Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
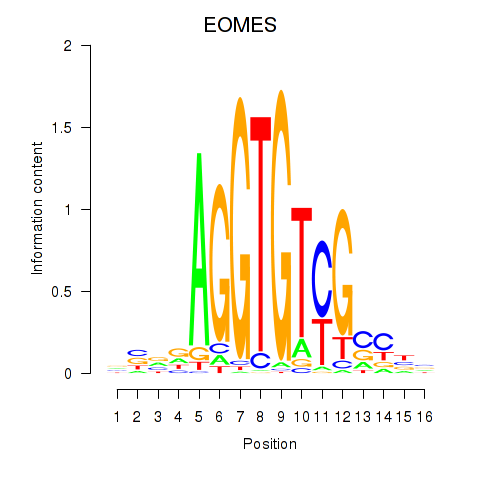
Results for EOMES
Z-value: 1.04
Transcription factors associated with EOMES
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EOMES
|
ENSG00000163508.8 | eomesodermin |
Activity profile of EOMES motif
Sorted Z-values of EOMES motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_55658252 | 24.21 |
ENST00000395425.2
|
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5 |
| chr9_+_91926103 | 23.75 |
ENST00000314355.6
|
CKS2
|
CDC28 protein kinase regulatory subunit 2 |
| chr14_-_55658323 | 20.83 |
ENST00000554067.1
ENST00000247191.2 |
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5 |
| chr7_-_96339132 | 19.10 |
ENST00000413065.1
|
SHFM1
|
split hand/foot malformation (ectrodactyly) type 1 |
| chr6_+_34204642 | 18.64 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chr1_-_167906277 | 16.99 |
ENST00000271373.4
|
MPC2
|
mitochondrial pyruvate carrier 2 |
| chr14_+_54863739 | 16.64 |
ENST00000541304.1
|
CDKN3
|
cyclin-dependent kinase inhibitor 3 |
| chr1_+_87170577 | 16.48 |
ENST00000482504.1
|
SH3GLB1
|
SH3-domain GRB2-like endophilin B1 |
| chr1_+_162531294 | 16.26 |
ENST00000367926.4
ENST00000271469.3 |
UAP1
|
UDP-N-acteylglucosamine pyrophosphorylase 1 |
| chr10_-_58120996 | 16.26 |
ENST00000361148.6
ENST00000395405.1 ENST00000373944.3 |
ZWINT
|
ZW10 interacting kinetochore protein |
| chr1_-_193029192 | 15.36 |
ENST00000417752.1
ENST00000367452.4 |
UCHL5
|
ubiquitin carboxyl-terminal hydrolase L5 |
| chr11_-_57103327 | 14.30 |
ENST00000529002.1
ENST00000278412.2 |
SSRP1
|
structure specific recognition protein 1 |
| chr11_+_64808675 | 13.99 |
ENST00000529996.1
|
SAC3D1
|
SAC3 domain containing 1 |
| chr2_-_161350305 | 13.89 |
ENST00000348849.3
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1 |
| chr7_-_99698338 | 13.59 |
ENST00000354230.3
ENST00000425308.1 |
MCM7
|
minichromosome maintenance complex component 7 |
| chr19_+_35645817 | 13.36 |
ENST00000423817.3
|
FXYD5
|
FXYD domain containing ion transport regulator 5 |
| chr3_-_167452262 | 12.88 |
ENST00000487947.2
|
PDCD10
|
programmed cell death 10 |
| chr7_+_87505544 | 12.00 |
ENST00000265728.1
|
DBF4
|
DBF4 homolog (S. cerevisiae) |
| chr11_-_66056596 | 11.64 |
ENST00000471387.2
ENST00000359461.6 ENST00000376901.4 |
YIF1A
|
Yip1 interacting factor homolog A (S. cerevisiae) |
| chr8_-_109260897 | 11.58 |
ENST00000521297.1
ENST00000519030.1 ENST00000521440.1 ENST00000518345.1 ENST00000519627.1 ENST00000220849.5 |
EIF3E
|
eukaryotic translation initiation factor 3, subunit E |
| chr12_+_54378923 | 11.31 |
ENST00000303460.4
|
HOXC10
|
homeobox C10 |
| chr19_+_35645618 | 11.28 |
ENST00000392218.2
ENST00000543307.1 ENST00000392219.2 ENST00000541435.2 ENST00000590686.1 ENST00000342879.3 ENST00000588699.1 |
FXYD5
|
FXYD domain containing ion transport regulator 5 |
| chr7_-_96339167 | 11.16 |
ENST00000444799.1
ENST00000417009.1 ENST00000248566.2 |
SHFM1
|
split hand/foot malformation (ectrodactyly) type 1 |
| chr3_-_167452298 | 10.68 |
ENST00000475915.2
ENST00000462725.2 ENST00000461494.1 |
PDCD10
|
programmed cell death 10 |
| chr3_-_182698381 | 10.48 |
ENST00000292782.4
|
DCUN1D1
|
DCN1, defective in cullin neddylation 1, domain containing 1 |
| chr13_+_28195988 | 10.42 |
ENST00000399697.3
ENST00000399696.1 |
POLR1D
|
polymerase (RNA) I polypeptide D, 16kDa |
| chr22_+_40742512 | 10.40 |
ENST00000454266.2
ENST00000342312.6 |
ADSL
|
adenylosuccinate lyase |
| chr11_-_102323489 | 10.40 |
ENST00000361236.3
|
TMEM123
|
transmembrane protein 123 |
| chr19_+_16178317 | 10.36 |
ENST00000344824.6
ENST00000538887.1 |
TPM4
|
tropomyosin 4 |
| chr19_+_49496705 | 10.10 |
ENST00000595090.1
|
RUVBL2
|
RuvB-like AAA ATPase 2 |
| chr11_-_107729887 | 9.99 |
ENST00000525815.1
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr7_-_93520259 | 9.91 |
ENST00000222543.5
|
TFPI2
|
tissue factor pathway inhibitor 2 |
| chr3_-_131221790 | 9.85 |
ENST00000512877.1
ENST00000264995.3 ENST00000511168.1 ENST00000425847.2 |
MRPL3
|
mitochondrial ribosomal protein L3 |
| chr5_+_32531893 | 9.82 |
ENST00000512913.1
|
SUB1
|
SUB1 homolog (S. cerevisiae) |
| chr1_+_43148625 | 9.62 |
ENST00000436427.1
|
YBX1
|
Y box binding protein 1 |
| chrX_+_21958814 | 9.61 |
ENST00000379404.1
ENST00000415881.2 |
SMS
|
spermine synthase |
| chr20_-_57617831 | 9.55 |
ENST00000371033.5
ENST00000355937.4 |
SLMO2
|
slowmo homolog 2 (Drosophila) |
| chrX_+_21958674 | 9.54 |
ENST00000404933.2
|
SMS
|
spermine synthase |
| chr11_+_64808368 | 9.45 |
ENST00000531072.1
ENST00000398846.1 |
SAC3D1
|
SAC3 domain containing 1 |
| chr1_+_43148059 | 9.29 |
ENST00000321358.7
ENST00000332220.6 |
YBX1
|
Y box binding protein 1 |
| chr22_+_40742497 | 9.29 |
ENST00000216194.7
|
ADSL
|
adenylosuccinate lyase |
| chr2_+_201936707 | 9.27 |
ENST00000433898.1
ENST00000454214.1 |
NDUFB3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3, 12kDa |
| chr18_-_47017956 | 9.18 |
ENST00000584895.1
ENST00000332968.6 ENST00000580210.1 ENST00000579408.1 |
RPL17-C18orf32
RPL17
|
RPL17-C18orf32 readthrough ribosomal protein L17 |
| chr13_+_28194873 | 8.80 |
ENST00000302979.3
|
POLR1D
|
polymerase (RNA) I polypeptide D, 16kDa |
| chr11_-_33757950 | 8.68 |
ENST00000533403.1
ENST00000528700.1 ENST00000527577.1 ENST00000395850.3 ENST00000351554.3 |
CD59
|
CD59 molecule, complement regulatory protein |
| chr1_+_19578033 | 8.65 |
ENST00000330263.4
|
MRTO4
|
mRNA turnover 4 homolog (S. cerevisiae) |
| chr2_+_201170770 | 8.58 |
ENST00000409988.3
ENST00000409385.1 |
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr4_-_183838596 | 8.54 |
ENST00000508994.1
ENST00000512766.1 |
DCTD
|
dCMP deaminase |
| chr19_+_49496782 | 8.39 |
ENST00000601968.1
ENST00000596837.1 |
RUVBL2
|
RuvB-like AAA ATPase 2 |
| chr19_+_33865218 | 8.29 |
ENST00000585933.2
|
CEBPG
|
CCAAT/enhancer binding protein (C/EBP), gamma |
| chr11_+_34073269 | 7.99 |
ENST00000389645.3
|
CAPRIN1
|
cell cycle associated protein 1 |
| chrX_-_114953669 | 7.95 |
ENST00000449327.1
|
RP1-241P17.4
|
Uncharacterized protein |
| chr2_+_201171242 | 7.91 |
ENST00000360760.5
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr11_+_65686728 | 7.88 |
ENST00000312515.2
ENST00000525501.1 |
DRAP1
|
DR1-associated protein 1 (negative cofactor 2 alpha) |
| chr8_-_124408652 | 7.88 |
ENST00000287394.5
|
ATAD2
|
ATPase family, AAA domain containing 2 |
| chr1_-_245027833 | 7.85 |
ENST00000444376.2
|
HNRNPU
|
heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A) |
| chr11_+_35201826 | 7.81 |
ENST00000531873.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr7_+_16685756 | 7.71 |
ENST00000415365.1
ENST00000258761.3 ENST00000433922.2 ENST00000452975.2 ENST00000405202.1 |
BZW2
|
basic leucine zipper and W2 domains 2 |
| chr9_+_110045537 | 7.69 |
ENST00000358015.3
|
RAD23B
|
RAD23 homolog B (S. cerevisiae) |
| chr5_+_52083730 | 7.67 |
ENST00000282588.6
ENST00000274311.2 |
ITGA1
PELO
|
integrin, alpha 1 pelota homolog (Drosophila) |
| chr13_-_113862401 | 7.66 |
ENST00000375459.1
|
PCID2
|
PCI domain containing 2 |
| chr11_+_111896320 | 7.64 |
ENST00000531306.1
ENST00000537636.1 |
DLAT
|
dihydrolipoamide S-acetyltransferase |
| chr6_-_131949200 | 7.57 |
ENST00000539158.1
ENST00000368058.1 |
MED23
|
mediator complex subunit 23 |
| chr7_-_93519471 | 7.56 |
ENST00000451238.1
|
TFPI2
|
tissue factor pathway inhibitor 2 |
| chr11_-_102323740 | 7.53 |
ENST00000398136.2
|
TMEM123
|
transmembrane protein 123 |
| chr1_-_167906020 | 7.49 |
ENST00000458574.1
|
MPC2
|
mitochondrial pyruvate carrier 2 |
| chr3_-_167452614 | 7.42 |
ENST00000392750.2
ENST00000464360.1 ENST00000492139.1 ENST00000471885.1 ENST00000470131.1 |
PDCD10
|
programmed cell death 10 |
| chr7_-_93520191 | 7.35 |
ENST00000545378.1
|
TFPI2
|
tissue factor pathway inhibitor 2 |
| chr7_+_156931606 | 7.29 |
ENST00000348165.5
|
UBE3C
|
ubiquitin protein ligase E3C |
| chr8_-_101734308 | 7.26 |
ENST00000519004.1
ENST00000519363.1 ENST00000520142.1 |
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr1_-_31769656 | 7.24 |
ENST00000446633.2
|
SNRNP40
|
small nuclear ribonucleoprotein 40kDa (U5) |
| chrX_+_118602363 | 7.17 |
ENST00000317881.8
|
SLC25A5
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 |
| chr4_+_17812525 | 7.12 |
ENST00000251496.2
|
NCAPG
|
non-SMC condensin I complex, subunit G |
| chr8_-_117778494 | 7.11 |
ENST00000276682.4
|
EIF3H
|
eukaryotic translation initiation factor 3, subunit H |
| chr16_+_30064444 | 7.05 |
ENST00000395248.1
ENST00000566897.1 ENST00000568435.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr11_+_65686802 | 7.02 |
ENST00000376991.2
|
DRAP1
|
DR1-associated protein 1 (negative cofactor 2 alpha) |
| chr2_-_69664549 | 7.01 |
ENST00000450796.2
ENST00000484177.1 |
NFU1
|
NFU1 iron-sulfur cluster scaffold homolog (S. cerevisiae) |
| chr11_+_34073757 | 6.92 |
ENST00000532820.1
|
CAPRIN1
|
cell cycle associated protein 1 |
| chr8_-_90996837 | 6.92 |
ENST00000519426.1
ENST00000265433.3 |
NBN
|
nibrin |
| chr2_-_9770706 | 6.87 |
ENST00000381844.4
|
YWHAQ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta |
| chr1_-_245027766 | 6.85 |
ENST00000283179.9
|
HNRNPU
|
heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A) |
| chr5_-_133968529 | 6.76 |
ENST00000402673.2
|
SAR1B
|
SAR1 homolog B (S. cerevisiae) |
| chr1_-_31769595 | 6.72 |
ENST00000263694.4
|
SNRNP40
|
small nuclear ribonucleoprotein 40kDa (U5) |
| chr3_-_167452703 | 6.70 |
ENST00000497056.2
ENST00000473645.2 |
PDCD10
|
programmed cell death 10 |
| chr8_-_103668114 | 6.68 |
ENST00000285407.6
|
KLF10
|
Kruppel-like factor 10 |
| chr11_+_112097069 | 6.56 |
ENST00000280362.3
ENST00000525803.1 |
PTS
|
6-pyruvoyltetrahydropterin synthase |
| chrX_+_150151752 | 6.45 |
ENST00000325307.7
|
HMGB3
|
high mobility group box 3 |
| chr3_+_196466710 | 6.45 |
ENST00000327134.3
|
PAK2
|
p21 protein (Cdc42/Rac)-activated kinase 2 |
| chr13_+_29233218 | 6.41 |
ENST00000380842.4
|
POMP
|
proteasome maturation protein |
| chr11_+_73498898 | 6.39 |
ENST00000535529.1
ENST00000497094.2 ENST00000411840.2 ENST00000535277.1 ENST00000398483.3 ENST00000542303.1 |
MRPL48
|
mitochondrial ribosomal protein L48 |
| chr16_-_67693846 | 6.34 |
ENST00000602850.1
|
ACD
|
adrenocortical dysplasia homolog (mouse) |
| chr6_-_131277510 | 6.33 |
ENST00000525193.1
ENST00000527659.1 |
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr1_-_246729544 | 6.29 |
ENST00000544618.1
ENST00000366514.4 |
TFB2M
|
transcription factor B2, mitochondrial |
| chr7_+_94285637 | 6.26 |
ENST00000482108.1
ENST00000488574.1 |
PEG10
|
paternally expressed 10 |
| chr22_-_18257178 | 6.24 |
ENST00000342111.5
|
BID
|
BH3 interacting domain death agonist |
| chr2_-_179315786 | 6.23 |
ENST00000457633.1
ENST00000438687.3 ENST00000325748.4 |
PRKRA
|
protein kinase, interferon-inducible double stranded RNA dependent activator |
| chr1_+_218458625 | 6.14 |
ENST00000366932.3
|
RRP15
|
ribosomal RNA processing 15 homolog (S. cerevisiae) |
| chr11_+_111896090 | 6.14 |
ENST00000393051.1
|
DLAT
|
dihydrolipoamide S-acetyltransferase |
| chr3_-_123710893 | 6.13 |
ENST00000467907.1
ENST00000459660.1 ENST00000495093.1 ENST00000460743.1 ENST00000405845.3 ENST00000484329.1 ENST00000479867.1 ENST00000496145.1 |
ROPN1
|
rhophilin associated tail protein 1 |
| chr2_+_201170596 | 6.04 |
ENST00000439084.1
ENST00000409718.1 |
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr1_-_244615425 | 6.03 |
ENST00000366535.3
|
ADSS
|
adenylosuccinate synthase |
| chr10_+_75910960 | 6.02 |
ENST00000539909.1
ENST00000286621.2 |
ADK
|
adenosine kinase |
| chr22_+_38071615 | 6.00 |
ENST00000215909.5
|
LGALS1
|
lectin, galactoside-binding, soluble, 1 |
| chr15_+_48623208 | 5.92 |
ENST00000559935.1
ENST00000559416.1 |
DUT
|
deoxyuridine triphosphatase |
| chr12_-_56709674 | 5.92 |
ENST00000551286.1
ENST00000549318.1 |
CNPY2
RP11-977G19.10
|
canopy FGF signaling regulator 2 Uncharacterized protein |
| chr4_+_169418195 | 5.91 |
ENST00000261509.6
ENST00000335742.7 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr2_-_179315453 | 5.87 |
ENST00000432031.2
|
PRKRA
|
protein kinase, interferon-inducible double stranded RNA dependent activator |
| chr17_+_33914460 | 5.82 |
ENST00000537622.2
|
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr2_-_69664586 | 5.81 |
ENST00000303698.3
ENST00000394305.1 ENST00000410022.2 |
NFU1
|
NFU1 iron-sulfur cluster scaffold homolog (S. cerevisiae) |
| chr2_-_179315490 | 5.73 |
ENST00000487082.1
|
PRKRA
|
protein kinase, interferon-inducible double stranded RNA dependent activator |
| chr17_-_61850894 | 5.67 |
ENST00000403162.3
ENST00000582252.1 ENST00000225726.5 |
CCDC47
|
coiled-coil domain containing 47 |
| chr22_-_39096661 | 5.61 |
ENST00000216039.5
|
JOSD1
|
Josephin domain containing 1 |
| chr11_-_62439012 | 5.56 |
ENST00000532208.1
ENST00000377954.2 ENST00000415855.2 ENST00000431002.2 ENST00000354588.3 |
C11orf48
|
chromosome 11 open reading frame 48 |
| chr3_+_125687987 | 5.56 |
ENST00000514116.1
ENST00000251776.4 ENST00000504401.1 ENST00000513830.1 ENST00000508088.1 |
ROPN1B
|
rhophilin associated tail protein 1B |
| chr3_-_127842612 | 5.52 |
ENST00000417360.1
ENST00000322623.5 |
RUVBL1
|
RuvB-like AAA ATPase 1 |
| chr2_+_201171372 | 5.52 |
ENST00000409140.3
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr15_-_90777277 | 5.49 |
ENST00000328649.6
|
CIB1
|
calcium and integrin binding 1 (calmyrin) |
| chr16_+_30064411 | 5.46 |
ENST00000338110.5
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr1_-_33283754 | 5.42 |
ENST00000373477.4
|
YARS
|
tyrosyl-tRNA synthetase |
| chr1_-_207095212 | 5.41 |
ENST00000420007.2
|
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr9_+_115983808 | 5.40 |
ENST00000374210.6
ENST00000374212.4 |
SLC31A1
|
solute carrier family 31 (copper transporter), member 1 |
| chr18_-_47018869 | 5.38 |
ENST00000583036.1
ENST00000580261.1 |
RPL17
|
ribosomal protein L17 |
| chr13_-_31191642 | 5.38 |
ENST00000405805.1
|
HMGB1
|
high mobility group box 1 |
| chr15_-_56285742 | 5.25 |
ENST00000435532.3
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4, E3 ubiquitin protein ligase |
| chr6_-_35888824 | 5.21 |
ENST00000361690.3
ENST00000512445.1 |
SRPK1
|
SRSF protein kinase 1 |
| chr1_-_207095324 | 5.17 |
ENST00000530505.1
ENST00000367091.3 ENST00000442471.2 |
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr16_+_29817841 | 5.15 |
ENST00000322945.6
ENST00000562337.1 ENST00000566906.2 ENST00000563402.1 ENST00000219782.6 |
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chrX_+_131157322 | 5.14 |
ENST00000481105.1
ENST00000354719.6 ENST00000394335.2 |
MST4
|
Serine/threonine-protein kinase MST4 |
| chrX_+_150151824 | 5.07 |
ENST00000455596.1
ENST00000448905.2 |
HMGB3
|
high mobility group box 3 |
| chrX_+_131157290 | 5.07 |
ENST00000394334.2
|
MST4
|
Serine/threonine-protein kinase MST4 |
| chr10_+_106014468 | 4.99 |
ENST00000369710.4
ENST00000369713.5 ENST00000445155.1 |
GSTO1
|
glutathione S-transferase omega 1 |
| chr2_-_170430277 | 4.97 |
ENST00000438035.1
ENST00000453929.2 |
FASTKD1
|
FAST kinase domains 1 |
| chr3_-_125239010 | 4.95 |
ENST00000536067.1
ENST00000251775.4 |
SNX4
|
sorting nexin 4 |
| chrX_+_77359726 | 4.86 |
ENST00000442431.1
|
PGK1
|
phosphoglycerate kinase 1 |
| chr11_+_34073195 | 4.85 |
ENST00000341394.4
|
CAPRIN1
|
cell cycle associated protein 1 |
| chr17_+_33914276 | 4.79 |
ENST00000592545.1
ENST00000538556.1 ENST00000312678.8 ENST00000589344.1 |
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr18_-_19284724 | 4.77 |
ENST00000580981.1
ENST00000289119.2 |
ABHD3
|
abhydrolase domain containing 3 |
| chr15_+_48623600 | 4.75 |
ENST00000558813.1
ENST00000331200.3 ENST00000558472.1 |
DUT
|
deoxyuridine triphosphatase |
| chr8_-_90996459 | 4.69 |
ENST00000517337.1
ENST00000409330.1 |
NBN
|
nibrin |
| chr20_+_1099233 | 4.63 |
ENST00000246015.4
ENST00000335877.6 ENST00000438768.2 |
PSMF1
|
proteasome (prosome, macropain) inhibitor subunit 1 (PI31) |
| chr20_-_34252759 | 4.57 |
ENST00000414711.1
ENST00000416778.1 ENST00000397442.1 ENST00000440240.1 ENST00000412056.1 ENST00000352393.4 ENST00000458038.1 ENST00000420363.1 ENST00000434795.1 ENST00000437100.1 ENST00000414664.1 ENST00000359646.1 ENST00000424458.1 ENST00000374104.3 ENST00000374114.3 |
CPNE1
RBM12
|
copine I RNA binding motif protein 12 |
| chr2_-_219134343 | 4.57 |
ENST00000447885.1
ENST00000420660.1 |
AAMP
|
angio-associated, migratory cell protein |
| chr6_-_75953484 | 4.53 |
ENST00000472311.2
ENST00000460985.1 ENST00000377978.3 ENST00000509698.1 ENST00000230459.4 ENST00000370089.2 |
COX7A2
|
cytochrome c oxidase subunit VIIa polypeptide 2 (liver) |
| chr1_-_153949751 | 4.48 |
ENST00000428469.1
|
JTB
|
jumping translocation breakpoint |
| chr8_+_144099896 | 4.43 |
ENST00000292494.6
ENST00000429120.2 |
LY6E
|
lymphocyte antigen 6 complex, locus E |
| chr17_-_1588101 | 4.32 |
ENST00000577001.1
ENST00000572621.1 ENST00000304992.6 |
PRPF8
|
pre-mRNA processing factor 8 |
| chr6_-_109703634 | 4.32 |
ENST00000324953.5
ENST00000310786.4 ENST00000275080.7 ENST00000413644.2 |
CD164
|
CD164 molecule, sialomucin |
| chr7_+_55177416 | 4.28 |
ENST00000450046.1
ENST00000454757.2 |
EGFR
|
epidermal growth factor receptor |
| chr10_+_85899196 | 4.28 |
ENST00000372134.3
|
GHITM
|
growth hormone inducible transmembrane protein |
| chr22_-_42486747 | 4.25 |
ENST00000602404.1
|
NDUFA6
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 6, 14kDa |
| chr1_+_167906056 | 4.25 |
ENST00000367840.3
|
DCAF6
|
DDB1 and CUL4 associated factor 6 |
| chr15_+_89182178 | 4.23 |
ENST00000559876.1
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr4_-_54930790 | 4.18 |
ENST00000263921.3
|
CHIC2
|
cysteine-rich hydrophobic domain 2 |
| chr2_+_99953816 | 4.17 |
ENST00000289371.6
|
EIF5B
|
eukaryotic translation initiation factor 5B |
| chr8_-_117768023 | 4.15 |
ENST00000518949.1
ENST00000522453.1 ENST00000521861.1 ENST00000518995.1 |
EIF3H
|
eukaryotic translation initiation factor 3, subunit H |
| chr8_+_144099914 | 4.13 |
ENST00000521699.1
ENST00000520531.1 ENST00000520466.1 ENST00000521003.1 ENST00000522528.1 ENST00000522971.1 ENST00000519611.1 ENST00000521182.1 ENST00000519546.1 ENST00000523847.1 ENST00000522024.1 |
LY6E
|
lymphocyte antigen 6 complex, locus E |
| chr15_-_85259294 | 4.08 |
ENST00000558217.1
ENST00000558196.1 ENST00000558134.1 |
SEC11A
|
SEC11 homolog A (S. cerevisiae) |
| chr19_+_19303008 | 4.08 |
ENST00000353145.1
ENST00000421262.3 ENST00000303088.4 ENST00000456252.3 ENST00000593273.1 |
RFXANK
|
regulatory factor X-associated ankyrin-containing protein |
| chr11_+_63706444 | 4.03 |
ENST00000377793.4
ENST00000456907.2 ENST00000539656.1 |
NAA40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
| chr19_+_13261216 | 4.00 |
ENST00000587885.1
ENST00000292433.3 |
IER2
|
immediate early response 2 |
| chr14_-_24682652 | 3.99 |
ENST00000556387.1
ENST00000530611.1 ENST00000609024.1 ENST00000530996.1 |
TM9SF1
TM9SF1
CHMP4A
|
transmembrane 9 superfamily member 1 Transmembrane 9 superfamily member 1 charged multivesicular body protein 4A |
| chr1_+_186344883 | 3.98 |
ENST00000367470.3
|
C1orf27
|
chromosome 1 open reading frame 27 |
| chr1_+_186344945 | 3.96 |
ENST00000419367.3
ENST00000287859.6 |
C1orf27
|
chromosome 1 open reading frame 27 |
| chrX_-_102757802 | 3.94 |
ENST00000372633.1
|
RAB40A
|
RAB40A, member RAS oncogene family |
| chr19_-_2427536 | 3.91 |
ENST00000591871.1
|
TIMM13
|
translocase of inner mitochondrial membrane 13 homolog (yeast) |
| chr1_-_205744205 | 3.89 |
ENST00000446390.2
|
RAB7L1
|
RAB7, member RAS oncogene family-like 1 |
| chr9_-_117853297 | 3.83 |
ENST00000542877.1
ENST00000537320.1 ENST00000341037.4 |
TNC
|
tenascin C |
| chr3_+_133292574 | 3.79 |
ENST00000264993.3
|
CDV3
|
CDV3 homolog (mouse) |
| chr6_-_74233480 | 3.77 |
ENST00000455918.1
|
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1 |
| chr14_-_51135036 | 3.77 |
ENST00000324679.4
|
SAV1
|
salvador homolog 1 (Drosophila) |
| chr16_+_202686 | 3.73 |
ENST00000252951.2
|
HBZ
|
hemoglobin, zeta |
| chr10_-_94333784 | 3.72 |
ENST00000265986.6
|
IDE
|
insulin-degrading enzyme |
| chr11_+_69455855 | 3.72 |
ENST00000227507.2
ENST00000536559.1 |
CCND1
|
cyclin D1 |
| chr17_-_15587602 | 3.68 |
ENST00000416464.2
ENST00000578237.1 ENST00000581200.1 |
TRIM16
|
tripartite motif containing 16 |
| chr19_+_34856141 | 3.66 |
ENST00000586425.1
|
GPI
|
glucose-6-phosphate isomerase |
| chr4_-_144940477 | 3.53 |
ENST00000513128.1
ENST00000429670.2 ENST00000502664.1 |
GYPB
|
glycophorin B (MNS blood group) |
| chr6_-_111927062 | 3.52 |
ENST00000359831.4
|
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr13_+_73302047 | 3.52 |
ENST00000377814.2
ENST00000377815.3 ENST00000390667.5 |
BORA
|
bora, aurora kinase A activator |
| chr19_+_9938562 | 3.52 |
ENST00000586895.1
ENST00000358666.3 ENST00000590068.1 ENST00000593087.1 |
UBL5
|
ubiquitin-like 5 |
| chr1_-_184943610 | 3.48 |
ENST00000367511.3
|
FAM129A
|
family with sequence similarity 129, member A |
| chr9_+_140172200 | 3.47 |
ENST00000357503.2
|
TOR4A
|
torsin family 4, member A |
| chr15_-_85259330 | 3.46 |
ENST00000560266.1
|
SEC11A
|
SEC11 homolog A (S. cerevisiae) |
| chr15_-_85259384 | 3.45 |
ENST00000455959.3
|
SEC11A
|
SEC11 homolog A (S. cerevisiae) |
| chr2_-_65357225 | 3.44 |
ENST00000398529.3
ENST00000409751.1 ENST00000356214.7 ENST00000409892.1 ENST00000409784.3 |
RAB1A
|
RAB1A, member RAS oncogene family |
| chr1_-_153950098 | 3.38 |
ENST00000356648.1
|
JTB
|
jumping translocation breakpoint |
| chr5_+_96212185 | 3.37 |
ENST00000379904.4
|
ERAP2
|
endoplasmic reticulum aminopeptidase 2 |
| chr2_-_54197915 | 3.31 |
ENST00000404125.1
|
PSME4
|
proteasome (prosome, macropain) activator subunit 4 |
| chr17_-_77813186 | 3.29 |
ENST00000448310.1
ENST00000269397.4 |
CBX4
|
chromobox homolog 4 |
| chr19_-_3061397 | 3.22 |
ENST00000586839.1
|
AES
|
amino-terminal enhancer of split |
| chr1_-_205744574 | 3.21 |
ENST00000367139.3
ENST00000235932.4 ENST00000437324.2 ENST00000414729.1 |
RAB7L1
|
RAB7, member RAS oncogene family-like 1 |
| chr7_+_129710350 | 3.21 |
ENST00000335420.5
ENST00000463413.1 |
KLHDC10
|
kelch domain containing 10 |
| chr6_-_109702885 | 3.16 |
ENST00000504373.1
|
CD164
|
CD164 molecule, sialomucin |
| chr20_-_34252847 | 3.15 |
ENST00000317619.3
ENST00000397446.1 ENST00000397445.1 ENST00000397443.1 ENST00000430570.1 ENST00000439806.2 ENST00000437340.1 ENST00000435161.1 ENST00000431148.1 |
CPNE1
RBM12
|
copine I RNA binding motif protein 12 |
| chr9_+_37486005 | 3.14 |
ENST00000377792.3
|
POLR1E
|
polymerase (RNA) I polypeptide E, 53kDa |
| chr4_-_151936865 | 3.13 |
ENST00000535741.1
|
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr1_-_153950116 | 3.09 |
ENST00000368589.1
|
JTB
|
jumping translocation breakpoint |
| chr19_+_55897699 | 3.08 |
ENST00000558131.1
ENST00000558752.1 ENST00000458349.2 |
RPL28
|
ribosomal protein L28 |
| chr19_-_49496557 | 3.08 |
ENST00000323798.3
ENST00000541188.1 ENST00000544287.1 ENST00000540532.1 ENST00000263276.6 |
GYS1
|
glycogen synthase 1 (muscle) |
| chr17_+_48133459 | 3.07 |
ENST00000320031.8
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) |
| chr3_+_111805182 | 3.01 |
ENST00000430855.1
ENST00000431717.2 ENST00000264848.5 |
C3orf52
|
chromosome 3 open reading frame 52 |
Network of associatons between targets according to the STRING database.
First level regulatory network of EOMES
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.6 | 25.7 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 8.2 | 24.6 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 7.5 | 37.7 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 7.5 | 45.0 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 6.4 | 19.2 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 6.1 | 24.5 | GO:1902361 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 4.8 | 33.6 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 4.7 | 18.6 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 4.6 | 18.5 | GO:0090202 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 4.1 | 16.5 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 3.6 | 10.7 | GO:0046081 | dUTP metabolic process(GO:0046080) dUTP catabolic process(GO:0046081) |
| 3.1 | 9.3 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 3.1 | 9.2 | GO:0060380 | regulation of single-stranded telomeric DNA binding(GO:0060380) positive regulation of single-stranded telomeric DNA binding(GO:0060381) positive regulation of telomeric DNA binding(GO:1904744) |
| 2.9 | 11.6 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 2.5 | 5.0 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 2.2 | 8.7 | GO:0001971 | negative regulation of activation of membrane attack complex(GO:0001971) |
| 2.1 | 8.5 | GO:0006226 | dUMP biosynthetic process(GO:0006226) |
| 2.1 | 10.6 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 2.0 | 6.0 | GO:0044209 | AMP salvage(GO:0044209) |
| 1.8 | 5.5 | GO:0038163 | endomitotic cell cycle(GO:0007113) thrombopoietin-mediated signaling pathway(GO:0038163) |
| 1.6 | 6.3 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 1.6 | 4.7 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 1.6 | 14.0 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 1.5 | 7.7 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 1.5 | 6.0 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 1.4 | 4.3 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 1.4 | 17.8 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 1.3 | 5.4 | GO:0002424 | plasmacytoid dendritic cell activation(GO:0002270) T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) regulation of restriction endodeoxyribonuclease activity(GO:0032072) negative regulation of apoptotic cell clearance(GO:2000426) |
| 1.3 | 5.3 | GO:0044111 | development involved in symbiotic interaction(GO:0044111) |
| 1.3 | 11.6 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 1.3 | 7.7 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 1.3 | 3.8 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 1.3 | 11.3 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 1.3 | 16.3 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 1.2 | 3.7 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 1.2 | 3.7 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) |
| 1.2 | 7.2 | GO:1901029 | adenine transport(GO:0015853) negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 1.1 | 4.5 | GO:0001188 | RNA polymerase I transcriptional preinitiation complex assembly(GO:0001188) RNA polymerase I transcriptional preinitiation complex assembly at the promoter for the nuclear large rRNA transcript(GO:0001189) |
| 1.1 | 17.9 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 1.1 | 5.3 | GO:0072719 | cellular response to cisplatin(GO:0072719) |
| 1.0 | 8.3 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 1.0 | 4.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 1.0 | 2.1 | GO:2000078 | positive regulation of type B pancreatic cell development(GO:2000078) |
| 1.0 | 7.8 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 1.0 | 15.4 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.9 | 8.5 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.8 | 5.0 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.8 | 7.3 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.8 | 4.0 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.8 | 2.3 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate catabolic process(GO:0006533) |
| 0.7 | 2.2 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.7 | 6.6 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.7 | 2.1 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.7 | 7.0 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.7 | 10.5 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.7 | 2.1 | GO:0038188 | cholecystokinin signaling pathway(GO:0038188) |
| 0.7 | 2.7 | GO:0072183 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of nephron tubule epithelial cell differentiation(GO:0072183) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) negative regulation of epithelial cell differentiation involved in kidney development(GO:2000697) |
| 0.7 | 7.4 | GO:0006983 | ER overload response(GO:0006983) |
| 0.7 | 2.0 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.7 | 11.8 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.7 | 5.9 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.6 | 5.8 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.6 | 7.7 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.6 | 19.8 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.6 | 1.9 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) renal inner medulla development(GO:0072053) renal outer medulla development(GO:0072054) |
| 0.6 | 13.8 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.6 | 2.4 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.6 | 27.5 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.6 | 18.2 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.6 | 12.8 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.6 | 1.7 | GO:0006404 | RNA import into nucleus(GO:0006404) |
| 0.5 | 1.6 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.5 | 3.1 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.5 | 3.6 | GO:0010918 | positive regulation of mitochondrial membrane potential(GO:0010918) |
| 0.5 | 3.0 | GO:0061083 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.5 | 19.2 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.5 | 2.4 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.5 | 1.4 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.5 | 1.9 | GO:0098838 | methotrexate transport(GO:0051958) reduced folate transmembrane transport(GO:0098838) |
| 0.5 | 1.9 | GO:0010728 | regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 0.5 | 1.8 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.5 | 6.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.5 | 6.3 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.4 | 7.9 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.4 | 1.8 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.4 | 3.9 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.4 | 3.7 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.4 | 1.6 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.4 | 5.2 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.4 | 8.7 | GO:0000717 | nucleotide-excision repair, DNA duplex unwinding(GO:0000717) |
| 0.3 | 1.4 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.3 | 2.8 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.3 | 2.7 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.3 | 2.5 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.3 | 5.6 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.3 | 7.1 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.3 | 1.5 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.3 | 0.9 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.3 | 3.9 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.3 | 3.2 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.3 | 3.7 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.3 | 0.8 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.3 | 22.2 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.3 | 16.6 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) |
| 0.3 | 6.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.3 | 10.1 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) |
| 0.3 | 1.1 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.3 | 2.3 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.3 | 2.8 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.2 | 15.1 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.2 | 4.9 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.2 | 3.6 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.2 | 1.9 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.2 | 3.8 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.2 | 3.8 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.2 | 2.4 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.2 | 11.5 | GO:0032392 | DNA geometric change(GO:0032392) |
| 0.2 | 1.2 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.2 | 1.2 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.2 | 2.5 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.2 | 16.2 | GO:0070126 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 0.2 | 2.2 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.2 | 1.8 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.2 | 2.6 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.2 | 1.4 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.2 | 3.4 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.2 | 0.8 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.2 | 5.2 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.2 | 5.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 4.0 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.2 | 1.6 | GO:0070828 | heterochromatin organization(GO:0070828) |
| 0.2 | 0.5 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.2 | 15.5 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.2 | 19.4 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.2 | 0.9 | GO:0014004 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.2 | 2.6 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.2 | 3.5 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.1 | 3.5 | GO:0001783 | B cell apoptotic process(GO:0001783) |
| 0.1 | 1.5 | GO:1903543 | positive regulation of exosomal secretion(GO:1903543) |
| 0.1 | 12.4 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.1 | 1.4 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.1 | 2.5 | GO:0036260 | 7-methylguanosine RNA capping(GO:0009452) RNA capping(GO:0036260) |
| 0.1 | 7.5 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 2.3 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.1 | 3.1 | GO:0097062 | dendritic spine maintenance(GO:0097062) renal filtration(GO:0097205) |
| 0.1 | 0.2 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.1 | 1.2 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.1 | 2.5 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 1.5 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.1 | 1.1 | GO:0014041 | regulation of neuron maturation(GO:0014041) |
| 0.1 | 0.8 | GO:0086073 | cardiac muscle cell-cardiac muscle cell adhesion(GO:0086042) bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.1 | 0.8 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.1 | 1.8 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 2.8 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 6.5 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.1 | 11.3 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.1 | 4.8 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.1 | 0.3 | GO:0061743 | motor learning(GO:0061743) |
| 0.1 | 0.3 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.1 | 0.6 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.1 | 11.8 | GO:0000070 | mitotic sister chromatid segregation(GO:0000070) |
| 0.1 | 1.4 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.1 | 8.2 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 4.0 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 | 1.5 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 6.9 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 0.4 | GO:0034343 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.1 | 6.2 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) |
| 0.1 | 3.9 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.1 | 0.3 | GO:0048073 | regulation of eye pigmentation(GO:0048073) |
| 0.1 | 0.6 | GO:0016255 | polyol transport(GO:0015791) attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 2.5 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
| 0.1 | 1.1 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.1 | 1.0 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 3.7 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.1 | 0.4 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.1 | 1.8 | GO:0002360 | T cell lineage commitment(GO:0002360) |
| 0.1 | 1.8 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.1 | 0.7 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.1 | 0.5 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.1 | 0.5 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 3.9 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 1.8 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.1 | 0.3 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.1 | 0.6 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.1 | 4.7 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.1 | 3.2 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.1 | 0.9 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 0.5 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 0.8 | GO:2000352 | negative regulation of endothelial cell apoptotic process(GO:2000352) |
| 0.0 | 2.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 0.0 | 0.0 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 2.6 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.8 | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
| 0.0 | 0.3 | GO:0051024 | positive regulation of immunoglobulin secretion(GO:0051024) |
| 0.0 | 1.5 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 0.2 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 1.2 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 3.6 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 1.9 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.1 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 1.8 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 1.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:1905066 | arterial endothelial cell fate commitment(GO:0060844) canonical Wnt signaling pathway involved in heart development(GO:0061316) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) positive regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061419) blood vessel lumenization(GO:0072554) positive regulation of ephrin receptor signaling pathway(GO:1901189) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 1.1 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.6 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 1.7 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.8 | GO:0003016 | respiratory system process(GO:0003016) |
| 0.0 | 2.4 | GO:0045333 | cellular respiration(GO:0045333) |
| 0.0 | 0.3 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.2 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.2 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.0 | 1.2 | GO:0071774 | response to fibroblast growth factor(GO:0071774) |
| 0.0 | 5.7 | GO:0006260 | DNA replication(GO:0006260) |
| 0.0 | 0.2 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.0 | 1.9 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 11.3 | GO:0051301 | cell division(GO:0051301) |
| 0.0 | 0.8 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.4 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.0 | 0.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.4 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:0032962 | regulation of inositol trisphosphate biosynthetic process(GO:0032960) positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.0 | 0.2 | GO:0031055 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.5 | GO:0006915 | apoptotic process(GO:0006915) |
| 0.0 | 0.1 | GO:1901642 | nucleoside transmembrane transport(GO:1901642) |
| 0.0 | 0.3 | GO:0042177 | negative regulation of protein catabolic process(GO:0042177) |
| 0.0 | 0.2 | GO:0090527 | actin filament reorganization(GO:0090527) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 13.8 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 3.4 | 33.6 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 3.1 | 3.1 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 2.6 | 45.0 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 2.6 | 7.7 | GO:0034665 | integrin alpha1-beta1 complex(GO:0034665) |
| 2.6 | 25.5 | GO:0097255 | R2TP complex(GO:0097255) |
| 2.0 | 13.8 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 1.7 | 18.6 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 1.7 | 11.6 | GO:0030870 | Mre11 complex(GO:0030870) |
| 1.4 | 7.2 | GO:0071817 | MMXD complex(GO:0071817) |
| 1.4 | 4.3 | GO:0097489 | multivesicular body, internal vesicle lumen(GO:0097489) |
| 1.3 | 23.7 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 1.3 | 11.3 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 1.3 | 11.3 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 1.1 | 9.2 | GO:0070187 | telosome(GO:0070187) |
| 1.1 | 7.8 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 1.1 | 5.4 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 1.0 | 16.8 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 1.0 | 13.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.9 | 12.3 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.9 | 24.5 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.8 | 2.4 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.7 | 11.6 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.7 | 7.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.7 | 2.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.6 | 14.0 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.6 | 2.4 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.6 | 3.0 | GO:0089701 | U2AF(GO:0089701) |
| 0.6 | 10.6 | GO:0030122 | AP-2 adaptor complex(GO:0030122) endolysosome membrane(GO:0036020) |
| 0.5 | 10.4 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.5 | 8.7 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.5 | 20.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.5 | 1.5 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.5 | 27.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.5 | 3.9 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.5 | 1.8 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.5 | 6.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.5 | 7.7 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.4 | 3.1 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.4 | 27.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.4 | 13.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.4 | 3.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.4 | 16.2 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.4 | 3.8 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.4 | 13.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.4 | 2.2 | GO:0001740 | Barr body(GO:0001740) |
| 0.3 | 3.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.3 | 2.8 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.3 | 15.1 | GO:0031430 | M band(GO:0031430) |
| 0.3 | 2.3 | GO:0032059 | bleb(GO:0032059) |
| 0.3 | 5.5 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.3 | 6.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.3 | 3.7 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.3 | 1.1 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.3 | 2.6 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.3 | 3.6 | GO:0036038 | MKS complex(GO:0036038) |
| 0.3 | 1.8 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.2 | 3.7 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.2 | 15.1 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.2 | 6.7 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.2 | 6.1 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.2 | 5.9 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.2 | 20.6 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.2 | 4.6 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.2 | 10.1 | GO:0015030 | Cajal body(GO:0015030) |
| 0.2 | 6.5 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.2 | 5.2 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.2 | 7.3 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.1 | 7.1 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 15.1 | GO:0000779 | condensed chromosome, centromeric region(GO:0000779) |
| 0.1 | 4.8 | GO:0032592 | integral component of mitochondrial membrane(GO:0032592) |
| 0.1 | 1.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 1.2 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 1.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 7.3 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 1.8 | GO:0000786 | nucleosome(GO:0000786) DNA packaging complex(GO:0044815) |
| 0.1 | 1.6 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 2.7 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 8.1 | GO:0005694 | chromosome(GO:0005694) |
| 0.1 | 4.3 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 30.5 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 1.5 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.1 | 2.3 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 3.5 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 0.5 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 0.8 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.6 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 5.4 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.1 | 4.5 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 5.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 13.7 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 2.1 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 3.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 2.0 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 3.4 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 1.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 1.4 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 6.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.9 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.2 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 5.2 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 1.0 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 3.6 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 10.6 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 8.0 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 1.0 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 2.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 13.8 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 4.0 | 24.0 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 3.6 | 10.7 | GO:0004170 | dUTP diphosphatase activity(GO:0004170) |
| 3.4 | 23.8 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 3.1 | 9.3 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 3.1 | 24.5 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 3.0 | 24.4 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 2.2 | 6.6 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 2.1 | 8.5 | GO:0004132 | dCMP deaminase activity(GO:0004132) |
| 2.0 | 6.0 | GO:0048030 | disaccharide binding(GO:0048030) |
| 2.0 | 17.8 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 1.8 | 10.9 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 1.8 | 7.2 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 1.7 | 5.0 | GO:0045174 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 1.6 | 4.9 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 1.5 | 7.7 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 1.5 | 16.3 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 1.4 | 4.3 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 1.3 | 18.6 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 1.3 | 23.7 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 1.3 | 2.5 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 1.3 | 6.3 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 1.2 | 3.7 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 1.2 | 3.7 | GO:0004347 | glucose-6-phosphate isomerase activity(GO:0004347) |
| 1.1 | 23.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 1.1 | 11.8 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 1.0 | 5.0 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 1.0 | 4.8 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.9 | 4.3 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.8 | 4.0 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.8 | 2.3 | GO:0070546 | phosphatidylserine decarboxylase activity(GO:0004609) L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.8 | 5.3 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.7 | 2.9 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.7 | 2.1 | GO:0004951 | cholecystokinin receptor activity(GO:0004951) |
| 0.7 | 2.0 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.6 | 25.2 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.6 | 19.8 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.6 | 6.3 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.6 | 2.5 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.6 | 3.0 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.6 | 2.3 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.6 | 10.5 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.5 | 1.6 | GO:0052871 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.5 | 5.4 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.5 | 5.4 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.5 | 6.0 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.5 | 2.4 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.5 | 1.9 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.5 | 9.2 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.5 | 10.6 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.5 | 1.4 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.5 | 1.9 | GO:0008518 | reduced folate carrier activity(GO:0008518) methotrexate transporter activity(GO:0015350) |
| 0.5 | 3.7 | GO:0043559 | insulin binding(GO:0043559) |
| 0.4 | 69.4 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.4 | 50.1 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.4 | 2.6 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.4 | 9.7 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.4 | 11.9 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.4 | 3.6 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.4 | 2.4 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 0.4 | 21.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.4 | 1.8 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.3 | 10.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.3 | 6.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.3 | 2.7 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.3 | 12.8 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.3 | 7.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.3 | 3.5 | GO:0031386 | protein tag(GO:0031386) |
| 0.3 | 15.4 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.3 | 3.7 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.3 | 1.4 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.3 | 2.7 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.3 | 24.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.3 | 2.6 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.3 | 3.1 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.3 | 8.7 | GO:0001848 | complement binding(GO:0001848) |
| 0.3 | 3.0 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.2 | 7.0 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.2 | 4.4 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.2 | 4.1 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.2 | 6.5 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 2.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.2 | 5.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.2 | 1.5 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.2 | 1.5 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.2 | 3.7 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
| 0.2 | 7.7 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.2 | 2.1 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.2 | 9.2 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.2 | 15.4 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.2 | 2.9 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.2 | 9.3 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.2 | 0.8 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.2 | 6.2 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.2 | 5.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.2 | 0.5 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.2 | 3.9 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 2.8 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 3.7 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 12.5 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.1 | 3.1 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 4.2 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 29.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 23.2 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.1 | 10.4 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 1.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 1.2 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.1 | 2.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 4.2 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.1 | 2.8 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 1.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 1.7 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 1.8 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 5.0 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.1 | 0.2 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.1 | 0.6 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 2.7 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.1 | 1.6 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.1 | 1.4 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.1 | 0.2 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 1.8 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 0.9 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 1.8 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 2.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 0.2 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 3.1 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.1 | 1.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.7 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 1.3 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.3 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 6.3 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 10.8 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 8.3 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 1.6 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 1.1 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.1 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.0 | 2.2 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 1.0 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 1.5 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.4 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.2 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 0.0 | 10.9 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 2.6 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 2.6 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.8 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.3 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.6 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.1 | GO:0005329 | dopamine transmembrane transporter activity(GO:0005329) |
| 0.0 | 0.3 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.4 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 41.3 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.6 | 89.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.4 | 6.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.4 | 8.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.3 | 17.1 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.3 | 1.9 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.3 | 3.7 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.3 | 1.5 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.3 | 22.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.3 | 14.0 | PID ATR PATHWAY | ATR signaling pathway |
| 0.2 | 2.7 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.2 | 8.6 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.2 | 23.2 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.2 | 7.8 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.2 | 3.9 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.2 | 5.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 3.3 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.2 | 8.6 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.2 | 12.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.2 | 7.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.2 | 7.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 1.8 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 9.0 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 3.7 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 4.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 2.8 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 3.7 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 3.0 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 4.1 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 1.6 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 2.0 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.1 | 0.9 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 1.6 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.8 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 3.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 2.3 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 1.1 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 1.9 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 2.6 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.8 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.4 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.8 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 1.2 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.4 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.3 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.8 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 18.6 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 1.5 | 25.7 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.9 | 21.5 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.8 | 13.6 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.8 | 19.2 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.8 | 13.8 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.7 | 13.3 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.7 | 12.0 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.6 | 24.0 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.6 | 2.4 | REACTOME PROCESSING OF CAPPED INTRON CONTAINING PRE MRNA | Genes involved in Processing of Capped Intron-Containing Pre-mRNA |
| 0.5 | 19.2 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.5 | 10.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.5 | 10.6 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.5 | 7.9 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.4 | 8.4 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.4 | 7.8 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.4 | 15.4 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.4 | 52.1 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.3 | 17.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.3 | 8.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.3 | 6.6 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.3 | 7.7 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.3 | 7.7 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.3 | 6.6 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.3 | 6.8 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.3 | 2.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.3 | 28.9 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 0.3 | 6.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.3 | 5.5 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.3 | 6.0 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.2 | 4.9 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.2 | 24.4 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.2 | 9.6 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.2 | 5.2 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.2 | 5.4 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.2 | 11.0 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.2 | 3.8 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.2 | 3.8 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.2 | 16.5 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.2 | 12.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.2 | 10.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.2 | 15.9 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.2 | 15.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.2 | 1.6 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.2 | 7.4 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.2 | 2.5 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.2 | 5.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 1.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 5.0 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 2.6 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 9.3 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 7.9 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.1 | 2.8 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 0.8 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 2.7 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.1 | 4.2 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.1 | 1.8 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 3.0 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 3.1 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 1.8 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 3.9 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 6.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 5.3 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.1 | 2.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.7 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 2.0 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 2.6 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 1.1 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.1 | 0.6 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.1 | 1.1 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 2.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.8 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.5 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 1.0 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.5 | REACTOME SIGNALING BY NOTCH1 | Genes involved in Signaling by NOTCH1 |
| 0.0 | 2.3 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.2 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 1.2 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.1 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.2 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |