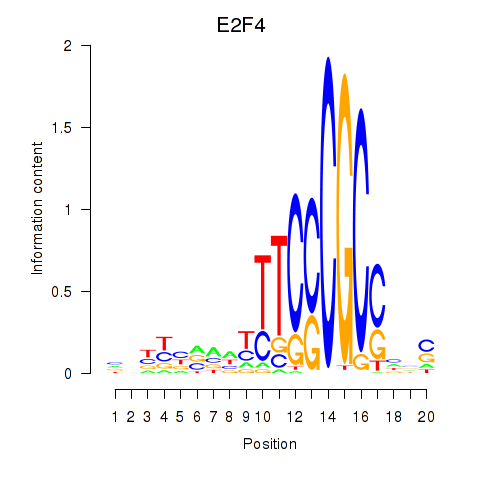
Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for E2F4
Z-value: 1.00
Transcription factors associated with E2F4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2F4
|
ENSG00000205250.4 | E2F transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2F4 | hg19_v2_chr16_+_67226019_67226127 | 0.21 | 2.1e-03 | Click! |
Activity profile of E2F4 motif
Sorted Z-values of E2F4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_172778952 | 25.70 |
ENST00000392584.1
ENST00000264108.4 |
HAT1
|
histone acetyltransferase 1 |
| chr2_+_10262857 | 24.14 |
ENST00000304567.5
|
RRM2
|
ribonucleotide reductase M2 |
| chr14_-_55658323 | 23.60 |
ENST00000554067.1
ENST00000247191.2 |
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5 |
| chr1_-_197115818 | 22.80 |
ENST00000367409.4
ENST00000294732.7 |
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr4_-_185655278 | 21.31 |
ENST00000281453.5
|
MLF1IP
|
centromere protein U |
| chr19_+_16187085 | 19.71 |
ENST00000300933.4
|
TPM4
|
tropomyosin 4 |
| chr3_+_180630444 | 19.18 |
ENST00000491062.1
ENST00000468861.1 ENST00000445140.2 ENST00000484958.1 |
FXR1
|
fragile X mental retardation, autosomal homolog 1 |
| chr4_-_1714037 | 19.16 |
ENST00000488267.1
ENST00000429429.2 ENST00000480936.1 |
SLBP
|
stem-loop binding protein |
| chr17_-_76183111 | 19.13 |
ENST00000405273.1
ENST00000590862.1 ENST00000590430.1 ENST00000586613.1 |
TK1
|
thymidine kinase 1, soluble |
| chr9_-_113018746 | 15.84 |
ENST00000374515.5
|
TXN
|
thioredoxin |
| chr6_-_52149475 | 14.49 |
ENST00000419835.2
ENST00000229854.7 ENST00000596288.1 |
MCM3
|
minichromosome maintenance complex component 3 |
| chr7_-_99698338 | 14.33 |
ENST00000354230.3
ENST00000425308.1 |
MCM7
|
minichromosome maintenance complex component 7 |
| chr4_-_1713977 | 14.28 |
ENST00000318386.4
|
SLBP
|
stem-loop binding protein |
| chr8_+_128748308 | 13.85 |
ENST00000377970.2
|
MYC
|
v-myc avian myelocytomatosis viral oncogene homolog |
| chr5_-_79950371 | 13.54 |
ENST00000511032.1
ENST00000504396.1 ENST00000505337.1 |
DHFR
|
dihydrofolate reductase |
| chr6_-_17706618 | 13.48 |
ENST00000262077.2
ENST00000537253.1 |
NUP153
|
nucleoporin 153kDa |
| chr8_+_128748466 | 13.31 |
ENST00000524013.1
ENST00000520751.1 |
MYC
|
v-myc avian myelocytomatosis viral oncogene homolog |
| chr13_+_34392185 | 12.39 |
ENST00000380071.3
|
RFC3
|
replication factor C (activator 1) 3, 38kDa |
| chr2_+_48010312 | 12.24 |
ENST00000540021.1
|
MSH6
|
mutS homolog 6 |
| chr3_+_180630090 | 12.07 |
ENST00000357559.4
ENST00000305586.7 |
FXR1
|
fragile X mental retardation, autosomal homolog 1 |
| chr1_-_8939265 | 11.93 |
ENST00000489867.1
|
ENO1
|
enolase 1, (alpha) |
| chr2_+_48010221 | 11.58 |
ENST00000234420.5
|
MSH6
|
mutS homolog 6 |
| chr11_+_58910201 | 11.48 |
ENST00000528737.1
|
FAM111A
|
family with sequence similarity 111, member A |
| chr3_+_52719936 | 11.47 |
ENST00000418458.1
ENST00000394799.2 |
GNL3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr15_+_48624300 | 11.47 |
ENST00000455976.2
ENST00000559540.1 |
DUT
|
deoxyuridine triphosphatase |
| chr21_-_33651324 | 11.46 |
ENST00000290130.3
|
MIS18A
|
MIS18 kinetochore protein A |
| chr12_-_102513843 | 11.41 |
ENST00000551744.2
ENST00000552283.1 |
NUP37
|
nucleoporin 37kDa |
| chr5_+_892745 | 11.25 |
ENST00000166345.3
|
TRIP13
|
thyroid hormone receptor interactor 13 |
| chr14_+_105219437 | 11.21 |
ENST00000329967.6
ENST00000347067.5 ENST00000553810.1 |
SIVA1
|
SIVA1, apoptosis-inducing factor |
| chr19_-_1095330 | 11.02 |
ENST00000586746.1
|
POLR2E
|
polymerase (RNA) II (DNA directed) polypeptide E, 25kDa |
| chr3_+_179280668 | 10.99 |
ENST00000429709.2
ENST00000450518.2 ENST00000392662.1 ENST00000490364.1 |
ACTL6A
|
actin-like 6A |
| chr3_+_160117418 | 10.95 |
ENST00000465903.1
ENST00000485645.1 ENST00000360111.2 ENST00000472991.1 ENST00000467468.1 ENST00000469762.1 ENST00000489573.1 ENST00000462787.1 ENST00000490207.1 ENST00000485867.1 |
SMC4
|
structural maintenance of chromosomes 4 |
| chr5_+_177631523 | 10.66 |
ENST00000506339.1
ENST00000355836.5 ENST00000514633.1 ENST00000515193.1 ENST00000506259.1 ENST00000504898.1 |
HNRNPAB
|
heterogeneous nuclear ribonucleoprotein A/B |
| chr11_+_58910295 | 10.61 |
ENST00000420244.1
|
FAM111A
|
family with sequence similarity 111, member A |
| chr3_+_133502877 | 10.60 |
ENST00000466490.2
|
SRPRB
|
signal recognition particle receptor, B subunit |
| chr8_+_48873453 | 10.43 |
ENST00000523944.1
|
MCM4
|
minichromosome maintenance complex component 4 |
| chr20_+_47662805 | 10.31 |
ENST00000262982.2
ENST00000542325.1 |
CSE1L
|
CSE1 chromosome segregation 1-like (yeast) |
| chr8_-_124408652 | 10.28 |
ENST00000287394.5
|
ATAD2
|
ATPase family, AAA domain containing 2 |
| chr8_+_48873479 | 10.23 |
ENST00000262105.2
|
MCM4
|
minichromosome maintenance complex component 4 |
| chr17_-_38804061 | 10.02 |
ENST00000474246.1
ENST00000377808.4 ENST00000578044.1 ENST00000580419.1 ENST00000400122.3 ENST00000580654.1 ENST00000577721.1 ENST00000478349.2 ENST00000431889.2 |
SMARCE1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 |
| chr6_+_30687978 | 9.53 |
ENST00000327892.8
ENST00000435534.1 |
TUBB
|
tubulin, beta class I |
| chr19_-_8070474 | 9.38 |
ENST00000407627.2
ENST00000593807.1 |
ELAVL1
|
ELAV like RNA binding protein 1 |
| chr22_+_24951949 | 9.25 |
ENST00000402849.1
|
SNRPD3
|
small nuclear ribonucleoprotein D3 polypeptide 18kDa |
| chr1_-_33283754 | 9.21 |
ENST00000373477.4
|
YARS
|
tyrosyl-tRNA synthetase |
| chr10_+_92631709 | 9.14 |
ENST00000413330.1
ENST00000277882.3 |
RPP30
|
ribonuclease P/MRP 30kDa subunit |
| chr1_+_28844648 | 9.10 |
ENST00000373832.1
ENST00000373831.3 |
RCC1
|
regulator of chromosome condensation 1 |
| chr6_+_135502466 | 8.92 |
ENST00000367814.4
|
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr7_-_102985288 | 8.86 |
ENST00000379263.3
|
DNAJC2
|
DnaJ (Hsp40) homolog, subfamily C, member 2 |
| chr22_-_42342692 | 8.76 |
ENST00000404067.1
ENST00000402338.1 |
CENPM
|
centromere protein M |
| chr6_+_135502408 | 8.54 |
ENST00000341911.5
ENST00000442647.2 ENST00000316528.8 |
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr2_-_38978492 | 8.43 |
ENST00000409276.1
ENST00000446327.2 ENST00000313117.6 |
SRSF7
|
serine/arginine-rich splicing factor 7 |
| chr2_+_47630255 | 8.38 |
ENST00000406134.1
|
MSH2
|
mutS homolog 2 |
| chr22_-_42343117 | 8.37 |
ENST00000407253.3
ENST00000215980.5 |
CENPM
|
centromere protein M |
| chr1_+_33116743 | 8.34 |
ENST00000414241.3
ENST00000373493.5 |
RBBP4
|
retinoblastoma binding protein 4 |
| chrX_+_109245863 | 8.26 |
ENST00000372072.3
|
TMEM164
|
transmembrane protein 164 |
| chr12_+_16064258 | 8.13 |
ENST00000524480.1
ENST00000531803.1 ENST00000532964.1 |
DERA
|
deoxyribose-phosphate aldolase (putative) |
| chr19_+_50180317 | 7.98 |
ENST00000534465.1
|
PRMT1
|
protein arginine methyltransferase 1 |
| chr15_+_91260552 | 7.79 |
ENST00000355112.3
ENST00000560509.1 |
BLM
|
Bloom syndrome, RecQ helicase-like |
| chrX_+_109246285 | 7.72 |
ENST00000372073.1
ENST00000372068.2 ENST00000288381.4 |
TMEM164
|
transmembrane protein 164 |
| chr7_-_102985035 | 7.69 |
ENST00000426036.2
ENST00000249270.7 ENST00000454277.1 ENST00000412522.1 |
DNAJC2
|
DnaJ (Hsp40) homolog, subfamily C, member 2 |
| chr2_+_47630108 | 7.68 |
ENST00000233146.2
ENST00000454849.1 ENST00000543555.1 |
MSH2
|
mutS homolog 2 |
| chr4_-_157892498 | 7.60 |
ENST00000502773.1
|
PDGFC
|
platelet derived growth factor C |
| chr7_-_73668692 | 7.58 |
ENST00000352131.3
ENST00000055077.3 |
RFC2
|
replication factor C (activator 1) 2, 40kDa |
| chr7_+_138145145 | 7.55 |
ENST00000415680.2
|
TRIM24
|
tripartite motif containing 24 |
| chr9_-_35080013 | 7.38 |
ENST00000378643.3
|
FANCG
|
Fanconi anemia, complementation group G |
| chr12_-_57146095 | 7.36 |
ENST00000550770.1
ENST00000338193.6 |
PRIM1
|
primase, DNA, polypeptide 1 (49kDa) |
| chr1_+_91966656 | 7.33 |
ENST00000428239.1
ENST00000426137.1 |
CDC7
|
cell division cycle 7 |
| chr7_+_116166331 | 7.27 |
ENST00000393468.1
ENST00000393467.1 |
CAV1
|
caveolin 1, caveolae protein, 22kDa |
| chr9_-_113018835 | 7.25 |
ENST00000374517.5
|
TXN
|
thioredoxin |
| chr7_+_116165754 | 7.13 |
ENST00000405348.1
|
CAV1
|
caveolin 1, caveolae protein, 22kDa |
| chr14_-_50154921 | 7.06 |
ENST00000553805.2
ENST00000554396.1 ENST00000216367.5 ENST00000539565.2 |
POLE2
|
polymerase (DNA directed), epsilon 2, accessory subunit |
| chr22_+_19467261 | 6.96 |
ENST00000455750.1
ENST00000437685.2 ENST00000263201.1 ENST00000404724.3 |
CDC45
|
cell division cycle 45 |
| chr1_+_91966384 | 6.93 |
ENST00000430031.2
ENST00000234626.6 |
CDC7
|
cell division cycle 7 |
| chr8_-_67525473 | 6.88 |
ENST00000522677.3
|
MYBL1
|
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
| chr4_+_88928777 | 6.86 |
ENST00000237596.2
|
PKD2
|
polycystic kidney disease 2 (autosomal dominant) |
| chrX_+_133594168 | 6.80 |
ENST00000298556.7
|
HPRT1
|
hypoxanthine phosphoribosyltransferase 1 |
| chr10_-_88281494 | 6.71 |
ENST00000298767.5
|
WAPAL
|
wings apart-like homolog (Drosophila) |
| chr10_-_43892279 | 6.71 |
ENST00000443950.2
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F |
| chr15_-_50647347 | 6.63 |
ENST00000220429.8
ENST00000429662.2 |
GABPB1
|
GA binding protein transcription factor, beta subunit 1 |
| chr11_+_34073269 | 6.51 |
ENST00000389645.3
|
CAPRIN1
|
cell cycle associated protein 1 |
| chr12_-_56843161 | 6.41 |
ENST00000554616.1
ENST00000553532.1 ENST00000229201.4 |
TIMELESS
|
timeless circadian clock |
| chr16_-_67693846 | 6.41 |
ENST00000602850.1
|
ACD
|
adrenocortical dysplasia homolog (mouse) |
| chr8_-_95907423 | 6.20 |
ENST00000396133.3
ENST00000308108.4 |
CCNE2
|
cyclin E2 |
| chr11_-_118972575 | 6.11 |
ENST00000432443.2
|
DPAGT1
|
dolichyl-phosphate (UDP-N-acetylglucosamine) N-acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase) |
| chr4_+_113558272 | 6.11 |
ENST00000509061.1
ENST00000508577.1 ENST00000513553.1 |
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr9_-_21994344 | 6.09 |
ENST00000530628.2
ENST00000361570.3 |
CDKN2A
|
cyclin-dependent kinase inhibitor 2A |
| chr3_-_185655795 | 5.99 |
ENST00000342294.4
ENST00000382191.4 ENST00000453386.2 |
TRA2B
|
transformer 2 beta homolog (Drosophila) |
| chr16_-_8962853 | 5.98 |
ENST00000565287.1
ENST00000311052.5 |
CARHSP1
|
calcium regulated heat stable protein 1, 24kDa |
| chr3_-_10028366 | 5.94 |
ENST00000429759.1
|
EMC3
|
ER membrane protein complex subunit 3 |
| chr5_+_36152091 | 5.92 |
ENST00000274254.5
|
SKP2
|
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr1_-_26185844 | 5.81 |
ENST00000538789.1
ENST00000374298.3 |
AUNIP
|
aurora kinase A and ninein interacting protein |
| chr11_+_34073195 | 5.79 |
ENST00000341394.4
|
CAPRIN1
|
cell cycle associated protein 1 |
| chr11_+_17298297 | 5.73 |
ENST00000529010.1
|
NUCB2
|
nucleobindin 2 |
| chr20_+_42295745 | 5.70 |
ENST00000396863.4
ENST00000217026.4 |
MYBL2
|
v-myb avian myeloblastosis viral oncogene homolog-like 2 |
| chr1_+_10490779 | 5.64 |
ENST00000477755.1
|
APITD1
|
apoptosis-inducing, TAF9-like domain 1 |
| chr8_-_17104356 | 5.62 |
ENST00000361272.4
ENST00000523917.1 |
CNOT7
|
CCR4-NOT transcription complex, subunit 7 |
| chr3_+_133292574 | 5.61 |
ENST00000264993.3
|
CDV3
|
CDV3 homolog (mouse) |
| chr19_+_10982189 | 5.60 |
ENST00000327064.4
ENST00000588947.1 |
CARM1
|
coactivator-associated arginine methyltransferase 1 |
| chr7_-_30544405 | 5.57 |
ENST00000409390.1
ENST00000409144.1 ENST00000005374.6 ENST00000409436.1 ENST00000275428.4 |
GGCT
|
gamma-glutamylcyclotransferase |
| chr1_+_28844778 | 5.53 |
ENST00000411533.1
|
RCC1
|
regulator of chromosome condensation 1 |
| chr19_+_13049413 | 5.52 |
ENST00000316448.5
ENST00000588454.1 |
CALR
|
calreticulin |
| chr7_+_138145076 | 5.51 |
ENST00000343526.4
|
TRIM24
|
tripartite motif containing 24 |
| chr16_+_29817399 | 5.50 |
ENST00000545521.1
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr3_-_148804275 | 5.36 |
ENST00000392912.2
ENST00000465259.1 ENST00000310053.5 ENST00000494055.1 |
HLTF
|
helicase-like transcription factor |
| chr11_+_17298255 | 5.34 |
ENST00000531172.1
ENST00000533738.2 ENST00000323688.6 |
NUCB2
|
nucleobindin 2 |
| chr12_+_102513950 | 5.23 |
ENST00000378128.3
ENST00000327680.2 ENST00000541394.1 ENST00000543784.1 |
PARPBP
|
PARP1 binding protein |
| chr6_+_167412835 | 5.22 |
ENST00000349556.4
|
FGFR1OP
|
FGFR1 oncogene partner |
| chr2_+_71357434 | 5.21 |
ENST00000244230.2
|
MPHOSPH10
|
M-phase phosphoprotein 10 (U3 small nucleolar ribonucleoprotein) |
| chr10_-_43892668 | 5.18 |
ENST00000544000.1
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F |
| chr5_+_36152163 | 5.04 |
ENST00000274255.6
|
SKP2
|
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr1_+_63833261 | 5.00 |
ENST00000371108.4
|
ALG6
|
ALG6, alpha-1,3-glucosyltransferase |
| chr13_-_113862948 | 4.95 |
ENST00000375457.2
ENST00000375477.1 ENST00000246505.5 ENST00000337344.4 ENST00000375479.2 |
PCID2
|
PCI domain containing 2 |
| chr2_+_27440229 | 4.95 |
ENST00000264705.4
ENST00000403525.1 |
CAD
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr8_+_95732095 | 4.93 |
ENST00000414645.2
|
DPY19L4
|
dpy-19-like 4 (C. elegans) |
| chr1_-_241683001 | 4.82 |
ENST00000366560.3
|
FH
|
fumarate hydratase |
| chr17_+_73008755 | 4.82 |
ENST00000584208.1
ENST00000301585.5 |
ICT1
|
immature colon carcinoma transcript 1 |
| chr5_+_141348640 | 4.73 |
ENST00000540015.1
ENST00000506938.1 ENST00000394514.2 ENST00000512565.1 ENST00000394515.3 |
RNF14
|
ring finger protein 14 |
| chr4_-_39367949 | 4.71 |
ENST00000503784.1
ENST00000349703.2 ENST00000381897.1 |
RFC1
|
replication factor C (activator 1) 1, 145kDa |
| chr2_+_68384976 | 4.65 |
ENST00000263657.2
|
PNO1
|
partner of NOB1 homolog (S. cerevisiae) |
| chr22_+_20105012 | 4.57 |
ENST00000331821.3
ENST00000411892.1 |
RANBP1
|
RAN binding protein 1 |
| chr11_+_85955787 | 4.51 |
ENST00000528180.1
|
EED
|
embryonic ectoderm development |
| chr14_-_71067360 | 4.50 |
ENST00000554963.1
ENST00000430055.2 ENST00000440435.2 ENST00000256379.5 |
MED6
|
mediator complex subunit 6 |
| chr19_+_797443 | 4.41 |
ENST00000394601.4
ENST00000589575.1 |
PTBP1
|
polypyrimidine tract binding protein 1 |
| chr19_+_34919257 | 4.39 |
ENST00000246548.4
ENST00000590048.2 |
UBA2
|
ubiquitin-like modifier activating enzyme 2 |
| chr12_+_49717019 | 4.36 |
ENST00000549275.1
ENST00000551245.1 ENST00000380327.5 ENST00000548311.1 ENST00000550346.1 ENST00000550709.1 ENST00000549534.1 ENST00000257909.3 |
TROAP
|
trophinin associated protein |
| chr12_+_16064106 | 4.29 |
ENST00000428559.2
|
DERA
|
deoxyribose-phosphate aldolase (putative) |
| chr1_+_27248203 | 4.29 |
ENST00000321265.5
|
NUDC
|
nudC nuclear distribution protein |
| chr7_+_130794846 | 4.24 |
ENST00000421797.2
|
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr3_+_37903432 | 4.22 |
ENST00000443503.2
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
| chr10_-_35104185 | 4.08 |
ENST00000374789.3
ENST00000374788.3 ENST00000346874.4 ENST00000374794.3 ENST00000350537.4 ENST00000374790.3 ENST00000374776.1 ENST00000374773.1 ENST00000545693.1 ENST00000545260.1 ENST00000340077.5 |
PARD3
|
par-3 family cell polarity regulator |
| chr5_-_134734901 | 4.05 |
ENST00000312469.4
ENST00000423969.2 |
H2AFY
|
H2A histone family, member Y |
| chr12_-_57472522 | 4.04 |
ENST00000379391.3
ENST00000300128.4 |
TMEM194A
|
transmembrane protein 194A |
| chr4_+_178230985 | 4.01 |
ENST00000264596.3
|
NEIL3
|
nei endonuclease VIII-like 3 (E. coli) |
| chr15_-_50647370 | 3.99 |
ENST00000558970.1
ENST00000396464.3 ENST00000560825.1 |
GABPB1
|
GA binding protein transcription factor, beta subunit 1 |
| chr17_-_17184605 | 3.98 |
ENST00000268717.5
|
COPS3
|
COP9 signalosome subunit 3 |
| chr3_-_55521323 | 3.97 |
ENST00000264634.4
|
WNT5A
|
wingless-type MMTV integration site family, member 5A |
| chr11_-_78285804 | 3.92 |
ENST00000281038.5
ENST00000529571.1 |
NARS2
|
asparaginyl-tRNA synthetase 2, mitochondrial (putative) |
| chr7_-_86849883 | 3.90 |
ENST00000433078.1
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chrX_+_70752917 | 3.87 |
ENST00000373719.3
|
OGT
|
O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr1_+_10490441 | 3.84 |
ENST00000470413.2
ENST00000309048.3 |
APITD1-CORT
APITD1
|
APITD1-CORT readthrough apoptosis-inducing, TAF9-like domain 1 |
| chr20_+_23331373 | 3.83 |
ENST00000254998.2
|
NXT1
|
NTF2-like export factor 1 |
| chr5_-_137548997 | 3.83 |
ENST00000505120.1
ENST00000394886.2 ENST00000394884.3 |
CDC23
|
cell division cycle 23 |
| chr7_+_77428149 | 3.81 |
ENST00000415251.2
ENST00000275575.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr5_+_72794233 | 3.81 |
ENST00000335895.8
ENST00000380591.3 ENST00000507081.2 |
BTF3
|
basic transcription factor 3 |
| chr17_-_41277467 | 3.74 |
ENST00000494123.1
ENST00000346315.3 ENST00000309486.4 ENST00000468300.1 ENST00000354071.3 ENST00000352993.3 ENST00000471181.2 |
BRCA1
|
breast cancer 1, early onset |
| chr19_+_797392 | 3.71 |
ENST00000350092.4
ENST00000349038.4 ENST00000586481.1 ENST00000585535.1 |
PTBP1
|
polypyrimidine tract binding protein 1 |
| chr9_-_21994597 | 3.66 |
ENST00000579755.1
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A |
| chr8_-_103668114 | 3.62 |
ENST00000285407.6
|
KLF10
|
Kruppel-like factor 10 |
| chr6_-_27114577 | 3.58 |
ENST00000356950.1
ENST00000396891.4 |
HIST1H2BK
|
histone cluster 1, H2bk |
| chr12_-_15942309 | 3.57 |
ENST00000544064.1
ENST00000543523.1 ENST00000536793.1 |
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr5_+_65222299 | 3.54 |
ENST00000284037.5
|
ERBB2IP
|
erbb2 interacting protein |
| chr15_-_50647274 | 3.50 |
ENST00000543881.1
|
GABPB1
|
GA binding protein transcription factor, beta subunit 1 |
| chr9_-_139304979 | 3.48 |
ENST00000357365.3
ENST00000371723.4 |
SDCCAG3
|
serologically defined colon cancer antigen 3 |
| chr1_-_229761717 | 3.40 |
ENST00000366675.3
ENST00000258281.2 ENST00000366674.1 |
TAF5L
|
TAF5-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor, 65kDa |
| chr19_+_18284477 | 3.38 |
ENST00000407280.3
|
IFI30
|
interferon, gamma-inducible protein 30 |
| chr12_-_15942503 | 3.35 |
ENST00000281172.5
|
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr20_-_32274179 | 3.30 |
ENST00000343380.5
|
E2F1
|
E2F transcription factor 1 |
| chr5_-_43313574 | 3.25 |
ENST00000325110.6
ENST00000433297.2 |
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr13_-_111358504 | 3.23 |
ENST00000257347.4
|
CARS2
|
cysteinyl-tRNA synthetase 2, mitochondrial (putative) |
| chr5_+_138629417 | 3.22 |
ENST00000510056.1
ENST00000511249.1 ENST00000503811.1 ENST00000511378.1 |
MATR3
|
matrin 3 |
| chrX_-_15511438 | 3.21 |
ENST00000380420.5
|
PIR
|
pirin (iron-binding nuclear protein) |
| chr15_+_52121822 | 3.14 |
ENST00000558455.1
ENST00000308580.7 |
TMOD3
|
tropomodulin 3 (ubiquitous) |
| chr17_-_41277370 | 3.02 |
ENST00000476777.1
ENST00000491747.2 ENST00000478531.1 ENST00000477152.1 ENST00000357654.3 ENST00000493795.1 ENST00000493919.1 |
BRCA1
|
breast cancer 1, early onset |
| chr17_-_41277317 | 3.01 |
ENST00000497488.1
ENST00000489037.1 ENST00000470026.1 ENST00000586385.1 ENST00000591534.1 ENST00000591849.1 |
BRCA1
|
breast cancer 1, early onset |
| chr7_+_77166592 | 2.98 |
ENST00000248594.6
|
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chr7_-_42971759 | 2.93 |
ENST00000538645.1
ENST00000445517.1 ENST00000223321.4 |
PSMA2
|
proteasome (prosome, macropain) subunit, alpha type, 2 |
| chr3_-_124774802 | 2.89 |
ENST00000311127.4
|
HEG1
|
heart development protein with EGF-like domains 1 |
| chr6_-_136610911 | 2.86 |
ENST00000530767.1
ENST00000527759.1 ENST00000527536.1 ENST00000529826.1 ENST00000531224.1 ENST00000353331.4 |
BCLAF1
|
BCL2-associated transcription factor 1 |
| chr18_-_54318353 | 2.80 |
ENST00000590954.1
ENST00000540155.1 |
TXNL1
|
thioredoxin-like 1 |
| chr6_-_100016678 | 2.80 |
ENST00000523799.1
ENST00000520429.1 |
CCNC
|
cyclin C |
| chr8_-_41754231 | 2.76 |
ENST00000265709.8
|
ANK1
|
ankyrin 1, erythrocytic |
| chr1_-_28241226 | 2.76 |
ENST00000373912.3
ENST00000373909.3 |
RPA2
|
replication protein A2, 32kDa |
| chr7_+_77428066 | 2.75 |
ENST00000422959.2
ENST00000307305.8 ENST00000424760.1 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr16_+_2510081 | 2.73 |
ENST00000361837.4
ENST00000569496.1 ENST00000567489.1 ENST00000563531.1 ENST00000483320.1 |
C16orf59
|
chromosome 16 open reading frame 59 |
| chr2_-_46844242 | 2.72 |
ENST00000281382.6
|
PIGF
|
phosphatidylinositol glycan anchor biosynthesis, class F |
| chr17_-_73781567 | 2.71 |
ENST00000586607.1
|
H3F3B
|
H3 histone, family 3B (H3.3B) |
| chr2_-_128785688 | 2.68 |
ENST00000259234.6
|
SAP130
|
Sin3A-associated protein, 130kDa |
| chr16_-_67694129 | 2.67 |
ENST00000602320.1
|
ACD
|
adrenocortical dysplasia homolog (mouse) |
| chr9_-_35732362 | 2.67 |
ENST00000314888.9
ENST00000540444.1 |
TLN1
|
talin 1 |
| chrX_-_20159934 | 2.66 |
ENST00000379593.1
ENST00000379607.5 |
EIF1AX
|
eukaryotic translation initiation factor 1A, X-linked |
| chr11_-_73694346 | 2.66 |
ENST00000310473.3
|
UCP2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr1_-_36615051 | 2.66 |
ENST00000373163.1
|
TRAPPC3
|
trafficking protein particle complex 3 |
| chr19_-_48673580 | 2.64 |
ENST00000427526.2
|
LIG1
|
ligase I, DNA, ATP-dependent |
| chrX_-_119763835 | 2.61 |
ENST00000371313.2
ENST00000304661.5 |
C1GALT1C1
|
C1GALT1-specific chaperone 1 |
| chr2_-_11484710 | 2.60 |
ENST00000315872.6
|
ROCK2
|
Rho-associated, coiled-coil containing protein kinase 2 |
| chr2_-_203103185 | 2.59 |
ENST00000409205.1
|
SUMO1
|
small ubiquitin-like modifier 1 |
| chr5_-_127873659 | 2.59 |
ENST00000262464.4
|
FBN2
|
fibrillin 2 |
| chr2_-_46844159 | 2.57 |
ENST00000474980.1
ENST00000306465.4 |
PIGF
|
phosphatidylinositol glycan anchor biosynthesis, class F |
| chr19_+_50887585 | 2.56 |
ENST00000440232.2
ENST00000601098.1 ENST00000599857.1 ENST00000593887.1 |
POLD1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr6_-_34855773 | 2.55 |
ENST00000420584.2
ENST00000361288.4 |
TAF11
|
TAF11 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 28kDa |
| chr16_+_30064444 | 2.52 |
ENST00000395248.1
ENST00000566897.1 ENST00000568435.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr5_+_141348721 | 2.50 |
ENST00000507163.1
ENST00000394519.1 |
RNF14
|
ring finger protein 14 |
| chr11_-_95657231 | 2.45 |
ENST00000409459.1
ENST00000352297.7 ENST00000393223.3 ENST00000346299.5 |
MTMR2
|
myotubularin related protein 2 |
| chr19_-_6424783 | 2.44 |
ENST00000398148.3
|
KHSRP
|
KH-type splicing regulatory protein |
| chr17_-_12921270 | 2.41 |
ENST00000578071.1
ENST00000426905.3 ENST00000395962.2 ENST00000583371.1 ENST00000338034.4 |
ELAC2
|
elaC ribonuclease Z 2 |
| chr6_+_57182400 | 2.39 |
ENST00000607273.1
|
PRIM2
|
primase, DNA, polypeptide 2 (58kDa) |
| chr6_+_31633011 | 2.38 |
ENST00000375885.4
|
CSNK2B
|
casein kinase 2, beta polypeptide |
| chr16_+_30064411 | 2.38 |
ENST00000338110.5
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr19_+_41770349 | 2.36 |
ENST00000602130.1
|
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr20_-_61493115 | 2.36 |
ENST00000335351.3
ENST00000217162.5 |
TCFL5
|
transcription factor-like 5 (basic helix-loop-helix) |
Network of associatons between targets according to the STRING database.
First level regulatory network of E2F4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.8 | 6.8 | GO:0043103 | hypoxanthine salvage(GO:0043103) |
| 6.7 | 33.4 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 6.6 | 19.8 | GO:1990922 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 5.5 | 16.6 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 4.8 | 14.4 | GO:1903609 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 4.8 | 19.1 | GO:0046104 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 4.5 | 27.2 | GO:0072133 | kidney mesenchyme morphogenesis(GO:0072131) metanephric mesenchyme morphogenesis(GO:0072133) |
| 4.5 | 40.7 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 4.0 | 20.0 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 3.9 | 23.6 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 3.8 | 11.5 | GO:0046080 | dUTP metabolic process(GO:0046080) dUTP catabolic process(GO:0046081) |
| 3.8 | 22.8 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 3.7 | 29.5 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 3.7 | 11.0 | GO:0060381 | regulation of single-stranded telomeric DNA binding(GO:0060380) positive regulation of single-stranded telomeric DNA binding(GO:0060381) positive regulation of telomeric DNA binding(GO:1904744) |
| 3.4 | 13.5 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 3.3 | 32.8 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 3.3 | 9.8 | GO:0070512 | regulation of histone H4-K20 methylation(GO:0070510) positive regulation of histone H4-K20 methylation(GO:0070512) |
| 3.2 | 9.7 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 2.6 | 23.1 | GO:0030091 | protein repair(GO:0030091) |
| 2.5 | 12.4 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 2.4 | 14.3 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 2.3 | 6.9 | GO:0031587 | detection of endogenous stimulus(GO:0009726) positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) |
| 2.3 | 11.4 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 2.0 | 10.1 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 1.9 | 13.1 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 1.7 | 6.9 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 1.7 | 11.9 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 1.7 | 5.0 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 1.6 | 4.9 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 1.6 | 4.8 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 1.6 | 6.4 | GO:0072719 | cellular response to cisplatin(GO:0072719) |
| 1.5 | 6.0 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 1.4 | 4.2 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 1.4 | 11.0 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 1.4 | 13.6 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 1.4 | 4.1 | GO:1901837 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 1.3 | 4.0 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 1.3 | 4.0 | GO:1904933 | cardiac right atrium morphogenesis(GO:0003213) negative regulation of melanin biosynthetic process(GO:0048022) positive regulation of anagen(GO:0051885) mediolateral intercalation(GO:0060031) cervix development(GO:0060067) lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) planar cell polarity pathway involved in gastrula mediolateral intercalation(GO:0060775) negative regulation of secondary metabolite biosynthetic process(GO:1900377) regulation of cell proliferation in midbrain(GO:1904933) |
| 1.3 | 38.8 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 1.2 | 37.5 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 1.2 | 2.5 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 1.2 | 6.1 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 1.1 | 10.3 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 1.1 | 11.3 | GO:0007144 | female meiosis I(GO:0007144) |
| 1.1 | 29.9 | GO:0060148 | positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 1.1 | 3.2 | GO:0006423 | cysteinyl-tRNA aminoacylation(GO:0006423) |
| 1.0 | 13.5 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 1.0 | 4.0 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 1.0 | 5.0 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 1.0 | 9.8 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) |
| 1.0 | 5.8 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.9 | 5.5 | GO:0002501 | peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.8 | 14.7 | GO:1904816 | positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.8 | 27.8 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.8 | 2.4 | GO:0000965 | mitochondrial RNA 3'-end processing(GO:0000965) |
| 0.8 | 5.6 | GO:0060339 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.7 | 3.3 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.6 | 3.7 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.6 | 3.0 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.6 | 9.4 | GO:0032201 | telomere maintenance via semi-conservative replication(GO:0032201) |
| 0.6 | 1.7 | GO:0006535 | cysteine biosynthetic process from serine(GO:0006535) |
| 0.5 | 1.6 | GO:0003099 | positive regulation of the force of heart contraction by chemical signal(GO:0003099) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.5 | 7.6 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.5 | 8.1 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.5 | 3.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.5 | 2.6 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.5 | 9.1 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.5 | 9.2 | GO:0008334 | histone mRNA metabolic process(GO:0008334) |
| 0.5 | 6.9 | GO:0060965 | negative regulation of gene silencing by miRNA(GO:0060965) |
| 0.5 | 1.8 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.4 | 1.3 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.4 | 1.3 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.4 | 1.2 | GO:0002353 | kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 0.4 | 21.0 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.4 | 12.3 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.4 | 2.8 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.4 | 6.1 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.4 | 22.4 | GO:0060964 | regulation of gene silencing by miRNA(GO:0060964) |
| 0.3 | 2.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.3 | 3.2 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.3 | 1.9 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.3 | 3.7 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.3 | 5.6 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.3 | 1.5 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.3 | 2.4 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.3 | 7.6 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.3 | 2.4 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.3 | 7.1 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.3 | 22.1 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.3 | 2.4 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.3 | 4.1 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.3 | 2.6 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.3 | 3.1 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.3 | 1.4 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.3 | 3.0 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.3 | 2.7 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.3 | 5.3 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.2 | 4.0 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.2 | 19.7 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.2 | 1.4 | GO:0045144 | meiotic sister chromatid segregation(GO:0045144) |
| 0.2 | 2.0 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.2 | 4.6 | GO:0044764 | multi-organism cellular process(GO:0044764) |
| 0.2 | 2.9 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.2 | 2.1 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.2 | 2.8 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.2 | 1.2 | GO:0033504 | floor plate development(GO:0033504) |
| 0.2 | 3.8 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.2 | 5.2 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.2 | 0.7 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.2 | 9.2 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.2 | 1.2 | GO:1904951 | positive regulation of establishment of protein localization(GO:1904951) |
| 0.2 | 2.8 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.2 | 3.5 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.1 | 0.6 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.1 | 7.0 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.1 | 2.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 3.6 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
| 0.1 | 1.3 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.1 | 1.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 11.0 | GO:0071349 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.1 | 1.4 | GO:0043032 | positive regulation of macrophage activation(GO:0043032) |
| 0.1 | 1.8 | GO:0010668 | ectodermal cell differentiation(GO:0010668) nail development(GO:0035878) |
| 0.1 | 3.9 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.1 | 8.3 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 5.4 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.1 | 1.7 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 2.9 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.1 | 2.6 | GO:0036344 | platelet morphogenesis(GO:0036344) |
| 0.1 | 11.6 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.1 | 3.2 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.1 | 0.3 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.1 | 11.4 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.1 | 3.2 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.1 | 4.0 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.1 | 0.3 | GO:0048627 | myoblast development(GO:0048627) |
| 0.1 | 2.2 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.1 | 2.1 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 | 0.3 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.1 | 5.2 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 0.6 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.1 | 1.4 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 1.6 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) modulation by virus of host process(GO:0019054) |
| 0.1 | 9.8 | GO:0046718 | viral entry into host cell(GO:0046718) |
| 0.1 | 0.9 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 4.4 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 0.3 | GO:0009233 | menaquinone metabolic process(GO:0009233) |
| 0.1 | 3.5 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.1 | 1.4 | GO:0070646 | protein deubiquitination(GO:0016579) protein modification by small protein removal(GO:0070646) |
| 0.1 | 2.3 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.1 | 3.8 | GO:0045087 | innate immune response(GO:0045087) |
| 0.1 | 0.7 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 0.8 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 1.1 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.0 | 0.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.4 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 2.9 | GO:0051439 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051439) |
| 0.0 | 2.6 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 3.7 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.2 | GO:0061441 | renal artery morphogenesis(GO:0061441) |
| 0.0 | 1.9 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.6 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.0 | 0.2 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 0.4 | GO:0010952 | positive regulation of peptidase activity(GO:0010952) |
| 0.0 | 3.4 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 1.0 | GO:0090004 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 | 0.3 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 1.4 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.0 | 0.5 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 3.1 | GO:0006413 | translational initiation(GO:0006413) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.0 | 39.9 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 6.0 | 24.1 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 4.2 | 33.4 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 4.0 | 24.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 3.5 | 24.7 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 3.3 | 23.3 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 2.9 | 35.0 | GO:0042555 | MCM complex(GO:0042555) |
| 2.6 | 13.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 2.4 | 9.8 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 2.3 | 7.0 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 2.1 | 8.3 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 2.0 | 11.9 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 1.9 | 5.7 | GO:0031523 | Myb complex(GO:0031523) |
| 1.8 | 9.2 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 1.7 | 7.0 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 1.7 | 23.6 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 1.6 | 6.2 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 1.5 | 9.2 | GO:0005683 | U7 snRNP(GO:0005683) |
| 1.4 | 14.4 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 1.4 | 7.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 1.4 | 5.6 | GO:0042025 | host cell nucleus(GO:0042025) host cell nuclear part(GO:0044094) |
| 1.3 | 9.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 1.3 | 8.8 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 1.1 | 13.5 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 1.1 | 11.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 1.1 | 11.0 | GO:0070187 | telosome(GO:0070187) |
| 1.1 | 3.3 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 1.1 | 11.0 | GO:0000796 | condensin complex(GO:0000796) |
| 1.0 | 19.7 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 1.0 | 6.9 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 1.0 | 13.3 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.9 | 8.4 | GO:0000800 | lateral element(GO:0000800) |
| 0.9 | 9.8 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.9 | 21.0 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.8 | 7.6 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.8 | 0.8 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.8 | 11.4 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.7 | 8.0 | GO:0034709 | methylosome(GO:0034709) |
| 0.7 | 2.8 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.7 | 5.5 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.6 | 2.6 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.6 | 9.5 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.6 | 11.0 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.6 | 6.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.6 | 1.8 | GO:0034676 | integrin alpha6-beta4 complex(GO:0034676) |
| 0.6 | 4.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.5 | 24.4 | GO:0043034 | costamere(GO:0043034) |
| 0.5 | 2.4 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.5 | 1.4 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.5 | 6.9 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.4 | 3.0 | GO:0070522 | nucleotide-excision repair factor 1 complex(GO:0000110) ERCC4-ERCC1 complex(GO:0070522) |
| 0.4 | 7.0 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.4 | 4.5 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.4 | 1.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.4 | 1.1 | GO:0031213 | RSF complex(GO:0031213) |
| 0.4 | 49.5 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.3 | 4.8 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.3 | 2.9 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.3 | 1.3 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.3 | 3.3 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.2 | 2.4 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.2 | 2.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.2 | 11.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.2 | 5.0 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.2 | 10.9 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.2 | 2.8 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.2 | 20.1 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 8.2 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.1 | 2.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 3.5 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 5.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 19.8 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 3.8 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 11.9 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.1 | 1.9 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.1 | 2.6 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 1.4 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 2.7 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.1 | 3.1 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.1 | 4.0 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 2.0 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 5.6 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 11.2 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.1 | 3.9 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 27.8 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.1 | 3.3 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 12.8 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 0.6 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 7.7 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 3.3 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.1 | 4.0 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.1 | 1.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 2.4 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 4.7 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.3 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 2.6 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 2.8 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 2.4 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 3.6 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 1.6 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 0.5 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 5.4 | GO:1990904 | ribonucleoprotein complex(GO:1990904) |
| 0.0 | 1.4 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 2.4 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 9.3 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.0 | 39.9 | GO:0032143 | single thymine insertion binding(GO:0032143) |
| 8.5 | 42.7 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 6.4 | 19.1 | GO:0004797 | thymidine kinase activity(GO:0004797) |
| 6.0 | 24.1 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 4.8 | 14.4 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 4.1 | 12.4 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 3.8 | 11.5 | GO:0004170 | dUTP diphosphatase activity(GO:0004170) |
| 3.7 | 25.9 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 3.5 | 10.6 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 3.5 | 24.7 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 2.9 | 32.4 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 2.8 | 31.3 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 2.8 | 16.6 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 2.4 | 2.4 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 2.3 | 13.5 | GO:0051870 | methotrexate binding(GO:0051870) |
| 2.1 | 14.6 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 2.0 | 6.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 2.0 | 11.9 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 1.8 | 7.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 1.7 | 8.6 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 1.7 | 6.9 | GO:0048763 | HLH domain binding(GO:0043398) calcium-induced calcium release activity(GO:0048763) |
| 1.7 | 5.0 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 1.4 | 4.2 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 1.4 | 13.6 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 1.3 | 5.3 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 1.3 | 25.4 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 1.2 | 13.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 1.2 | 2.3 | GO:0009055 | electron carrier activity(GO:0009055) |
| 1.2 | 7.0 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 1.2 | 7.0 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 1.1 | 3.2 | GO:0004817 | cysteine-tRNA ligase activity(GO:0004817) |
| 0.9 | 10.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.8 | 35.0 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.8 | 4.0 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.8 | 33.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.7 | 5.5 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.7 | 13.5 | GO:0043495 | protein anchor(GO:0043495) |
| 0.6 | 3.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.6 | 5.6 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.6 | 11.0 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.6 | 9.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.6 | 9.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.6 | 11.5 | GO:0004549 | tRNA-specific ribonuclease activity(GO:0004549) |
| 0.6 | 1.7 | GO:0098809 | cystathionine beta-synthase activity(GO:0004122) oxidoreductase activity, acting on other nitrogenous compounds as donors, cytochrome as acceptor(GO:0016662) nitrite reductase (NO-forming) activity(GO:0050421) carbon monoxide binding(GO:0070025) nitrite reductase activity(GO:0098809) |
| 0.5 | 9.5 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.5 | 27.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.4 | 0.9 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.4 | 1.3 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.4 | 12.0 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.4 | 5.6 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.4 | 3.0 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.4 | 4.0 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.4 | 2.3 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.4 | 2.7 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.4 | 8.4 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.4 | 1.8 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.4 | 11.5 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.3 | 11.9 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.3 | 1.6 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.3 | 2.6 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.3 | 14.1 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.3 | 1.9 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.3 | 2.4 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.3 | 18.3 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.3 | 5.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.3 | 4.9 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.3 | 10.0 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.3 | 9.8 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.3 | 5.0 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.3 | 4.8 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 2.7 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 7.4 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.2 | 3.0 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.2 | 4.0 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.2 | 9.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.2 | 19.7 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.2 | 3.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.2 | 4.6 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.2 | 0.9 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.2 | 2.6 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.1 | 1.8 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 2.1 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 1.4 | GO:0036459 | cysteine-type peptidase activity(GO:0008234) ubiquitin-like protein-specific protease activity(GO:0019783) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.1 | 3.2 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 7.3 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 3.1 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 14.2 | GO:0004386 | helicase activity(GO:0004386) |
| 0.1 | 24.9 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.1 | 1.4 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 2.9 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 3.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 4.1 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 0.6 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.1 | 4.0 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 4.5 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 4.8 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.1 | 0.3 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.1 | 0.6 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 1.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 2.5 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 0.5 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 2.7 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 2.6 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.1 | 3.9 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 0.7 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 1.1 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 1.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 4.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 1.6 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 2.6 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 2.4 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 1.4 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 1.1 | GO:0016887 | ATPase activity(GO:0016887) |
| 0.0 | 4.3 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 1.4 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.7 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 3.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.5 | GO:0016667 | oxidoreductase activity, acting on a sulfur group of donors(GO:0016667) |
| 0.0 | 0.2 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 3.2 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 0.2 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.0 | 2.5 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.6 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.0 | 0.1 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.8 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.2 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 2.6 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.6 | GO:0033613 | activating transcription factor binding(GO:0033613) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 59.3 | PID MYC PATHWAY | C-MYC pathway |
| 0.9 | 102.6 | PID E2F PATHWAY | E2F transcription factor network |
| 0.6 | 27.9 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.5 | 17.7 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.4 | 10.0 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.4 | 22.4 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.4 | 16.8 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.4 | 23.6 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.4 | 13.6 | PID ATR PATHWAY | ATR signaling pathway |
| 0.4 | 14.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.4 | 37.9 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.3 | 32.1 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.3 | 2.6 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.3 | 3.8 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.3 | 11.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.2 | 12.6 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.2 | 11.0 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.2 | 25.7 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 4.0 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 4.1 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 5.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 1.6 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.1 | 6.8 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 0.7 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.1 | 1.8 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 2.7 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 4.9 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 4.2 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 1.0 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.6 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 1.4 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.6 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.6 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 3.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.6 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 56.4 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 2.7 | 42.7 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 2.4 | 2.4 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 2.2 | 32.3 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 2.0 | 4.0 | REACTOME LAGGING STRAND SYNTHESIS | Genes involved in Lagging Strand Synthesis |
| 1.7 | 41.0 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 1.3 | 9.2 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 1.2 | 23.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 1.1 | 35.5 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 1.1 | 16.6 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 1.0 | 14.3 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.7 | 27.8 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.6 | 28.4 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.6 | 10.1 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.5 | 50.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.5 | 11.0 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.5 | 18.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.5 | 8.6 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.4 | 14.4 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.4 | 5.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.4 | 22.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.4 | 8.4 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.3 | 3.0 | REACTOME TRANSCRIPTION COUPLED NER TC NER | Genes involved in Transcription-coupled NER (TC-NER) |
| 0.3 | 7.8 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.3 | 6.8 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.3 | 21.7 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.3 | 9.2 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.3 | 20.4 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.2 | 7.2 | REACTOME MEIOSIS | Genes involved in Meiosis |
| 0.2 | 22.8 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.2 | 5.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.2 | 9.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 4.8 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.2 | 3.3 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.2 | 11.8 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.2 | 3.8 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.2 | 3.7 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.2 | 17.3 | REACTOME CHROMOSOME MAINTENANCE | Genes involved in Chromosome Maintenance |
| 0.2 | 5.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 9.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 2.6 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 0.1 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.1 | 1.8 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 4.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 3.3 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 3.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 2.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 1.9 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 4.4 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 1.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 2.6 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 0.8 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 5.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 2.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 4.0 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 2.6 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 4.8 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 1.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |