Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
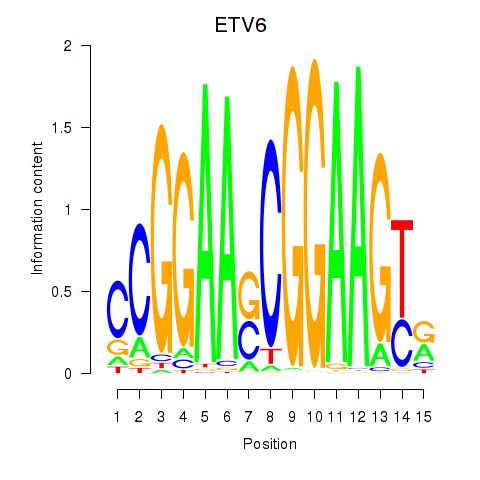
Results for ETV6
Z-value: 1.75
Transcription factors associated with ETV6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ETV6
|
ENSG00000139083.6 | ETS variant transcription factor 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ETV6 | hg19_v2_chr12_+_11802753_11802834 | -0.26 | 1.0e-04 | Click! |
Activity profile of ETV6 motif
Sorted Z-values of ETV6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_167913450 | 73.77 |
ENST00000231572.3
ENST00000538719.1 |
RARS
|
arginyl-tRNA synthetase |
| chr2_+_65454926 | 43.13 |
ENST00000542850.1
ENST00000377982.4 |
ACTR2
|
ARP2 actin-related protein 2 homolog (yeast) |
| chr2_+_65454863 | 41.84 |
ENST00000260641.5
|
ACTR2
|
ARP2 actin-related protein 2 homolog (yeast) |
| chr2_-_96971259 | 40.94 |
ENST00000349783.5
|
SNRNP200
|
small nuclear ribonucleoprotein 200kDa (U5) |
| chr11_+_65770227 | 36.93 |
ENST00000527348.1
|
BANF1
|
barrier to autointegration factor 1 |
| chr2_-_96971232 | 33.85 |
ENST00000323853.5
|
SNRNP200
|
small nuclear ribonucleoprotein 200kDa (U5) |
| chr1_+_203830703 | 33.42 |
ENST00000414487.2
|
SNRPE
|
small nuclear ribonucleoprotein polypeptide E |
| chr20_-_49575058 | 33.17 |
ENST00000371584.4
ENST00000371583.5 ENST00000413082.1 |
DPM1
|
dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit |
| chr11_+_65769550 | 31.73 |
ENST00000312175.2
ENST00000445560.2 ENST00000530204.1 |
BANF1
|
barrier to autointegration factor 1 |
| chr1_-_19811996 | 31.71 |
ENST00000264203.3
ENST00000401084.2 |
CAPZB
|
capping protein (actin filament) muscle Z-line, beta |
| chr3_+_52740094 | 31.29 |
ENST00000602728.1
|
SPCS1
|
signal peptidase complex subunit 1 homolog (S. cerevisiae) |
| chr1_-_149900122 | 30.42 |
ENST00000271628.8
|
SF3B4
|
splicing factor 3b, subunit 4, 49kDa |
| chr12_+_108079664 | 30.15 |
ENST00000541166.1
|
PWP1
|
PWP1 homolog (S. cerevisiae) |
| chr7_+_99006550 | 30.03 |
ENST00000222969.5
|
BUD31
|
BUD31 homolog (S. cerevisiae) |
| chr11_+_65769946 | 29.10 |
ENST00000533166.1
|
BANF1
|
barrier to autointegration factor 1 |
| chr2_+_201936707 | 28.46 |
ENST00000433898.1
ENST00000454214.1 |
NDUFB3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3, 12kDa |
| chr19_+_49497121 | 27.98 |
ENST00000413176.2
|
RUVBL2
|
RuvB-like AAA ATPase 2 |
| chr12_+_10658201 | 26.93 |
ENST00000322446.3
|
EIF2S3L
|
Putative eukaryotic translation initiation factor 2 subunit 3-like protein |
| chr7_-_54826920 | 26.42 |
ENST00000395535.3
ENST00000352861.4 |
SEC61G
|
Sec61 gamma subunit |
| chr20_-_33872518 | 26.25 |
ENST00000374436.3
|
EIF6
|
eukaryotic translation initiation factor 6 |
| chr22_+_40742497 | 26.22 |
ENST00000216194.7
|
ADSL
|
adenylosuccinate lyase |
| chr1_+_84944926 | 25.83 |
ENST00000370656.1
ENST00000370654.5 |
RPF1
|
ribosome production factor 1 homolog (S. cerevisiae) |
| chr20_-_33872548 | 25.80 |
ENST00000374443.3
|
EIF6
|
eukaryotic translation initiation factor 6 |
| chr1_+_32687971 | 25.41 |
ENST00000373586.1
|
EIF3I
|
eukaryotic translation initiation factor 3, subunit I |
| chr15_-_65809581 | 25.39 |
ENST00000341861.5
|
DPP8
|
dipeptidyl-peptidase 8 |
| chr3_+_127771212 | 24.99 |
ENST00000243253.3
ENST00000481210.1 |
SEC61A1
|
Sec61 alpha 1 subunit (S. cerevisiae) |
| chr2_+_122494676 | 24.76 |
ENST00000455432.1
|
TSN
|
translin |
| chr19_+_49496705 | 24.53 |
ENST00000595090.1
|
RUVBL2
|
RuvB-like AAA ATPase 2 |
| chr8_-_121457332 | 24.52 |
ENST00000518918.1
|
MRPL13
|
mitochondrial ribosomal protein L13 |
| chr7_+_99006232 | 24.52 |
ENST00000403633.2
|
BUD31
|
BUD31 homolog (S. cerevisiae) |
| chr20_+_16710606 | 24.00 |
ENST00000377943.5
ENST00000246071.6 |
SNRPB2
|
small nuclear ribonucleoprotein polypeptide B |
| chr14_-_24616426 | 23.97 |
ENST00000216802.5
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr22_+_40742512 | 23.65 |
ENST00000454266.2
ENST00000342312.6 |
ADSL
|
adenylosuccinate lyase |
| chr15_-_65282232 | 23.35 |
ENST00000416889.2
|
SPG21
|
spastic paraplegia 21 (autosomal recessive, Mast syndrome) |
| chr8_-_117768023 | 23.30 |
ENST00000518949.1
ENST00000522453.1 ENST00000521861.1 ENST00000518995.1 |
EIF3H
|
eukaryotic translation initiation factor 3, subunit H |
| chr3_-_49142178 | 23.18 |
ENST00000452739.1
ENST00000414533.1 ENST00000417025.1 |
QARS
|
glutaminyl-tRNA synthetase |
| chr15_-_65810042 | 23.06 |
ENST00000321147.6
|
DPP8
|
dipeptidyl-peptidase 8 |
| chr11_+_47600562 | 23.06 |
ENST00000263774.4
ENST00000529276.1 ENST00000528192.1 ENST00000530295.1 ENST00000534208.1 ENST00000534716.2 |
NDUFS3
|
NADH dehydrogenase (ubiquinone) Fe-S protein 3, 30kDa (NADH-coenzyme Q reductase) |
| chr11_-_6633799 | 22.95 |
ENST00000299424.4
|
TAF10
|
TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 30kDa |
| chr15_-_65809991 | 22.80 |
ENST00000559526.1
ENST00000358939.4 ENST00000560665.1 ENST00000321118.7 ENST00000339244.5 ENST00000300141.6 |
DPP8
|
dipeptidyl-peptidase 8 |
| chr3_-_10362725 | 22.63 |
ENST00000397109.3
ENST00000428626.1 ENST00000445064.1 ENST00000431352.1 ENST00000397117.1 ENST00000337354.4 ENST00000383801.2 ENST00000432213.1 ENST00000350697.3 |
SEC13
|
SEC13 homolog (S. cerevisiae) |
| chr3_+_23847394 | 22.25 |
ENST00000306627.3
|
UBE2E1
|
ubiquitin-conjugating enzyme E2E 1 |
| chr15_-_65282274 | 21.76 |
ENST00000204566.2
|
SPG21
|
spastic paraplegia 21 (autosomal recessive, Mast syndrome) |
| chr1_+_151372010 | 21.48 |
ENST00000290541.6
|
PSMB4
|
proteasome (prosome, macropain) subunit, beta type, 4 |
| chr1_-_165738072 | 21.25 |
ENST00000481278.1
|
TMCO1
|
transmembrane and coiled-coil domains 1 |
| chr8_-_124054362 | 21.03 |
ENST00000405944.3
|
DERL1
|
derlin 1 |
| chr13_+_28194873 | 21.02 |
ENST00000302979.3
|
POLR1D
|
polymerase (RNA) I polypeptide D, 16kDa |
| chr17_+_49337881 | 20.86 |
ENST00000225298.7
|
UTP18
|
UTP18 small subunit (SSU) processome component homolog (yeast) |
| chr14_+_35452104 | 20.69 |
ENST00000216774.6
ENST00000546080.1 |
SRP54
|
signal recognition particle 54kDa |
| chr16_+_74330673 | 20.61 |
ENST00000219313.4
ENST00000540379.1 ENST00000567958.1 ENST00000568615.2 |
PSMD7
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 7 |
| chr1_-_150207017 | 20.27 |
ENST00000369119.3
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr7_+_100482595 | 20.08 |
ENST00000448764.1
|
SRRT
|
serrate RNA effector molecule homolog (Arabidopsis) |
| chr15_-_85259294 | 20.08 |
ENST00000558217.1
ENST00000558196.1 ENST00000558134.1 |
SEC11A
|
SEC11 homolog A (S. cerevisiae) |
| chrX_+_69509927 | 20.08 |
ENST00000374403.3
|
KIF4A
|
kinesin family member 4A |
| chr8_-_124054484 | 19.92 |
ENST00000419562.2
|
DERL1
|
derlin 1 |
| chr4_+_1723197 | 19.91 |
ENST00000485989.2
ENST00000313288.4 |
TACC3
|
transforming, acidic coiled-coil containing protein 3 |
| chr6_-_13814663 | 19.71 |
ENST00000359495.2
ENST00000379170.4 |
MCUR1
|
mitochondrial calcium uniporter regulator 1 |
| chr1_-_19811962 | 19.62 |
ENST00000375142.1
ENST00000264202.6 |
CAPZB
|
capping protein (actin filament) muscle Z-line, beta |
| chr9_-_2844058 | 19.44 |
ENST00000397885.2
|
KIAA0020
|
KIAA0020 |
| chr10_+_43278217 | 19.39 |
ENST00000374518.5
|
BMS1
|
BMS1 ribosome biogenesis factor |
| chr3_-_49142504 | 19.39 |
ENST00000306125.6
ENST00000420147.2 |
QARS
|
glutaminyl-tRNA synthetase |
| chr13_+_29233218 | 19.38 |
ENST00000380842.4
|
POMP
|
proteasome maturation protein |
| chr19_-_40336969 | 19.21 |
ENST00000599134.1
ENST00000597634.1 ENST00000598417.1 ENST00000601274.1 ENST00000594309.1 ENST00000221801.3 |
FBL
|
fibrillarin |
| chr15_-_65281775 | 19.12 |
ENST00000433215.2
ENST00000558415.1 ENST00000557795.1 |
SPG21
|
spastic paraplegia 21 (autosomal recessive, Mast syndrome) |
| chr19_+_49496782 | 18.88 |
ENST00000601968.1
ENST00000596837.1 |
RUVBL2
|
RuvB-like AAA ATPase 2 |
| chr19_-_59066327 | 18.85 |
ENST00000596708.1
ENST00000601220.1 ENST00000597848.1 |
CHMP2A
|
charged multivesicular body protein 2A |
| chr15_-_85259330 | 18.81 |
ENST00000560266.1
|
SEC11A
|
SEC11 homolog A (S. cerevisiae) |
| chr14_+_21458127 | 18.80 |
ENST00000382985.4
ENST00000556670.2 ENST00000553564.1 ENST00000554751.1 ENST00000554283.1 ENST00000555670.1 |
METTL17
|
methyltransferase like 17 |
| chr19_+_9938562 | 18.79 |
ENST00000586895.1
ENST00000358666.3 ENST00000590068.1 ENST00000593087.1 |
UBL5
|
ubiquitin-like 5 |
| chr9_+_127631399 | 18.32 |
ENST00000259477.6
|
ARPC5L
|
actin related protein 2/3 complex, subunit 5-like |
| chrX_+_24072833 | 18.30 |
ENST00000253039.4
|
EIF2S3
|
eukaryotic translation initiation factor 2, subunit 3 gamma, 52kDa |
| chr3_+_23847432 | 18.23 |
ENST00000346855.3
|
UBE2E1
|
ubiquitin-conjugating enzyme E2E 1 |
| chr11_-_18548426 | 18.19 |
ENST00000357193.3
ENST00000536719.1 |
TSG101
|
tumor susceptibility 101 |
| chr17_+_61904766 | 18.15 |
ENST00000581842.1
ENST00000582130.1 ENST00000584320.1 ENST00000585123.1 ENST00000580864.1 |
PSMC5
|
proteasome (prosome, macropain) 26S subunit, ATPase, 5 |
| chr11_+_58910201 | 18.13 |
ENST00000528737.1
|
FAM111A
|
family with sequence similarity 111, member A |
| chr12_-_120907374 | 18.13 |
ENST00000550458.1
|
SRSF9
|
serine/arginine-rich splicing factor 9 |
| chr1_-_43855444 | 18.12 |
ENST00000372455.4
|
MED8
|
mediator complex subunit 8 |
| chr19_-_9546177 | 17.83 |
ENST00000592292.1
ENST00000588221.1 |
ZNF266
|
zinc finger protein 266 |
| chr6_-_170862322 | 17.82 |
ENST00000262193.6
|
PSMB1
|
proteasome (prosome, macropain) subunit, beta type, 1 |
| chr19_-_10230562 | 17.73 |
ENST00000587146.1
ENST00000588709.1 ENST00000253108.4 |
EIF3G
|
eukaryotic translation initiation factor 3, subunit G |
| chr14_+_35452169 | 17.64 |
ENST00000555557.1
|
SRP54
|
signal recognition particle 54kDa |
| chr6_+_31633902 | 17.48 |
ENST00000375865.2
ENST00000375866.2 |
CSNK2B
|
casein kinase 2, beta polypeptide |
| chr15_-_85259384 | 17.41 |
ENST00000455959.3
|
SEC11A
|
SEC11 homolog A (S. cerevisiae) |
| chr20_-_49575081 | 17.40 |
ENST00000371588.5
ENST00000371582.4 |
DPM1
|
dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit |
| chr14_-_67826486 | 17.31 |
ENST00000555431.1
ENST00000554236.1 ENST00000555474.1 |
ATP6V1D
|
ATPase, H+ transporting, lysosomal 34kDa, V1 subunit D |
| chr11_-_60674037 | 17.29 |
ENST00000541371.1
ENST00000227524.4 |
PRPF19
|
pre-mRNA processing factor 19 |
| chr19_-_59066452 | 17.21 |
ENST00000312547.2
|
CHMP2A
|
charged multivesicular body protein 2A |
| chr2_+_201936458 | 17.19 |
ENST00000237889.4
|
NDUFB3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3, 12kDa |
| chr4_-_10118348 | 17.14 |
ENST00000502702.1
|
WDR1
|
WD repeat domain 1 |
| chr21_-_46340884 | 16.95 |
ENST00000302347.5
ENST00000517819.1 |
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr6_-_75953484 | 16.88 |
ENST00000472311.2
ENST00000460985.1 ENST00000377978.3 ENST00000509698.1 ENST00000230459.4 ENST00000370089.2 |
COX7A2
|
cytochrome c oxidase subunit VIIa polypeptide 2 (liver) |
| chr12_-_9102549 | 16.84 |
ENST00000000412.3
|
M6PR
|
mannose-6-phosphate receptor (cation dependent) |
| chr14_+_55518349 | 16.81 |
ENST00000395468.4
|
MAPK1IP1L
|
mitogen-activated protein kinase 1 interacting protein 1-like |
| chr2_+_201981527 | 16.68 |
ENST00000441224.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr19_-_10514184 | 16.67 |
ENST00000589629.1
ENST00000222005.2 |
CDC37
|
cell division cycle 37 |
| chr22_+_24951436 | 16.63 |
ENST00000215829.3
|
SNRPD3
|
small nuclear ribonucleoprotein D3 polypeptide 18kDa |
| chr4_-_10118573 | 16.38 |
ENST00000382452.2
ENST00000382451.2 |
WDR1
|
WD repeat domain 1 |
| chr19_-_10230540 | 16.36 |
ENST00000589454.1
|
EIF3G
|
eukaryotic translation initiation factor 3, subunit G |
| chr16_-_69373396 | 16.32 |
ENST00000562595.1
ENST00000562081.1 ENST00000306875.4 |
COG8
|
component of oligomeric golgi complex 8 |
| chr8_-_101962777 | 16.23 |
ENST00000395951.3
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr20_+_60962143 | 16.23 |
ENST00000343986.4
|
RPS21
|
ribosomal protein S21 |
| chr17_-_38574169 | 16.10 |
ENST00000423485.1
|
TOP2A
|
topoisomerase (DNA) II alpha 170kDa |
| chr11_+_58910295 | 16.02 |
ENST00000420244.1
|
FAM111A
|
family with sequence similarity 111, member A |
| chr1_-_43638168 | 15.99 |
ENST00000431635.2
|
EBNA1BP2
|
EBNA1 binding protein 2 |
| chr8_-_121457608 | 15.96 |
ENST00000306185.3
|
MRPL13
|
mitochondrial ribosomal protein L13 |
| chr20_-_524415 | 15.95 |
ENST00000400217.2
|
CSNK2A1
|
casein kinase 2, alpha 1 polypeptide |
| chr5_-_31532160 | 15.90 |
ENST00000511367.2
ENST00000513349.1 |
DROSHA
|
drosha, ribonuclease type III |
| chr19_-_9546227 | 15.79 |
ENST00000361451.2
ENST00000361151.1 |
ZNF266
|
zinc finger protein 266 |
| chr3_-_196669248 | 15.71 |
ENST00000447325.1
|
NCBP2
|
nuclear cap binding protein subunit 2, 20kDa |
| chr7_+_44240520 | 15.70 |
ENST00000496112.1
ENST00000223369.2 |
YKT6
|
YKT6 v-SNARE homolog (S. cerevisiae) |
| chr21_-_46340770 | 15.70 |
ENST00000397854.3
|
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr5_+_36152091 | 15.63 |
ENST00000274254.5
|
SKP2
|
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr8_-_48872686 | 15.37 |
ENST00000314191.2
ENST00000338368.3 |
PRKDC
|
protein kinase, DNA-activated, catalytic polypeptide |
| chr19_+_55897699 | 15.28 |
ENST00000558131.1
ENST00000558752.1 ENST00000458349.2 |
RPL28
|
ribosomal protein L28 |
| chr5_+_96079240 | 15.20 |
ENST00000515663.1
|
CAST
|
calpastatin |
| chr19_+_19303008 | 14.86 |
ENST00000353145.1
ENST00000421262.3 ENST00000303088.4 ENST00000456252.3 ENST00000593273.1 |
RFXANK
|
regulatory factor X-associated ankyrin-containing protein |
| chr19_-_13044494 | 14.85 |
ENST00000593021.1
ENST00000587981.1 ENST00000423140.2 ENST00000314606.4 |
FARSA
|
phenylalanyl-tRNA synthetase, alpha subunit |
| chr1_+_15736359 | 14.84 |
ENST00000375980.4
|
EFHD2
|
EF-hand domain family, member D2 |
| chr6_-_144416737 | 14.81 |
ENST00000367569.2
|
SF3B5
|
splicing factor 3b, subunit 5, 10kDa |
| chr6_-_31620149 | 14.57 |
ENST00000435080.1
ENST00000375976.4 ENST00000441054.1 |
BAG6
|
BCL2-associated athanogene 6 |
| chr12_-_120907459 | 14.37 |
ENST00000229390.3
|
SRSF9
|
serine/arginine-rich splicing factor 9 |
| chr2_+_96931834 | 14.31 |
ENST00000488633.1
|
CIAO1
|
cytosolic iron-sulfur protein assembly 1 |
| chr6_+_31633833 | 14.25 |
ENST00000375882.2
ENST00000375880.2 |
CSNK2B
CSNK2B-LY6G5B-1181
|
casein kinase 2, beta polypeptide Uncharacterized protein |
| chr19_-_16770915 | 14.22 |
ENST00000593459.1
ENST00000358726.6 ENST00000597711.1 ENST00000487416.2 |
CTC-429P9.4
SMIM7
|
Small integral membrane protein 7; Uncharacterized protein small integral membrane protein 7 |
| chr8_+_145137489 | 14.21 |
ENST00000355091.4
ENST00000525087.1 ENST00000361036.6 ENST00000524418.1 |
GPAA1
|
glycosylphosphatidylinositol anchor attachment 1 |
| chr8_-_117778494 | 14.20 |
ENST00000276682.4
|
EIF3H
|
eukaryotic translation initiation factor 3, subunit H |
| chr3_-_16555150 | 14.06 |
ENST00000334133.4
|
RFTN1
|
raftlin, lipid raft linker 1 |
| chr19_-_46195029 | 13.80 |
ENST00000588599.1
ENST00000585392.1 ENST00000590212.1 ENST00000587367.1 ENST00000391932.3 |
SNRPD2
|
small nuclear ribonucleoprotein D2 polypeptide 16.5kDa |
| chr19_+_54619125 | 13.75 |
ENST00000445811.1
ENST00000419967.1 ENST00000445124.1 ENST00000447810.1 |
PRPF31
|
pre-mRNA processing factor 31 |
| chr1_+_26644441 | 13.67 |
ENST00000374213.2
|
CD52
|
CD52 molecule |
| chr22_+_32871224 | 13.63 |
ENST00000452138.1
ENST00000382058.3 ENST00000397426.1 |
FBXO7
|
F-box protein 7 |
| chr3_-_196669298 | 13.60 |
ENST00000411704.1
ENST00000452404.2 |
NCBP2
|
nuclear cap binding protein subunit 2, 20kDa |
| chr15_-_85259360 | 13.48 |
ENST00000559729.1
|
SEC11A
|
SEC11 homolog A (S. cerevisiae) |
| chr19_+_49375649 | 13.46 |
ENST00000200453.5
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A |
| chr16_+_30075783 | 13.35 |
ENST00000412304.2
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr14_+_75348592 | 13.32 |
ENST00000334220.4
|
DLST
|
dihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) |
| chr6_-_31697255 | 13.30 |
ENST00000436437.1
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr6_-_108395907 | 13.18 |
ENST00000193322.3
|
OSTM1
|
osteopetrosis associated transmembrane protein 1 |
| chr16_+_30075463 | 13.11 |
ENST00000562168.1
ENST00000569545.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr20_+_3190006 | 13.06 |
ENST00000380113.3
ENST00000455664.2 ENST00000399838.3 |
ITPA
|
inosine triphosphatase (nucleoside triphosphate pyrophosphatase) |
| chr9_+_36190853 | 12.95 |
ENST00000433436.2
ENST00000538225.1 ENST00000540080.1 |
CLTA
|
clathrin, light chain A |
| chr1_-_43637915 | 12.93 |
ENST00000236051.2
|
EBNA1BP2
|
EBNA1 binding protein 2 |
| chr17_-_30228678 | 12.87 |
ENST00000261708.4
|
UTP6
|
UTP6, small subunit (SSU) processome component, homolog (yeast) |
| chr10_+_22605374 | 12.85 |
ENST00000448361.1
|
COMMD3
|
COMM domain containing 3 |
| chr3_-_33481835 | 12.79 |
ENST00000283629.3
|
UBP1
|
upstream binding protein 1 (LBP-1a) |
| chr17_+_79650962 | 12.72 |
ENST00000329138.4
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate |
| chrX_+_148622513 | 12.58 |
ENST00000393985.3
ENST00000423421.1 ENST00000423540.2 ENST00000434353.2 ENST00000514208.1 |
CXorf40A
|
chromosome X open reading frame 40A |
| chr19_+_39109735 | 12.52 |
ENST00000593149.1
ENST00000248342.4 ENST00000538434.1 ENST00000588934.1 ENST00000545173.2 ENST00000589307.1 ENST00000586513.1 ENST00000591409.1 ENST00000592558.1 |
EIF3K
|
eukaryotic translation initiation factor 3, subunit K |
| chr5_+_31532373 | 12.51 |
ENST00000325366.9
ENST00000355907.3 ENST00000507818.2 |
C5orf22
|
chromosome 5 open reading frame 22 |
| chr16_+_30075595 | 12.47 |
ENST00000563060.2
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chrX_-_100872911 | 12.38 |
ENST00000361910.4
ENST00000539247.1 ENST00000538627.1 |
ARMCX6
|
armadillo repeat containing, X-linked 6 |
| chr8_-_131028869 | 12.38 |
ENST00000518283.1
ENST00000519110.1 |
FAM49B
|
family with sequence similarity 49, member B |
| chr9_+_36190905 | 12.36 |
ENST00000345519.5
ENST00000470744.1 ENST00000242285.6 ENST00000466396.1 ENST00000396603.2 |
CLTA
|
clathrin, light chain A |
| chr14_-_75179774 | 12.25 |
ENST00000555249.1
ENST00000556202.1 ENST00000356357.4 ENST00000338772.5 |
AREL1
AC007956.1
|
apoptosis resistant E3 ubiquitin protein ligase 1 Full-length cDNA 5-PRIME end of clone CS0CAP004YO05 of Thymus of Homo sapiens (human); Uncharacterized protein |
| chr12_+_53693812 | 12.21 |
ENST00000549488.1
|
C12orf10
|
chromosome 12 open reading frame 10 |
| chr15_+_41624892 | 12.08 |
ENST00000260359.6
ENST00000450318.1 ENST00000450592.2 ENST00000559596.1 ENST00000414849.2 ENST00000560747.1 ENST00000560177.1 |
NUSAP1
|
nucleolar and spindle associated protein 1 |
| chr10_-_5855350 | 11.92 |
ENST00000456041.1
ENST00000380181.3 ENST00000418688.1 ENST00000380132.4 ENST00000609712.1 ENST00000380191.4 |
GDI2
|
GDP dissociation inhibitor 2 |
| chr1_-_235324530 | 11.90 |
ENST00000447801.1
ENST00000366606.3 ENST00000429912.1 |
RBM34
|
RNA binding motif protein 34 |
| chr19_+_50919056 | 11.85 |
ENST00000599632.1
|
CTD-2545M3.6
|
CTD-2545M3.6 |
| chr1_-_20987889 | 11.77 |
ENST00000415136.2
|
DDOST
|
dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit (non-catalytic) |
| chr1_-_20987851 | 11.77 |
ENST00000464364.1
ENST00000602624.2 |
DDOST
|
dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit (non-catalytic) |
| chr14_+_23340822 | 11.76 |
ENST00000359591.4
|
LRP10
|
low density lipoprotein receptor-related protein 10 |
| chr22_-_18111499 | 11.64 |
ENST00000413576.1
ENST00000399796.2 ENST00000399798.2 ENST00000253413.5 |
ATP6V1E1
|
ATPase, H+ transporting, lysosomal 31kDa, V1 subunit E1 |
| chr11_+_60681346 | 11.53 |
ENST00000227525.3
|
TMEM109
|
transmembrane protein 109 |
| chr6_+_30539153 | 11.22 |
ENST00000326195.8
ENST00000376545.3 ENST00000396515.4 ENST00000441867.1 ENST00000468958.1 |
ABCF1
|
ATP-binding cassette, sub-family F (GCN20), member 1 |
| chr10_+_7830125 | 11.12 |
ENST00000335698.4
ENST00000541227.1 |
ATP5C1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| chr6_+_133135580 | 11.09 |
ENST00000230050.3
|
RPS12
|
ribosomal protein S12 |
| chr4_-_10118469 | 11.05 |
ENST00000499869.2
|
WDR1
|
WD repeat domain 1 |
| chr17_-_79876010 | 10.99 |
ENST00000328666.6
|
SIRT7
|
sirtuin 7 |
| chr6_-_149969829 | 10.93 |
ENST00000367411.2
|
KATNA1
|
katanin p60 (ATPase containing) subunit A 1 |
| chr11_+_8008867 | 10.92 |
ENST00000309828.4
ENST00000449102.2 |
EIF3F
|
eukaryotic translation initiation factor 3, subunit F |
| chr3_+_101292939 | 10.90 |
ENST00000265260.3
ENST00000469941.1 ENST00000296024.5 |
PCNP
|
PEST proteolytic signal containing nuclear protein |
| chr12_+_27863706 | 10.88 |
ENST00000081029.3
ENST00000538315.1 ENST00000542791.1 |
MRPS35
|
mitochondrial ribosomal protein S35 |
| chr18_-_45456930 | 10.87 |
ENST00000262160.6
ENST00000587269.1 |
SMAD2
|
SMAD family member 2 |
| chr8_-_104427313 | 10.82 |
ENST00000297578.4
|
SLC25A32
|
solute carrier family 25 (mitochondrial folate carrier), member 32 |
| chr12_+_120875910 | 10.77 |
ENST00000551806.1
|
AL021546.6
|
Glutamyl-tRNA(Gln) amidotransferase subunit C, mitochondrial |
| chr3_-_196669371 | 10.70 |
ENST00000427641.2
ENST00000321256.5 |
NCBP2
|
nuclear cap binding protein subunit 2, 20kDa |
| chr2_+_64069459 | 10.68 |
ENST00000445915.2
ENST00000475462.1 |
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr5_+_36152163 | 10.60 |
ENST00000274255.6
|
SKP2
|
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr6_-_127664736 | 10.57 |
ENST00000368291.2
ENST00000309620.9 ENST00000454859.3 |
ECHDC1
|
enoyl CoA hydratase domain containing 1 |
| chrX_+_100646190 | 10.56 |
ENST00000471855.1
|
RPL36A
|
ribosomal protein L36a |
| chrX_+_100645812 | 10.48 |
ENST00000427805.2
ENST00000553110.3 ENST00000392994.3 ENST00000409338.1 ENST00000409170.3 |
RPL36A
RPL36A-HNRNPH2
|
ribosomal protein L36a RPL36A-HNRNPH2 readthrough |
| chr15_+_75074385 | 10.39 |
ENST00000220003.9
|
CSK
|
c-src tyrosine kinase |
| chr15_+_75074410 | 10.32 |
ENST00000439220.2
|
CSK
|
c-src tyrosine kinase |
| chr1_+_150293921 | 10.30 |
ENST00000324862.6
|
PRPF3
|
pre-mRNA processing factor 3 |
| chr14_+_96000930 | 10.29 |
ENST00000331334.4
|
GLRX5
|
glutaredoxin 5 |
| chr8_-_124054587 | 10.29 |
ENST00000259512.4
|
DERL1
|
derlin 1 |
| chr16_-_67694129 | 10.22 |
ENST00000602320.1
|
ACD
|
adrenocortical dysplasia homolog (mouse) |
| chr2_-_27632390 | 10.19 |
ENST00000350803.4
ENST00000344034.4 |
PPM1G
|
protein phosphatase, Mg2+/Mn2+ dependent, 1G |
| chr6_-_127664683 | 10.14 |
ENST00000528402.1
ENST00000454591.2 |
ECHDC1
|
enoyl CoA hydratase domain containing 1 |
| chr6_-_31697563 | 10.05 |
ENST00000375789.2
ENST00000416410.1 |
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr11_-_67205538 | 10.04 |
ENST00000326294.3
|
PTPRCAP
|
protein tyrosine phosphatase, receptor type, C-associated protein |
| chr9_+_33264861 | 10.04 |
ENST00000223500.8
|
CHMP5
|
charged multivesicular body protein 5 |
| chr19_-_50143452 | 10.04 |
ENST00000246792.3
|
RRAS
|
related RAS viral (r-ras) oncogene homolog |
| chr13_-_31736027 | 9.90 |
ENST00000380406.5
ENST00000320027.5 ENST00000380405.4 |
HSPH1
|
heat shock 105kDa/110kDa protein 1 |
| chr10_+_22605304 | 9.88 |
ENST00000475460.2
ENST00000602390.1 ENST00000489125.2 ENST00000456711.1 ENST00000444869.1 |
COMMD3-BMI1
COMMD3
|
COMMD3-BMI1 readthrough COMM domain containing 3 |
| chr20_-_524455 | 9.88 |
ENST00000349736.5
ENST00000217244.3 |
CSNK2A1
|
casein kinase 2, alpha 1 polypeptide |
| chr18_+_21032781 | 9.87 |
ENST00000339486.3
|
RIOK3
|
RIO kinase 3 |
| chr1_+_150293973 | 9.78 |
ENST00000414970.2
ENST00000543398.1 |
PRPF3
|
pre-mRNA processing factor 3 |
| chr12_+_53693466 | 9.76 |
ENST00000267103.5
ENST00000548632.1 |
C12orf10
|
chromosome 12 open reading frame 10 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ETV6
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 28.3 | 85.0 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 24.9 | 74.8 | GO:0000354 | cis assembly of pre-catalytic spliceosome(GO:0000354) |
| 24.1 | 72.3 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 17.8 | 71.4 | GO:0090579 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 16.6 | 49.9 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 14.2 | 42.6 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 13.2 | 39.5 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 13.0 | 52.0 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 12.8 | 38.3 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 12.0 | 36.1 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 11.1 | 44.6 | GO:0016334 | morphogenesis of follicular epithelium(GO:0016333) establishment or maintenance of polarity of follicular epithelium(GO:0016334) establishment of planar polarity of follicular epithelium(GO:0042247) |
| 10.3 | 51.3 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 10.1 | 50.6 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 10.0 | 40.0 | GO:0046833 | snRNA export from nucleus(GO:0006408) positive regulation of RNA export from nucleus(GO:0046833) |
| 8.5 | 51.2 | GO:0051708 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 8.1 | 97.0 | GO:0015074 | DNA integration(GO:0015074) |
| 7.2 | 7.2 | GO:1903094 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 7.1 | 21.4 | GO:0060380 | regulation of single-stranded telomeric DNA binding(GO:0060380) positive regulation of single-stranded telomeric DNA binding(GO:0060381) positive regulation of telomeric DNA binding(GO:1904744) |
| 6.8 | 54.6 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 6.1 | 18.2 | GO:2000397 | ubiquitin-dependent endocytosis(GO:0070086) regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 5.6 | 16.8 | GO:0070902 | mitochondrial tRNA pseudouridine synthesis(GO:0070902) |
| 5.5 | 33.0 | GO:0048254 | snoRNA localization(GO:0048254) |
| 5.1 | 40.5 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 5.0 | 25.0 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 4.6 | 101.1 | GO:0006465 | signal peptide processing(GO:0006465) |
| 4.5 | 13.5 | GO:1902310 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 4.4 | 13.3 | GO:0019474 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 4.3 | 34.1 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 4.1 | 20.7 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 4.1 | 8.3 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 4.0 | 15.9 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 4.0 | 31.7 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 3.8 | 15.3 | GO:1904379 | maintenance of unfolded protein(GO:0036506) protein localization to cytosolic proteasome complex(GO:1904327) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 3.7 | 18.7 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 3.7 | 18.6 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 3.7 | 14.9 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 3.7 | 11.0 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 3.6 | 10.8 | GO:0015883 | FAD transport(GO:0015883) FAD transmembrane transport(GO:0035350) |
| 3.3 | 19.7 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 3.1 | 18.9 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 3.1 | 40.7 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 2.7 | 8.2 | GO:0002588 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen(GO:0002585) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 2.6 | 7.9 | GO:1901355 | response to rapamycin(GO:1901355) |
| 2.5 | 10.0 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 2.5 | 7.4 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 2.4 | 14.7 | GO:0051013 | microtubule severing(GO:0051013) |
| 2.4 | 23.5 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 2.3 | 6.9 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 2.2 | 44.4 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 2.2 | 28.9 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 2.2 | 28.5 | GO:0000479 | endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 2.2 | 17.4 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 2.2 | 38.9 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 2.1 | 12.4 | GO:2000568 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 2.0 | 6.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 2.0 | 14.2 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 2.0 | 31.4 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 1.9 | 15.4 | GO:0002326 | B cell lineage commitment(GO:0002326) immunoglobulin V(D)J recombination(GO:0033152) ectopic germ cell programmed cell death(GO:0035234) |
| 1.9 | 7.7 | GO:0043128 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 1.9 | 7.6 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 1.9 | 5.6 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 1.9 | 13.1 | GO:0009146 | purine nucleoside triphosphate catabolic process(GO:0009146) |
| 1.9 | 5.6 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 1.8 | 19.8 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 1.8 | 83.9 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 1.8 | 22.9 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 1.7 | 17.2 | GO:0045047 | protein targeting to ER(GO:0045047) |
| 1.7 | 16.8 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 1.7 | 21.5 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 1.6 | 8.1 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 1.6 | 16.1 | GO:0030263 | resolution of meiotic recombination intermediates(GO:0000712) apoptotic chromosome condensation(GO:0030263) |
| 1.6 | 4.8 | GO:0002881 | negative regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002881) |
| 1.6 | 30.4 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 1.6 | 9.4 | GO:1903750 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 1.6 | 32.6 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 1.5 | 18.1 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 1.5 | 6.0 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 1.4 | 5.7 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 1.4 | 25.3 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 1.4 | 12.6 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 1.4 | 16.7 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 1.3 | 20.1 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 1.3 | 5.2 | GO:1904502 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 1.3 | 16.6 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 1.3 | 5.1 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 1.3 | 13.9 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 1.3 | 29.0 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 1.2 | 22.5 | GO:1903204 | negative regulation of oxidative stress-induced neuron death(GO:1903204) |
| 1.2 | 3.7 | GO:0034184 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 1.2 | 22.0 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 1.2 | 19.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 1.2 | 3.6 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 1.1 | 8.0 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 1.1 | 10.3 | GO:0009249 | protein lipoylation(GO:0009249) |
| 1.1 | 71.8 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 1.1 | 7.7 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 1.1 | 15.2 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 1.1 | 32.5 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 1.1 | 16.2 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 1.0 | 3.9 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 1.0 | 12.7 | GO:1903543 | positive regulation of exosomal secretion(GO:1903543) |
| 1.0 | 4.9 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 1.0 | 4.8 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.9 | 20.7 | GO:1904886 | beta-catenin destruction complex disassembly(GO:1904886) |
| 0.9 | 65.6 | GO:0070125 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 0.9 | 7.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.9 | 33.4 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.9 | 9.5 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.9 | 27.4 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.9 | 23.0 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.8 | 10.9 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.8 | 5.0 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.8 | 6.6 | GO:0006983 | ER overload response(GO:0006983) |
| 0.8 | 8.0 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) positive regulation of translational elongation(GO:0045901) |
| 0.8 | 17.5 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.8 | 11.8 | GO:0048839 | inner ear development(GO:0048839) |
| 0.7 | 62.4 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.7 | 6.3 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.7 | 183.3 | GO:0006413 | translational initiation(GO:0006413) |
| 0.6 | 10.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.6 | 5.7 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.6 | 15.1 | GO:0043101 | purine-containing compound salvage(GO:0043101) |
| 0.6 | 8.4 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.6 | 1.8 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.6 | 7.0 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.6 | 18.6 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.6 | 3.5 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.6 | 16.5 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.6 | 1.1 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.6 | 6.6 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.5 | 16.7 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.5 | 7.9 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.5 | 9.0 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.5 | 10.5 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.5 | 8.7 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.5 | 25.3 | GO:0007091 | metaphase/anaphase transition of mitotic cell cycle(GO:0007091) |
| 0.5 | 1.5 | GO:0070945 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 0.5 | 1.5 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.5 | 34.1 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.5 | 5.2 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.5 | 7.9 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.5 | 21.3 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.5 | 1.8 | GO:0034398 | telomere tethering at nuclear periphery(GO:0034398) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.4 | 13.7 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.4 | 1.3 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.4 | 1.7 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.4 | 4.6 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.4 | 4.6 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.4 | 2.4 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.4 | 8.0 | GO:0009226 | nucleotide-sugar biosynthetic process(GO:0009226) |
| 0.4 | 4.7 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.4 | 2.3 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.4 | 29.0 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.4 | 11.8 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.3 | 19.6 | GO:0019079 | viral genome replication(GO:0019079) |
| 0.3 | 8.3 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.3 | 6.7 | GO:0017004 | cytochrome complex assembly(GO:0017004) |
| 0.3 | 4.6 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.3 | 4.8 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.3 | 19.2 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.3 | 15.7 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.3 | 1.1 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.3 | 1.4 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.3 | 15.1 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.3 | 2.2 | GO:0034128 | negative regulation of MyD88-independent toll-like receptor signaling pathway(GO:0034128) |
| 0.3 | 3.8 | GO:0034447 | very-low-density lipoprotein particle clearance(GO:0034447) |
| 0.3 | 5.5 | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.2 | 20.3 | GO:0000271 | polysaccharide biosynthetic process(GO:0000271) |
| 0.2 | 8.4 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.2 | 4.4 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.2 | 1.4 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.2 | 0.6 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.2 | 9.1 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.2 | 3.4 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.2 | 4.0 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.2 | 0.7 | GO:0030242 | pexophagy(GO:0030242) |
| 0.2 | 2.5 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.2 | 2.1 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.1 | 8.7 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.1 | 40.9 | GO:0050851 | antigen receptor-mediated signaling pathway(GO:0050851) |
| 0.1 | 2.0 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.1 | 1.0 | GO:0070236 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 13.2 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.1 | 6.6 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.1 | 5.9 | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage(GO:0008630) |
| 0.1 | 1.3 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 0.7 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 2.2 | GO:0006337 | nucleosome disassembly(GO:0006337) protein-DNA complex disassembly(GO:0032986) |
| 0.1 | 3.5 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.1 | 5.5 | GO:0043647 | inositol phosphate metabolic process(GO:0043647) |
| 0.1 | 20.2 | GO:0000398 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.1 | 0.3 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 18.8 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 0.1 | GO:1903599 | positive regulation of mitophagy(GO:1903599) |
| 0.0 | 3.1 | GO:0042113 | B cell activation(GO:0042113) |
| 0.0 | 0.9 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 14.0 | GO:0002283 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.0 | 1.6 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.7 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 1.3 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 1.7 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.5 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.0 | 0.1 | GO:0042791 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 16.9 | 50.6 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 14.4 | 101.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 12.8 | 51.2 | GO:0036502 | Derlin-1-VIMP complex(GO:0036502) |
| 10.0 | 40.0 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 8.9 | 44.6 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 8.9 | 26.6 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 8.2 | 73.8 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 8.0 | 63.9 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 7.3 | 51.3 | GO:0071203 | WASH complex(GO:0071203) |
| 7.1 | 71.4 | GO:0097255 | R2TP complex(GO:0097255) |
| 6.7 | 33.7 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 6.5 | 32.6 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 6.4 | 19.2 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 5.8 | 122.4 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 5.7 | 85.0 | GO:0030478 | actin cap(GO:0030478) |
| 5.4 | 16.1 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 5.3 | 37.2 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 5.2 | 15.7 | GO:0097441 | basilar dendrite(GO:0097441) |
| 4.8 | 38.3 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 4.7 | 14.2 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 4.3 | 120.3 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 4.2 | 12.7 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 4.0 | 24.0 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 4.0 | 15.9 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 3.9 | 46.8 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 3.8 | 15.3 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 3.7 | 22.5 | GO:1990037 | Lewy body core(GO:1990037) |
| 3.6 | 18.2 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 3.6 | 111.2 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 3.0 | 9.1 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 3.0 | 38.8 | GO:0005686 | U2 snRNP(GO:0005686) |
| 2.9 | 8.8 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 2.9 | 46.6 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 2.9 | 40.1 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 2.8 | 25.3 | GO:0071439 | clathrin complex(GO:0071439) |
| 2.8 | 36.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 2.7 | 18.6 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 2.6 | 18.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 2.6 | 15.4 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 2.5 | 22.9 | GO:0000125 | PCAF complex(GO:0000125) |
| 2.5 | 15.1 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 2.5 | 39.3 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 2.4 | 14.3 | GO:0071817 | MMXD complex(GO:0071817) |
| 2.3 | 6.9 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 2.1 | 64.2 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 2.0 | 8.0 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 1.9 | 13.3 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 1.7 | 12.0 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 1.7 | 10.0 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 1.6 | 6.6 | GO:0032044 | DSIF complex(GO:0032044) |
| 1.5 | 18.1 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 1.5 | 11.8 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 1.4 | 25.7 | GO:0097342 | ripoptosome(GO:0097342) |
| 1.4 | 1.4 | GO:0048500 | signal recognition particle(GO:0048500) |
| 1.4 | 8.2 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 1.3 | 5.2 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 1.3 | 51.4 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 1.3 | 84.4 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 1.3 | 17.6 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 1.2 | 7.4 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 1.2 | 19.4 | GO:0030686 | 90S preribosome(GO:0030686) |
| 1.2 | 21.0 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 1.2 | 4.6 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 1.0 | 20.6 | GO:0022624 | proteasome regulatory particle(GO:0005838) proteasome accessory complex(GO:0022624) |
| 0.9 | 3.7 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.8 | 15.8 | GO:0033176 | proton-transporting V-type ATPase complex(GO:0033176) |
| 0.8 | 42.4 | GO:0031430 | M band(GO:0031430) |
| 0.8 | 13.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.7 | 11.0 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.7 | 11.6 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.7 | 7.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.7 | 16.8 | GO:0030904 | retromer complex(GO:0030904) |
| 0.7 | 15.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.7 | 19.8 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.6 | 70.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.6 | 34.9 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.6 | 10.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.6 | 8.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.6 | 5.7 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.6 | 14.2 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.6 | 19.2 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.5 | 57.3 | GO:0005840 | ribosome(GO:0005840) |
| 0.4 | 1.3 | GO:0031251 | PAN complex(GO:0031251) |
| 0.4 | 12.1 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.4 | 21.6 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.4 | 11.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.4 | 9.6 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.4 | 4.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.3 | 9.4 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.3 | 23.6 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.3 | 3.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.3 | 3.5 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.3 | 47.4 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.3 | 58.2 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.3 | 40.2 | GO:0101002 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.3 | 3.0 | GO:0044754 | autolysosome(GO:0044754) |
| 0.2 | 1.5 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.2 | 1.5 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.2 | 3.6 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.2 | 2.0 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.2 | 3.8 | GO:0042627 | chylomicron(GO:0042627) |
| 0.2 | 9.4 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.2 | 4.8 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.2 | 1.8 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 22.6 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 7.7 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 3.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 0.9 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 3.9 | GO:0000776 | kinetochore(GO:0000776) |
| 0.1 | 4.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 0.5 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 12.4 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 2.1 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 8.4 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 7.9 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.1 | 0.5 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 2.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 13.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 15.4 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 30.3 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 1.5 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 10.1 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 5.5 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 19.9 | GO:0019866 | organelle inner membrane(GO:0019866) |
| 0.1 | 4.6 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 0.9 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 2.1 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 1.1 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 35.7 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 5.7 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 1.8 | GO:0042641 | actomyosin(GO:0042641) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 24.1 | 72.3 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 16.9 | 50.6 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 14.2 | 42.6 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 10.6 | 63.8 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 10.4 | 52.0 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 8.8 | 26.3 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 8.6 | 25.8 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 7.7 | 38.3 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 7.3 | 87.8 | GO:0070990 | snRNP binding(GO:0070990) |
| 6.7 | 40.5 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 6.3 | 31.4 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 6.2 | 18.7 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 5.6 | 16.8 | GO:0004730 | pseudouridylate synthase activity(GO:0004730) |
| 5.4 | 16.1 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 5.0 | 40.0 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 4.9 | 39.6 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 4.7 | 14.2 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 4.7 | 32.6 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 4.4 | 13.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 3.7 | 14.9 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 3.7 | 66.6 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 3.7 | 11.0 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 3.6 | 10.8 | GO:0015230 | FAD transmembrane transporter activity(GO:0015230) |
| 3.5 | 38.9 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 3.4 | 24.0 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 3.3 | 13.3 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 3.2 | 71.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 3.2 | 19.4 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 3.2 | 15.9 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 3.2 | 9.5 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 3.1 | 30.8 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 2.9 | 8.8 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 2.9 | 64.2 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 2.8 | 8.4 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 2.7 | 8.2 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 2.6 | 26.4 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 2.6 | 15.4 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 2.5 | 15.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 2.5 | 15.1 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 2.4 | 14.7 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 2.4 | 173.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 2.4 | 7.2 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 2.3 | 23.2 | GO:0046790 | virion binding(GO:0046790) |
| 2.3 | 6.9 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 2.1 | 12.4 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 2.0 | 8.0 | GO:0032143 | single thymine insertion binding(GO:0032143) |
| 1.9 | 5.8 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 1.9 | 11.6 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 1.9 | 19.2 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 1.7 | 5.2 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 1.7 | 5.0 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 1.6 | 18.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 1.6 | 8.1 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 1.6 | 4.8 | GO:0016495 | C-X3-C chemokine receptor activity(GO:0016495) |
| 1.6 | 84.4 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 1.6 | 6.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 1.6 | 18.8 | GO:0031386 | protein tag(GO:0031386) |
| 1.6 | 6.2 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 1.5 | 4.6 | GO:0051538 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 1.5 | 18.2 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 1.5 | 6.0 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 1.4 | 5.5 | GO:0052827 | inositol pentakisphosphate phosphatase activity(GO:0052827) |
| 1.4 | 5.4 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 1.3 | 8.0 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 1.3 | 39.3 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 1.2 | 36.1 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 1.2 | 6.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 1.2 | 16.7 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 1.2 | 21.0 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 1.1 | 14.4 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 1.1 | 33.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 1.1 | 74.8 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 1.0 | 13.9 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 1.0 | 7.9 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 1.0 | 16.8 | GO:0005537 | mannose binding(GO:0005537) |
| 0.8 | 6.7 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.8 | 23.5 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.8 | 10.0 | GO:0089720 | caspase binding(GO:0089720) |
| 0.8 | 20.7 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.8 | 198.6 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.8 | 8.8 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.8 | 22.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.8 | 21.4 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.8 | 3.8 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.7 | 11.8 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.7 | 112.2 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.7 | 7.8 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.7 | 56.7 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.7 | 18.6 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.7 | 10.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.7 | 19.2 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.7 | 3.4 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.7 | 12.9 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.7 | 7.4 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.7 | 8.0 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.6 | 7.7 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.6 | 139.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.6 | 33.5 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.6 | 7.9 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.6 | 2.4 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.6 | 3.5 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.6 | 5.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.6 | 25.3 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.6 | 1.1 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.5 | 9.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.5 | 11.2 | GO:0008135 | translation factor activity, RNA binding(GO:0008135) |
| 0.5 | 11.9 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.4 | 4.8 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.4 | 9.4 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.4 | 1.1 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.4 | 19.6 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.3 | 1.7 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.3 | 5.9 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.3 | 29.0 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.2 | 6.9 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.2 | 9.2 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.2 | 1.6 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.2 | 17.4 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.2 | 7.6 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.2 | 12.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.2 | 3.3 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.2 | 14.1 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.2 | 2.3 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.2 | 1.0 | GO:0043426 | MRF binding(GO:0043426) |
| 0.2 | 15.2 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.2 | 4.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.2 | 1.4 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 0.7 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.1 | 16.6 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.1 | 10.0 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 37.8 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.1 | 8.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 16.2 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 0.3 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.1 | 10.1 | GO:0015405 | primary active transmembrane transporter activity(GO:0015399) P-P-bond-hydrolysis-driven transmembrane transporter activity(GO:0015405) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.1 | 6.6 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 11.5 | GO:0022832 | voltage-gated ion channel activity(GO:0005244) voltage-gated channel activity(GO:0022832) |
| 0.1 | 5.9 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 1.5 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 4.8 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.1 | 2.5 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 0.5 | GO:0035325 | Toll-like receptor binding(GO:0035325) |
| 0.0 | 0.5 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.8 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 2.2 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 1.4 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 1.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.3 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 2.5 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 1.1 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 18.4 | GO:0003723 | RNA binding(GO:0003723) |
| 0.0 | 0.7 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 103.6 | PID MYC PATHWAY | C-MYC pathway |
| 1.9 | 52.5 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 1.7 | 96.9 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 1.0 | 13.5 | PID BMP PATHWAY | BMP receptor signaling |
| 0.9 | 23.5 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.8 | 32.6 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.7 | 19.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.4 | 23.1 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.4 | 28.9 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.4 | 10.5 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.4 | 31.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.4 | 42.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.4 | 7.0 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.3 | 7.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.3 | 14.9 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.3 | 5.1 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.2 | 10.0 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 13.5 | PID FOXO PATHWAY | FoxO family signaling |
| 0.2 | 16.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.2 | 12.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 8.3 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 2.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 4.7 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 3.3 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 4.5 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 2.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.6 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 2.6 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.1 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.9 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 1.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.4 | 102.8 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 5.9 | 101.1 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 5.6 | 90.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 4.0 | 145.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 3.4 | 159.8 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 2.9 | 49.9 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 2.7 | 66.5 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 2.6 | 41.7 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 2.3 | 174.3 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 2.0 | 37.8 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 2.0 | 15.9 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 1.9 | 72.9 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 1.8 | 40.5 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 1.7 | 53.8 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 1.7 | 18.6 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 1.7 | 147.7 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 1.4 | 25.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 1.3 | 6.6 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 1.0 | 27.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 1.0 | 22.9 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.9 | 72.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.8 | 97.8 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.8 | 23.5 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.8 | 18.6 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.8 | 20.1 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.8 | 20.7 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.8 | 9.1 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.7 | 19.4 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.7 | 13.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.7 | 47.4 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.7 | 18.7 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.7 | 21.0 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.7 | 37.2 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.6 | 18.2 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.6 | 15.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.6 | 16.7 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.5 | 8.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.5 | 34.0 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.5 | 15.9 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.5 | 40.3 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.5 | 7.9 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.5 | 4.6 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.5 | 16.8 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.4 | 10.0 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.3 | 7.6 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.3 | 9.9 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.3 | 5.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.3 | 12.3 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.3 | 44.6 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.3 | 6.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.3 | 8.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.3 | 19.6 | REACTOME PROCESSING OF CAPPED INTRON CONTAINING PRE MRNA | Genes involved in Processing of Capped Intron-Containing Pre-mRNA |
| 0.2 | 7.9 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.2 | 2.2 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.2 | 2.3 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.2 | 6.9 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.2 | 8.3 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.2 | 12.2 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.2 | 4.6 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.2 | 5.2 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.6 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 4.8 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 10.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 5.7 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.1 | 0.6 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 1.8 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 3.0 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 0.5 | REACTOME IL 2 SIGNALING | Genes involved in Interleukin-2 signaling |
| 0.1 | 1.1 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 4.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 1.1 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 1.1 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 4.7 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |