3.61 |
ENST00000405548.3
|
FAM118A
|
family with sequence similarity 118, member A |
| chr11_-_85376121 | 3.60 |
ENST00000527447.1
|
CREBZF
|
CREB/ATF bZIP transcription factor |
| chr15_-_26874230 | 3.60 |
ENST00000400188.3
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr7_-_14942283 | 3.59 |
ENST00000402815.1
|
DGKB
|
diacylglycerol kinase, beta 90kDa |
| chr3_-_120461378 | 3.57 |
ENST00000273375.3
|
RABL3
|
RAB, member of RAS oncogene family-like 3 |
| chr2_-_74570520 | 3.56 |
ENST00000394019.2
ENST00000346834.4 ENST00000359484.4 ENST00000423644.1 ENST00000377634.4 ENST00000436454.1 |
SLC4A5
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 5 |
| chr8_-_6914251 | 3.54 |
ENST00000330590.2
|
DEFA5
|
defensin, alpha 5, Paneth cell-specific |
| chr6_-_31125850 | 3.54 |
ENST00000507751.1
ENST00000448162.2 ENST00000502557.1 ENST00000503420.1 ENST00000507892.1 ENST00000507226.1 ENST00000513222.1 ENST00000503934.1 ENST00000396263.2 ENST00000508683.1 ENST00000428174.1 ENST00000448141.2 ENST00000507829.1 ENST00000455279.2 ENST00000376266.5 |
CCHCR1
|
coiled-coil alpha-helical rod protein 1 |
| chr2_+_60983361 | 3.52 |
ENST00000238714.3
|
PAPOLG
|
poly(A) polymerase gamma |
| chr5_+_140227048 | 3.51 |
ENST00000532602.1
|
PCDHA9
|
protocadherin alpha 9 |
| chr8_+_133879193 | 3.50 |
ENST00000377869.1
ENST00000220616.4 |
TG
|
thyroglobulin |
| chrX_-_72347916 | 3.45 |
ENST00000373518.1
|
NAP1L6
|
nucleosome assembly protein 1-like 6 |
| chr5_-_95297534 | 3.44 |
ENST00000513343.1
ENST00000431061.2 |
ELL2
|
elongation factor, RNA polymerase II, 2 |
| chr13_-_113242439 | 3.44 |
ENST00000375669.3
ENST00000261965.3 |
TUBGCP3
|
tubulin, gamma complex associated protein 3 |
| chr12_-_110434021 | 3.31 |
ENST00000355312.3
ENST00000551209.1 ENST00000550186.1 |
GIT2
|
G protein-coupled receptor kinase interacting ArfGAP 2 |
| chr1_-_12908578 | 3.30 |
ENST00000317869.6
|
HNRNPCL1
|
heterogeneous nuclear ribonucleoprotein C-like 1 |
| chr5_-_74326724 | 3.25 |
ENST00000322348.4
|
GCNT4
|
glucosaminyl (N-acetyl) transferase 4, core 2 |
| chr19_-_12833361 | 3.25 |
ENST00000592287.1
|
TNPO2
|
transportin 2 |
| chr21_+_34398153 | 3.22 |
ENST00000382357.3
ENST00000430860.1 ENST00000333337.3 |
OLIG2
|
oligodendrocyte lineage transcription factor 2 |
| chr12_-_6055398 | 3.18 |
ENST00000327087.8
ENST00000356134.5 ENST00000546188.1 |
ANO2
|
anoctamin 2 |
| chr11_+_121447469 | 3.16 |
ENST00000532694.1
ENST00000534286.1 |
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing |
| chr14_+_22615942 | 3.16 |
ENST00000390457.2
|
TRAV27
|
T cell receptor alpha variable 27 |
| chrX_+_37639302 | 3.13 |
ENST00000545017.1
ENST00000536160.1 |
CYBB
|
cytochrome b-245, beta polypeptide |
| chr19_-_51472031 | 3.12 |
ENST00000391808.1
|
KLK6
|
kallikrein-related peptidase 6 |
| chr1_+_117963209 | 3.08 |
ENST00000449370.2
|
MAN1A2
|
mannosidase, alpha, class 1A, member 2 |
| chr2_-_198062758 | 3.04 |
ENST00000328737.2
|
ANKRD44
|
ankyrin repeat domain 44 |
| chr6_+_167525277 | 3.03 |
ENST00000400926.2
|
CCR6
|
chemokine (C-C motif) receptor 6 |
| chr15_-_65426174 | 3.00 |
ENST00000204549.4
|
PDCD7
|
programmed cell death 7 |
| chr10_-_17171817 | 3.00 |
ENST00000377833.4
|
CUBN
|
cubilin (intrinsic factor-cobalamin receptor) |
| chr4_-_110651143 | 2.99 |
ENST00000243501.5
|
PLA2G12A
|
phospholipase A2, group XIIA |
| chr17_+_3118915 | 2.96 |
ENST00000304094.1
|
OR1A1
|
olfactory receptor, family 1, subfamily A, member 1 |
| chr12_-_110434183 | 2.95 |
ENST00000360185.4
ENST00000354574.4 ENST00000338373.5 ENST00000343646.5 ENST00000356259.4 ENST00000553118.1 |
GIT2
|
G protein-coupled receptor kinase interacting ArfGAP 2 |
| chr7_-_82792215 | 2.91 |
ENST00000333891.9
ENST00000423517.2 |
PCLO
|
piccolo presynaptic cytomatrix protein |
| chr6_-_26199471 | 2.91 |
ENST00000341023.1
|
HIST1H2AD
|
histone cluster 1, H2ad |
| chr12_-_112123524 | 2.91 |
ENST00000327551.6
|
BRAP
|
BRCA1 associated protein |
| chr15_+_62853562 | 2.86 |
ENST00000561311.1
|
TLN2
|
talin 2 |
| chr1_+_50569575 | 2.82 |
ENST00000371827.1
|
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr12_+_122688090 | 2.80 |
ENST00000324189.4
ENST00000546192.1 |
B3GNT4
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 4 |
| chr5_+_54455946 | 2.77 |
ENST00000503787.1
ENST00000296734.6 ENST00000515370.1 |
GPX8
|
glutathione peroxidase 8 (putative) |
| chr5_+_176853702 | 2.70 |
ENST00000507633.1
ENST00000393576.3 ENST00000355958.5 ENST00000528793.1 ENST00000512684.1 |
GRK6
|
G protein-coupled receptor kinase 6 |
| chr12_+_12878829 | 2.66 |
ENST00000326765.6
|
APOLD1
|
apolipoprotein L domain containing 1 |
| chr8_+_58890917 | 2.64 |
ENST00000522992.1
|
RP11-1112C15.1
|
RP11-1112C15.1 |
| chr5_-_75919253 | 2.63 |
ENST00000296641.4
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2 |
| chr21_+_43619796 | 2.60 |
ENST00000398457.2
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr15_+_71228826 | 2.60 |
ENST00000558456.1
ENST00000560158.2 ENST00000558808.1 ENST00000559806.1 ENST00000559069.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr1_-_13117736 | 2.60 |
ENST00000376192.5
ENST00000376182.1 |
PRAMEF6
|
PRAME family member 6 |
| chr12_-_118796910 | 2.60 |
ENST00000541186.1
ENST00000539872.1 |
TAOK3
|
TAO kinase 3 |
| chr5_+_54320078 | 2.59 |
ENST00000231009.2
|
GZMK
|
granzyme K (granzyme 3; tryptase II) |
| chr3_-_164914640 | 2.57 |
ENST00000241274.3
|
SLITRK3
|
SLIT and NTRK-like family, member 3 |
| chr6_-_133055896 | 2.57 |
ENST00000367927.5
ENST00000425515.2 ENST00000207771.3 ENST00000392393.3 ENST00000450865.2 ENST00000392394.2 |
VNN3
|
vanin 3 |
| chr4_-_130692631 | 2.54 |
ENST00000500092.2
ENST00000509105.1 |
RP11-519M16.1
|
RP11-519M16.1 |
| chr6_-_152639479 | 2.54 |
ENST00000356820.4
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr18_-_53253112 | 2.53 |
ENST00000568673.1
ENST00000562847.1 ENST00000568147.1 |
TCF4
|
transcription factor 4 |
| chr5_+_172571445 | 2.52 |
ENST00000231668.9
ENST00000351486.5 ENST00000352523.6 ENST00000393770.4 |
BNIP1
|
BCL2/adenovirus E1B 19kDa interacting protein 1 |
| chr1_+_17575584 | 2.51 |
ENST00000375460.3
|
PADI3
|
peptidyl arginine deiminase, type III |
| chr6_+_73076432 | 2.51 |
ENST00000414192.2
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr22_+_41956767 | 2.50 |
ENST00000306149.7
|
CSDC2
|
cold shock domain containing C2, RNA binding |
| chr11_-_89654935 | 2.49 |
ENST00000530311.2
|
TRIM49D1
|
tripartite motif containing 49D1 |
| chr5_-_75919217 | 2.48 |
ENST00000504899.1
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2 |
| chr1_-_92371839 | 2.48 |
ENST00000370399.2
|
TGFBR3
|
transforming growth factor, beta receptor III |
| chr17_-_39553844 | 2.48 |
ENST00000251645.2
|
KRT31
|
keratin 31 |
| chrX_+_11311533 | 2.48 |
ENST00000380714.3
ENST00000380712.3 ENST00000348912.4 |
AMELX
|
amelogenin, X-linked |
| chr11_+_68671310 | 2.46 |
ENST00000255078.3
ENST00000539224.1 |
IGHMBP2
|
immunoglobulin mu binding protein 2 |
| chr1_+_13641973 | 2.45 |
ENST00000330087.5
|
PRAMEF15
|
PRAME family member 15 |
| chr17_-_2996290 | 2.42 |
ENST00000331459.1
|
OR1D2
|
olfactory receptor, family 1, subfamily D, member 2 |
| chr5_-_150948414 | 2.41 |
ENST00000261800.5
|
FAT2
|
FAT atypical cadherin 2 |
| chr6_-_73935163 | 2.41 |
ENST00000370388.3
|
KHDC1L
|
KH homology domain containing 1-like |
| chr1_-_12958101 | 2.40 |
ENST00000235347.4
|
PRAMEF10
|
PRAME family member 10 |
| chr12_-_49259643 | 2.40 |
ENST00000309739.5
|
RND1
|
Rho family GTPase 1 |
| chr6_-_10838710 | 2.40 |
ENST00000313243.2
|
MAK
|
male germ cell-associated kinase |
| chr5_-_95297678 | 2.39 |
ENST00000237853.4
|
ELL2
|
elongation factor, RNA polymerase II, 2 |
| chr6_-_31939734 | 2.39 |
ENST00000375356.3
|
DXO
|
decapping exoribonuclease |
| chr4_+_76649797 | 2.37 |
ENST00000538159.1
ENST00000514213.2 |
USO1
|
USO1 vesicle transport factor |
| chr18_+_61554932 | 2.32 |
ENST00000299502.4
ENST00000457692.1 ENST00000413956.1 |
SERPINB2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
| chr18_-_53253323 | 2.30 |
ENST00000540999.1
ENST00000563888.2 |
TCF4
|
transcription factor 4 |
| chr2_-_208994548 | 2.29 |
ENST00000282141.3
|
CRYGC
|
crystallin, gamma C |
| chr5_+_140227357 | 2.28 |
ENST00000378122.3
|
PCDHA9
|
protocadherin alpha 9 |
| chr6_+_31553978 | 2.26 |
ENST00000376096.1
ENST00000376099.1 ENST00000376110.3 |
LST1
|
leukocyte specific transcript 1 |
| chr20_+_23471727 | 2.26 |
ENST00000449810.1
ENST00000246012.1 |
CST8
|
cystatin 8 (cystatin-related epididymal specific) |
| chr8_+_24151620 | 2.25 |
ENST00000437154.2
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr9_-_95186739 | 2.23 |
ENST00000375550.4
|
OMD
|
osteomodulin |
| chr1_+_66458072 | 2.21 |
ENST00000423207.2
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr3_+_93781728 | 2.20 |
ENST00000314622.4
|
NSUN3
|
NOP2/Sun domain family, member 3 |
| chr8_-_86290333 | 2.19 |
ENST00000521846.1
ENST00000523022.1 ENST00000524324.1 ENST00000519991.1 ENST00000520663.1 ENST00000517590.1 ENST00000522579.1 ENST00000522814.1 ENST00000522662.1 ENST00000523858.1 ENST00000519129.1 |
CA1
|
carbonic anhydrase I |
| chr16_+_67226019 | 2.15 |
ENST00000379378.3
|
E2F4
|
E2F transcription factor 4, p107/p130-binding |
| chr11_+_30253410 | 2.13 |
ENST00000533718.1
|
FSHB
|
follicle stimulating hormone, beta polypeptide |
| chr5_+_176853669 | 2.13 |
ENST00000355472.5
|
GRK6
|
G protein-coupled receptor kinase 6 |
| chr4_-_186732048 | 2.12 |
ENST00000448662.2
ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr2_-_166930131 | 2.08 |
ENST00000303395.4
ENST00000409050.1 ENST00000423058.2 ENST00000375405.3 |
SCN1A
|
sodium channel, voltage-gated, type I, alpha subunit |
| chr10_+_90424196 | 2.08 |
ENST00000394375.3
ENST00000608620.1 ENST00000238983.4 ENST00000355843.2 |
LIPF
|
lipase, gastric |
| chr2_+_169923504 | 2.07 |
ENST00000357546.2
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr17_-_59668550 | 2.07 |
ENST00000521764.1
|
NACA2
|
nascent polypeptide-associated complex alpha subunit 2 |
| chr4_+_646960 | 2.04 |
ENST00000488061.1
ENST00000429163.2 |
PDE6B
|
phosphodiesterase 6B, cGMP-specific, rod, beta |
| chr11_-_2924970 | 2.03 |
ENST00000533594.1
|
SLC22A18AS
|
solute carrier family 22 (organic cation transporter), member 18 antisense |
| chr15_+_81589254 | 2.01 |
ENST00000394652.2
|
IL16
|
interleukin 16 |
| chr8_-_91095099 | 2.01 |
ENST00000265431.3
|
CALB1
|
calbindin 1, 28kDa |
| chr6_+_26402517 | 2.00 |
ENST00000414912.2
|
BTN3A1
|
butyrophilin, subfamily 3, member A1 |
| chr17_-_73401567 | 1.98 |
ENST00000392562.1
|
GRB2
|
growth factor receptor-bound protein 2 |
| chrX_+_37639264 | 1.97 |
ENST00000378588.4
|
CYBB
|
cytochrome b-245, beta polypeptide |
| chr1_-_92351769 | 1.93 |
ENST00000212355.4
|
TGFBR3
|
transforming growth factor, beta receptor III |
| chr8_-_125577940 | 1.90 |
ENST00000519168.1
ENST00000395508.2 |
MTSS1
|
metastasis suppressor 1 |
| chr1_-_165414414 | 1.90 |
ENST00000359842.5
|
RXRG
|
retinoid X receptor, gamma |
| chr2_+_234600253 | 1.89 |
ENST00000373424.1
ENST00000441351.1 |
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr3_+_190333097 | 1.89 |
ENST00000412080.1
|
IL1RAP
|
interleukin 1 receptor accessory protein |
| chr19_-_14785622 | 1.87 |
ENST00000443157.2
|
EMR3
|
egf-like module containing, mucin-like, hormone receptor-like 3 |
| chr19_+_56187987 | 1.86 |
ENST00000411543.2
|
EPN1
|
epsin 1 |
| chr11_-_119991589 | 1.85 |
ENST00000526881.1
|
TRIM29
|
tripartite motif containing 29 |
| chr8_+_24151553 | 1.84 |
ENST00000265769.4
ENST00000540823.1 ENST00000397649.3 |
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr7_-_14942944 | 1.78 |
ENST00000403951.2
|
DGKB
|
diacylglycerol kinase, beta 90kDa |
| chr6_-_26199499 | 1.77 |
ENST00000377831.5
|
HIST1H3D
|
histone cluster 1, H3d |
| chrX_-_138724677 | 1.73 |
ENST00000370573.4
ENST00000338585.6 ENST00000370576.4 |
MCF2
|
MCF.2 cell line derived transforming sequence |
| chrX_+_15525426 | 1.72 |
ENST00000342014.6
|
BMX
|
BMX non-receptor tyrosine kinase |
| chr7_+_134832808 | 1.71 |
ENST00000275767.3
|
TMEM140
|
transmembrane protein 140 |
| chr1_+_12834984 | 1.70 |
ENST00000357726.4
|
PRAMEF12
|
PRAME family member 12 |
| chr6_+_55039050 | 1.66 |
ENST00000370862.3
|
HCRTR2
|
hypocretin (orexin) receptor 2 |
| chr12_-_81763127 | 1.66 |
ENST00000541017.1
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr12_-_23737534 | 1.65 |
ENST00000396007.2
|
SOX5
|
SRY (sex determining region Y)-box 5 |
| chr10_-_46620012 | 1.65 |
ENST00000508602.1
ENST00000374339.3 ENST00000502254.1 ENST00000437863.1 ENST00000374342.2 ENST00000395722.3 |
PTPN20A
|
protein tyrosine phosphatase, non-receptor type 20A |
| chr5_+_140723601 | 1.65 |
ENST00000253812.6
|
PCDHGA3
|
protocadherin gamma subfamily A, 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of DUXA
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 11.2 | GO:1904395 | retinal rod cell differentiation(GO:0060221) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 2.8 | 11.1 | GO:1904749 | regulation of protein localization to nucleolus(GO:1904749) |
| 1.8 | 5.5 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 1.6 | 4.9 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 1.5 | 18.2 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 1.5 | 4.4 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 1.4 | 11.5 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 1.4 | 4.2 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 1.3 | 4.0 | GO:0002881 | negative regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002881) |
| 1.1 | 6.8 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 1.1 | 3.2 | GO:0021779 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 1.1 | 3.2 | GO:1902997 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 1.0 | 5.1 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 1.0 | 3.0 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.9 | 3.8 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.9 | 2.8 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.9 | 3.6 | GO:1900827 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.9 | 0.9 | GO:0035378 | carbon dioxide transmembrane transport(GO:0035378) |
| 0.9 | 6.9 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.9 | 2.6 | GO:0009726 | detection of endogenous stimulus(GO:0009726) |
| 0.8 | 5.9 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.8 | 3.1 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.8 | 7.6 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.7 | 2.2 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.7 | 8.7 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.7 | 5.0 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.7 | 3.6 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.7 | 5.3 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.6 | 6.5 | GO:0006553 | lysine metabolic process(GO:0006553) lysine catabolic process(GO:0006554) |
| 0.6 | 5.2 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.5 | 1.6 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.5 | 4.3 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.5 | 3.7 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.5 | 3.1 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.5 | 6.1 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.5 | 2.5 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.5 | 10.9 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.5 | 1.5 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.5 | 3.4 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.4 | 1.3 | GO:0030264 | nuclear fragmentation involved in apoptotic nuclear change(GO:0030264) positive regulation of cardiac vascular smooth muscle cell differentiation(GO:2000724) |
| 0.4 | 7.7 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.4 | 4.2 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.4 | 2.5 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.4 | 3.6 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.4 | 3.5 | GO:0015705 | iodide transport(GO:0015705) |
| 0.4 | 13.0 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.4 | 1.1 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.4 | 3.3 | GO:0002118 | aggressive behavior(GO:0002118) |
| 0.4 | 5.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.3 | 1.0 | GO:0005989 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.3 | 2.4 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.3 | 1.7 | GO:0042746 | regulation of circadian sleep/wake cycle, wakefulness(GO:0010840) circadian sleep/wake cycle, wakefulness(GO:0042746) |
| 0.3 | 1.6 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.3 | 1.9 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.3 | 1.9 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.3 | 3.1 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.3 | 1.5 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 0.3 | 0.9 | GO:0050883 | negative regulation of sodium:proton antiporter activity(GO:0032416) musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.3 | 2.1 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.3 | 3.0 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.3 | 0.9 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.3 | 6.4 | GO:0016246 | RNA interference(GO:0016246) |
| 0.3 | 2.0 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.3 | 1.7 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.3 | 1.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.3 | 14.3 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.3 | 2.4 | GO:1902856 | negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.3 | 5.3 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.3 | 5.2 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.3 | 3.8 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.3 | 2.5 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.2 | 5.1 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.2 | 15.1 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.2 | 4.1 | GO:0002052 | positive regulation of neuroblast proliferation(GO:0002052) |
| 0.2 | 2.7 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.2 | 5.9 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.2 | 1.1 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.2 | 2.6 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.2 | 0.8 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.2 | 5.2 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.2 | 2.1 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.2 | 1.3 | GO:0050890 | cognition(GO:0050890) |
| 0.2 | 2.2 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.2 | 1.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.2 | 3.6 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 5.4 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.2 | 2.9 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.2 | 3.0 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.2 | 2.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.2 | 2.4 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.1 | 2.4 | GO:0071027 | nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.1 | 8.0 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 2.9 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 1.1 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 1.4 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 1.4 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.1 | 3.1 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.1 | 1.4 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 1.7 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 1.3 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.1 | 18.5 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.1 | 2.0 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.1 | 2.2 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.1 | 1.9 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.1 | 0.5 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.1 | 3.5 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.1 | 1.7 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.1 | 2.1 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 1.3 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.1 | 1.5 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.1 | 3.5 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.1 | 1.7 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.1 | 70.3 | GO:0045596 | negative regulation of cell differentiation(GO:0045596) |
| 0.1 | 0.7 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.1 | 27.1 | GO:0090305 | nucleic acid phosphodiester bond hydrolysis(GO:0090305) |
| 0.1 | 0.2 | GO:0031635 | adenylate cyclase-inhibiting opioid receptor signaling pathway(GO:0031635) |
| 0.1 | 2.8 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.1 | 2.2 | GO:0006884 | cell volume homeostasis(GO:0006884) motile cilium assembly(GO:0044458) |
| 0.1 | 2.3 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.1 | 6.3 | GO:0098869 | cellular oxidant detoxification(GO:0098869) |
| 0.1 | 0.7 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.1 | 0.9 | GO:0051775 | response to redox state(GO:0051775) |
| 0.1 | 4.4 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.1 | 9.4 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.1 | 1.3 | GO:0030220 | platelet formation(GO:0030220) |
| 0.1 | 0.4 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.1 | 4.3 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 2.5 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.1 | 3.1 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.1 | 7.3 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.1 | 9.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 5.5 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.1 | 3.5 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 1.9 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) |
| 0.1 | 0.1 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.1 | 0.6 | GO:0051647 | nucleus localization(GO:0051647) |
| 0.1 | 1.8 | GO:0006356 | regulation of transcription from RNA polymerase I promoter(GO:0006356) |
| 0.1 | 4.2 | GO:0045814 | negative regulation of gene expression, epigenetic(GO:0045814) |
| 0.0 | 1.1 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 2.5 | GO:0032508 | DNA duplex unwinding(GO:0032508) |
| 0.0 | 2.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.2 | GO:1904180 | negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.0 | 3.6 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 2.5 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.2 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.0 | 1.8 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.0 | 1.3 | GO:0009798 | axis specification(GO:0009798) |
| 0.0 | 2.6 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 1.0 | GO:0060348 | bone development(GO:0060348) |
| 0.0 | 2.3 | GO:2000117 | negative regulation of cysteine-type endopeptidase activity(GO:2000117) |
| 0.0 | 0.7 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.9 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 1.4 | GO:1902807 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 2.6 | GO:0016358 | dendrite development(GO:0016358) |
| 0.0 | 2.5 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 1.7 | GO:0007338 | single fertilization(GO:0007338) |
| 0.0 | 1.1 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
| 0.0 | 0.5 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 6.8 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 1.1 | 4.4 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 1.0 | 13.0 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.7 | 2.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.7 | 3.4 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.6 | 8.7 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.6 | 11.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.5 | 3.0 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.5 | 11.5 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.5 | 8.9 | GO:0031045 | dense core granule(GO:0031045) |
| 0.5 | 8.0 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.4 | 5.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.4 | 4.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.4 | 1.6 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.4 | 5.4 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.4 | 8.0 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.3 | 3.7 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.3 | 2.0 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.3 | 5.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.3 | 2.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.3 | 3.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.3 | 3.0 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.3 | 14.3 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.3 | 1.3 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.3 | 3.6 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.3 | 1.8 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.2 | 4.2 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.2 | 2.5 | GO:0032797 | SMN complex(GO:0032797) |
| 0.2 | 9.8 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.2 | 5.0 | GO:0042627 | chylomicron(GO:0042627) |
| 0.2 | 2.5 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.2 | 1.2 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 4.9 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 6.9 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 1.6 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.1 | 1.8 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 1.0 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.1 | 4.1 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 11.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 24.1 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 2.2 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 2.3 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.1 | 1.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 1.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 3.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 4.2 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 1.6 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 6.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 6.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 13.8 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 2.5 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 2.7 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 4.5 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 6.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.5 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 19.7 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 2.0 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.6 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 2.6 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 5.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 2.4 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 7.5 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.7 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.9 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 6.3 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 9.0 | GO:0044431 | Golgi apparatus part(GO:0044431) |
| 0.0 | 1.7 | GO:0005925 | cell-substrate adherens junction(GO:0005924) focal adhesion(GO:0005925) cell-substrate junction(GO:0030055) |
| 0.0 | 0.7 | GO:0043204 | perikaryon(GO:0043204) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 11.2 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 1.8 | 5.3 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 1.7 | 5.0 | GO:0017129 | triglyceride binding(GO:0017129) |
| 1.6 | 4.9 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 1.6 | 6.5 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 1.6 | 4.8 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 1.5 | 7.6 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 1.5 | 9.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 1.3 | 4.0 | GO:0016495 | C-X3-C chemokine receptor activity(GO:0016495) |
| 1.3 | 5.2 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 1.2 | 3.7 | GO:0070052 | collagen V binding(GO:0070052) |
| 1.2 | 5.9 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 1.2 | 6.9 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 1.1 | 3.3 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.9 | 3.8 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.9 | 2.8 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.9 | 3.7 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.9 | 5.4 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.9 | 8.0 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.9 | 2.6 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.9 | 5.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.8 | 2.5 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.8 | 2.4 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.7 | 2.2 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.7 | 4.2 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.7 | 9.3 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.6 | 6.9 | GO:0008430 | selenium binding(GO:0008430) |
| 0.6 | 11.1 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.5 | 2.5 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.5 | 11.5 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.5 | 1.9 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.5 | 5.2 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.4 | 4.4 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.4 | 2.2 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.4 | 3.5 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.4 | 1.3 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.4 | 13.0 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.4 | 1.7 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.4 | 2.5 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.4 | 1.6 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.4 | 4.8 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.4 | 1.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.3 | 2.0 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.3 | 1.9 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.3 | 1.5 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.3 | 43.3 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.3 | 3.0 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.3 | 5.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.3 | 0.9 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.3 | 3.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.3 | 3.6 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.3 | 1.4 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.3 | 27.6 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.3 | 2.2 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.3 | 3.0 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.3 | 1.3 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.3 | 3.4 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.3 | 1.0 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.3 | 5.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.3 | 1.5 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.2 | 0.7 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.2 | 3.0 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.2 | 0.9 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.2 | 3.2 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.2 | 3.6 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.2 | 3.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.2 | 6.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 4.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 2.1 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.2 | 2.9 | GO:0005522 | profilin binding(GO:0005522) |
| 0.2 | 1.4 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 2.0 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.1 | 4.2 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 3.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 1.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.1 | 1.4 | GO:0030882 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.1 | 3.2 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 0.9 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 2.1 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 1.3 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 10.0 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.1 | 2.0 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 2.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 1.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 1.5 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 1.8 | GO:0001163 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.1 | 8.0 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 2.8 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 2.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 21.5 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.1 | 0.8 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.1 | 3.0 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 4.2 | GO:0004114 | 3',5'-cyclic-nucleotide phosphodiesterase activity(GO:0004114) |
| 0.1 | 0.2 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.1 | 1.8 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 1.1 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 2.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.7 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.1 | 9.3 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 2.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 2.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 0.9 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 1.4 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 1.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 1.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 3.3 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 5.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.3 | GO:0008237 | metallopeptidase activity(GO:0008237) |
| 0.0 | 1.5 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 1.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 4.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 2.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 4.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.1 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.4 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 2.5 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 24.2 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 0.9 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 1.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 6.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.3 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.5 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.3 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 2.6 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 2.8 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 4.5 | GO:0005525 | GTP binding(GO:0005525) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 11.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.3 | 5.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.2 | 17.1 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.2 | 14.3 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.2 | 10.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 4.7 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 2.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 2.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 10.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 6.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 2.3 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 5.4 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 2.9 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 1.9 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 4.4 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 1.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 1.9 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.1 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 3.6 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 3.8 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 1.9 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 2.7 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.9 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 8.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.1 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 1.0 | NABA COLLAGENS | Genes encoding collagen proteins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 11.5 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.5 | 4.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.4 | 7.3 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.4 | 8.0 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.4 | 9.8 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.3 | 12.9 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.3 | 9.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.3 | 10.7 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.3 | 6.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 11.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.2 | 3.4 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.2 | 3.0 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.2 | 2.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.2 | 3.1 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.2 | 5.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.2 | 3.9 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.2 | 2.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 2.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 6.1 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 2.5 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.1 | 5.4 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.1 | 2.4 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 5.4 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 1.9 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 3.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 5.1 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 2.0 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 4.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 2.2 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 1.1 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 3.4 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.1 | 5.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 1.1 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 2.8 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 2.0 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.9 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 2.5 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.5 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.9 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 1.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 3.6 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.9 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.9 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 1.1 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.7 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 1.7 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 1.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.4 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 1.0 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |