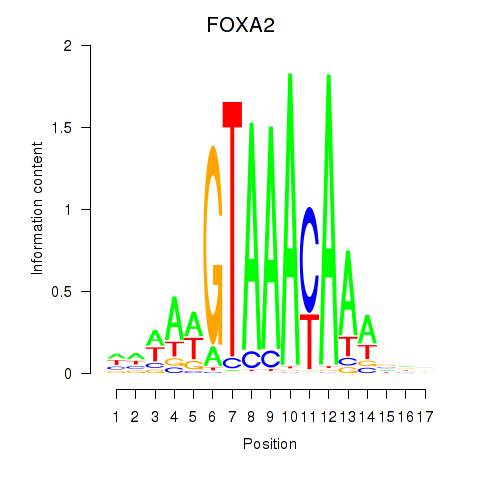
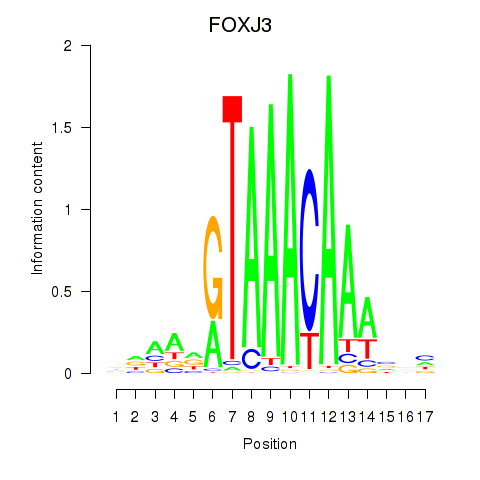
Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for FOXA2_FOXJ3
Z-value: 1.02
Transcription factors associated with FOXA2_FOXJ3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXA2
|
ENSG00000125798.10 | forkhead box A2 |
|
FOXJ3
|
ENSG00000198815.4 | forkhead box J3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXJ3 | hg19_v2_chr1_-_42800614_42800649, hg19_v2_chr1_-_42801540_42801562, hg19_v2_chr1_-_42800860_42800912 | 0.47 | 2.0e-13 | Click! |
| FOXA2 | hg19_v2_chr20_-_22565101_22565223, hg19_v2_chr20_-_22566089_22566097 | -0.38 | 4.3e-09 | Click! |
Activity profile of FOXA2_FOXJ3 motif
Sorted Z-values of FOXA2_FOXJ3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_70329118 | 32.32 |
ENST00000374188.3
|
IL2RG
|
interleukin 2 receptor, gamma |
| chr10_+_70847852 | 25.04 |
ENST00000242465.3
|
SRGN
|
serglycin |
| chr12_-_92539614 | 22.33 |
ENST00000256015.3
|
BTG1
|
B-cell translocation gene 1, anti-proliferative |
| chr4_-_174256276 | 18.92 |
ENST00000296503.5
|
HMGB2
|
high mobility group box 2 |
| chr5_-_98262240 | 18.58 |
ENST00000284049.3
|
CHD1
|
chromodomain helicase DNA binding protein 1 |
| chr1_-_207095324 | 16.85 |
ENST00000530505.1
ENST00000367091.3 ENST00000442471.2 |
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr7_-_37026108 | 16.75 |
ENST00000396045.3
|
ELMO1
|
engulfment and cell motility 1 |
| chr1_+_198608146 | 15.52 |
ENST00000367376.2
ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chr1_+_150122034 | 14.02 |
ENST00000025469.6
ENST00000369124.4 |
PLEKHO1
|
pleckstrin homology domain containing, family O member 1 |
| chr19_-_9546177 | 13.93 |
ENST00000592292.1
ENST00000588221.1 |
ZNF266
|
zinc finger protein 266 |
| chr2_+_58655461 | 13.62 |
ENST00000429095.1
ENST00000429664.1 ENST00000452840.1 |
AC007092.1
|
long intergenic non-protein coding RNA 1122 |
| chr22_-_43036607 | 13.61 |
ENST00000505920.1
|
ATP5L2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit G2 |
| chr1_+_73771844 | 13.20 |
ENST00000440762.1
ENST00000444827.1 ENST00000415686.1 ENST00000411903.1 |
RP4-598G3.1
|
RP4-598G3.1 |
| chr12_-_76462713 | 12.70 |
ENST00000552056.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr2_+_109223595 | 12.41 |
ENST00000410093.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr8_+_11666649 | 11.50 |
ENST00000528643.1
ENST00000525777.1 |
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr8_-_80993010 | 11.12 |
ENST00000537855.1
ENST00000520527.1 ENST00000517427.1 ENST00000448733.2 ENST00000379097.3 |
TPD52
|
tumor protein D52 |
| chr13_-_46756351 | 10.90 |
ENST00000323076.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr5_+_159436120 | 10.89 |
ENST00000522793.1
ENST00000231238.5 |
TTC1
|
tetratricopeptide repeat domain 1 |
| chr14_+_88471468 | 10.78 |
ENST00000267549.3
|
GPR65
|
G protein-coupled receptor 65 |
| chr1_-_207095212 | 10.73 |
ENST00000420007.2
|
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr19_-_39826639 | 10.20 |
ENST00000602185.1
ENST00000598034.1 ENST00000601387.1 ENST00000595636.1 ENST00000253054.8 ENST00000594700.1 ENST00000597595.1 |
GMFG
|
glia maturation factor, gamma |
| chrX_-_122756660 | 10.14 |
ENST00000441692.1
|
THOC2
|
THO complex 2 |
| chr13_-_99910673 | 9.96 |
ENST00000397473.2
ENST00000397470.2 |
GPR18
|
G protein-coupled receptor 18 |
| chr3_-_185641681 | 9.96 |
ENST00000259043.7
|
TRA2B
|
transformer 2 beta homolog (Drosophila) |
| chr2_+_12857043 | 9.48 |
ENST00000381465.2
|
TRIB2
|
tribbles pseudokinase 2 |
| chr18_+_21032781 | 9.15 |
ENST00000339486.3
|
RIOK3
|
RIO kinase 3 |
| chr2_+_68592305 | 9.05 |
ENST00000234313.7
|
PLEK
|
pleckstrin |
| chr10_-_73848086 | 8.91 |
ENST00000536168.1
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr7_+_26332645 | 8.48 |
ENST00000396376.1
|
SNX10
|
sorting nexin 10 |
| chrX_+_9431324 | 8.45 |
ENST00000407597.2
ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X
|
transducin (beta)-like 1X-linked |
| chr1_+_192544857 | 8.31 |
ENST00000367459.3
ENST00000469578.2 |
RGS1
|
regulator of G-protein signaling 1 |
| chr12_-_51718436 | 8.31 |
ENST00000544402.1
|
BIN2
|
bridging integrator 2 |
| chr14_-_60632162 | 8.06 |
ENST00000557185.1
|
DHRS7
|
dehydrogenase/reductase (SDR family) member 7 |
| chr20_-_62582475 | 8.05 |
ENST00000369908.5
|
UCKL1
|
uridine-cytidine kinase 1-like 1 |
| chr17_-_30228678 | 7.88 |
ENST00000261708.4
|
UTP6
|
UTP6, small subunit (SSU) processome component, homolog (yeast) |
| chr3_-_64009658 | 7.85 |
ENST00000394431.2
|
PSMD6
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 |
| chr9_+_70856397 | 7.76 |
ENST00000360171.6
|
CBWD3
|
COBW domain containing 3 |
| chr17_-_685493 | 7.72 |
ENST00000536578.1
ENST00000301328.5 ENST00000576419.1 |
GLOD4
|
glyoxalase domain containing 4 |
| chr16_+_84801852 | 7.53 |
ENST00000569925.1
ENST00000567526.1 |
USP10
|
ubiquitin specific peptidase 10 |
| chr14_-_60632011 | 7.46 |
ENST00000554101.1
ENST00000557137.1 |
DHRS7
|
dehydrogenase/reductase (SDR family) member 7 |
| chr10_-_103578182 | 7.38 |
ENST00000439817.1
|
MGEA5
|
meningioma expressed antigen 5 (hyaluronidase) |
| chr10_-_103578162 | 7.29 |
ENST00000361464.3
ENST00000357797.5 ENST00000370094.3 |
MGEA5
|
meningioma expressed antigen 5 (hyaluronidase) |
| chr5_+_54398463 | 7.11 |
ENST00000274306.6
|
GZMA
|
granzyme A (granzyme 1, cytotoxic T-lymphocyte-associated serine esterase 3) |
| chr6_-_42016385 | 7.10 |
ENST00000502771.1
ENST00000508143.1 ENST00000514588.1 ENST00000510503.1 ENST00000415497.2 ENST00000372988.4 |
CCND3
|
cyclin D3 |
| chr5_-_131132658 | 7.09 |
ENST00000514667.1
ENST00000511848.1 ENST00000510461.1 |
CTC-432M15.3
FNIP1
|
Folliculin-interacting protein 1 folliculin interacting protein 1 |
| chr5_+_162932554 | 7.06 |
ENST00000321757.6
ENST00000421814.2 ENST00000518095.1 |
MAT2B
|
methionine adenosyltransferase II, beta |
| chr16_-_28857677 | 7.02 |
ENST00000313511.3
|
TUFM
|
Tu translation elongation factor, mitochondrial |
| chr9_-_70490107 | 6.81 |
ENST00000377395.4
ENST00000429800.2 ENST00000430059.2 ENST00000377384.1 ENST00000382405.3 |
CBWD5
|
COBW domain containing 5 |
| chr5_+_147774275 | 6.75 |
ENST00000513826.1
|
FBXO38
|
F-box protein 38 |
| chr1_+_117297007 | 6.66 |
ENST00000369478.3
ENST00000369477.1 |
CD2
|
CD2 molecule |
| chr6_+_37012607 | 6.61 |
ENST00000423336.1
|
COX6A1P2
|
cytochrome c oxidase subunit VIa polypeptide 1 pseudogene 2 |
| chr20_+_36373032 | 6.59 |
ENST00000373473.1
|
CTNNBL1
|
catenin, beta like 1 |
| chr11_-_67374177 | 6.54 |
ENST00000333139.3
|
C11orf72
|
chromosome 11 open reading frame 72 |
| chr10_+_111985713 | 6.42 |
ENST00000239007.7
|
MXI1
|
MAX interactor 1, dimerization protein |
| chr15_-_40600026 | 6.35 |
ENST00000456256.2
ENST00000557821.1 |
PLCB2
|
phospholipase C, beta 2 |
| chr6_-_122792919 | 6.22 |
ENST00000339697.4
|
SERINC1
|
serine incorporator 1 |
| chr1_+_95616933 | 6.14 |
ENST00000604203.1
|
RP11-57H12.6
|
TMEM56-RWDD3 readthrough |
| chr16_+_30064444 | 6.12 |
ENST00000395248.1
ENST00000566897.1 ENST00000568435.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr8_-_101321584 | 6.04 |
ENST00000523167.1
|
RNF19A
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr4_+_147096837 | 6.04 |
ENST00000296581.5
ENST00000502781.1 |
LSM6
|
LSM6 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr5_+_176853702 | 6.01 |
ENST00000507633.1
ENST00000393576.3 ENST00000355958.5 ENST00000528793.1 ENST00000512684.1 |
GRK6
|
G protein-coupled receptor kinase 6 |
| chr7_+_106809406 | 5.95 |
ENST00000468410.1
ENST00000478930.1 ENST00000464009.1 ENST00000222574.4 |
HBP1
|
HMG-box transcription factor 1 |
| chr17_-_29641104 | 5.94 |
ENST00000577894.1
ENST00000330927.4 |
EVI2B
|
ecotropic viral integration site 2B |
| chr16_+_30064411 | 5.91 |
ENST00000338110.5
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr11_+_126173647 | 5.87 |
ENST00000263579.4
|
DCPS
|
decapping enzyme, scavenger |
| chr22_-_39268308 | 5.85 |
ENST00000407418.3
|
CBX6
|
chromobox homolog 6 |
| chr1_+_174844645 | 5.82 |
ENST00000486220.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr14_+_97263641 | 5.69 |
ENST00000216639.3
|
VRK1
|
vaccinia related kinase 1 |
| chr12_+_54892550 | 5.66 |
ENST00000545638.2
|
NCKAP1L
|
NCK-associated protein 1-like |
| chr5_+_176853669 | 5.65 |
ENST00000355472.5
|
GRK6
|
G protein-coupled receptor kinase 6 |
| chr4_-_140223614 | 5.63 |
ENST00000394223.1
|
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr12_-_68696652 | 5.62 |
ENST00000539972.1
|
MDM1
|
Mdm1 nuclear protein homolog (mouse) |
| chr2_-_170430277 | 5.58 |
ENST00000438035.1
ENST00000453929.2 |
FASTKD1
|
FAST kinase domains 1 |
| chr11_+_102217936 | 5.58 |
ENST00000532832.1
ENST00000530675.1 ENST00000533742.1 ENST00000227758.2 ENST00000532672.1 ENST00000531259.1 ENST00000527465.1 |
BIRC2
|
baculoviral IAP repeat containing 2 |
| chr4_-_164534657 | 5.52 |
ENST00000339875.5
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr5_+_32531893 | 5.51 |
ENST00000512913.1
|
SUB1
|
SUB1 homolog (S. cerevisiae) |
| chr15_+_42697065 | 5.39 |
ENST00000565559.1
|
CAPN3
|
calpain 3, (p94) |
| chr2_+_109204909 | 5.38 |
ENST00000393310.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr1_+_174846570 | 5.38 |
ENST00000392064.2
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr7_-_23571586 | 5.27 |
ENST00000538367.1
ENST00000392502.4 ENST00000297071.4 |
TRA2A
|
transformer 2 alpha homolog (Drosophila) |
| chr2_-_86422523 | 5.23 |
ENST00000442664.2
ENST00000409051.2 ENST00000449247.2 |
IMMT
|
inner membrane protein, mitochondrial |
| chr1_+_84609944 | 5.22 |
ENST00000370685.3
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr17_-_3595181 | 5.20 |
ENST00000552050.1
|
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr1_+_144811744 | 5.17 |
ENST00000338347.4
ENST00000440491.2 ENST00000375552.4 |
NBPF9
|
neuroblastoma breakpoint family, member 9 |
| chr16_-_24942411 | 5.14 |
ENST00000571843.1
|
ARHGAP17
|
Rho GTPase activating protein 17 |
| chr1_+_144811943 | 5.11 |
ENST00000281815.8
|
NBPF9
|
neuroblastoma breakpoint family, member 9 |
| chr4_-_170679024 | 5.02 |
ENST00000393381.2
|
C4orf27
|
chromosome 4 open reading frame 27 |
| chr6_-_32160622 | 4.95 |
ENST00000487761.1
ENST00000375040.3 |
GPSM3
|
G-protein signaling modulator 3 |
| chr12_+_59989918 | 4.93 |
ENST00000547379.1
ENST00000549465.1 |
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr16_-_28634874 | 4.92 |
ENST00000395609.1
ENST00000350842.4 |
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr2_+_102413726 | 4.90 |
ENST00000350878.4
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr16_-_2314222 | 4.87 |
ENST00000566397.1
|
RNPS1
|
RNA binding protein S1, serine-rich domain |
| chr19_+_18208603 | 4.77 |
ENST00000262811.6
|
MAST3
|
microtubule associated serine/threonine kinase 3 |
| chr7_-_144435985 | 4.75 |
ENST00000549981.1
|
TPK1
|
thiamin pyrophosphokinase 1 |
| chr1_+_158815588 | 4.74 |
ENST00000438394.1
|
MNDA
|
myeloid cell nuclear differentiation antigen |
| chr14_-_58893832 | 4.74 |
ENST00000556007.2
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr1_-_3566627 | 4.73 |
ENST00000419924.2
ENST00000270708.7 |
WRAP73
|
WD repeat containing, antisense to TP73 |
| chr2_+_114195268 | 4.59 |
ENST00000259199.4
ENST00000416503.2 ENST00000433343.2 |
CBWD2
|
COBW domain containing 2 |
| chr7_+_112063192 | 4.59 |
ENST00000005558.4
|
IFRD1
|
interferon-related developmental regulator 1 |
| chr1_-_241803679 | 4.57 |
ENST00000331838.5
|
OPN3
|
opsin 3 |
| chr14_-_58894223 | 4.55 |
ENST00000555593.1
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr12_-_49582978 | 4.55 |
ENST00000301071.7
|
TUBA1A
|
tubulin, alpha 1a |
| chr1_-_241803649 | 4.54 |
ENST00000366554.2
|
OPN3
|
opsin 3 |
| chr12_-_58240470 | 4.39 |
ENST00000548823.1
ENST00000398073.2 |
CTDSP2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr7_-_6098770 | 4.32 |
ENST00000536084.1
ENST00000446699.1 ENST00000199389.6 |
EIF2AK1
|
eukaryotic translation initiation factor 2-alpha kinase 1 |
| chr14_+_35747825 | 4.32 |
ENST00000540871.1
|
PSMA6
|
proteasome (prosome, macropain) subunit, alpha type, 6 |
| chr4_-_90756769 | 4.27 |
ENST00000345009.4
ENST00000505199.1 ENST00000502987.1 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr1_-_161193349 | 4.22 |
ENST00000469730.2
ENST00000463273.1 ENST00000464492.1 ENST00000367990.3 ENST00000470459.2 ENST00000468465.1 ENST00000463812.1 |
APOA2
|
apolipoprotein A-II |
| chr14_+_56127989 | 4.21 |
ENST00000555573.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr1_+_203830703 | 4.19 |
ENST00000414487.2
|
SNRPE
|
small nuclear ribonucleoprotein polypeptide E |
| chr3_+_69928256 | 4.16 |
ENST00000394355.2
|
MITF
|
microphthalmia-associated transcription factor |
| chr2_-_170430366 | 4.16 |
ENST00000453153.2
ENST00000445210.1 |
FASTKD1
|
FAST kinase domains 1 |
| chr14_+_102276209 | 4.16 |
ENST00000445439.3
ENST00000334743.5 ENST00000557095.1 |
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr4_-_90757364 | 4.13 |
ENST00000508895.1
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr8_+_31497271 | 4.05 |
ENST00000520407.1
|
NRG1
|
neuregulin 1 |
| chr1_-_3566590 | 3.99 |
ENST00000424367.1
ENST00000378322.3 |
WRAP73
|
WD repeat containing, antisense to TP73 |
| chr2_+_65283506 | 3.97 |
ENST00000377990.2
|
CEP68
|
centrosomal protein 68kDa |
| chr3_+_101292939 | 3.94 |
ENST00000265260.3
ENST00000469941.1 ENST00000296024.5 |
PCNP
|
PEST proteolytic signal containing nuclear protein |
| chr3_+_157828152 | 3.93 |
ENST00000476899.1
|
RSRC1
|
arginine/serine-rich coiled-coil 1 |
| chr5_-_88119580 | 3.92 |
ENST00000539796.1
|
MEF2C
|
myocyte enhancer factor 2C |
| chr4_+_57845024 | 3.90 |
ENST00000431623.2
ENST00000441246.2 |
POLR2B
|
polymerase (RNA) II (DNA directed) polypeptide B, 140kDa |
| chr1_+_74701062 | 3.83 |
ENST00000326637.3
|
TNNI3K
|
TNNI3 interacting kinase |
| chr1_+_207943667 | 3.81 |
ENST00000462968.2
|
CD46
|
CD46 molecule, complement regulatory protein |
| chr1_-_28559502 | 3.76 |
ENST00000263697.4
|
DNAJC8
|
DnaJ (Hsp40) homolog, subfamily C, member 8 |
| chr17_-_56350797 | 3.75 |
ENST00000577220.1
|
MPO
|
myeloperoxidase |
| chr2_+_65283529 | 3.73 |
ENST00000546106.1
ENST00000537589.1 ENST00000260569.4 |
CEP68
|
centrosomal protein 68kDa |
| chr1_+_145883868 | 3.71 |
ENST00000447947.2
|
GPR89C
|
G protein-coupled receptor 89C |
| chr15_+_64680003 | 3.67 |
ENST00000261884.3
|
TRIP4
|
thyroid hormone receptor interactor 4 |
| chr19_+_14491948 | 3.64 |
ENST00000358600.3
|
CD97
|
CD97 molecule |
| chr12_+_21525818 | 3.64 |
ENST00000240652.3
ENST00000542023.1 ENST00000537593.1 |
IAPP
|
islet amyloid polypeptide |
| chrX_-_106959631 | 3.59 |
ENST00000486554.1
ENST00000372390.4 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr8_-_101962777 | 3.55 |
ENST00000395951.3
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr4_+_154622652 | 3.53 |
ENST00000260010.6
|
TLR2
|
toll-like receptor 2 |
| chr7_-_115670804 | 3.47 |
ENST00000320239.7
|
TFEC
|
transcription factor EC |
| chr12_+_56435637 | 3.47 |
ENST00000356464.5
ENST00000552361.1 |
RPS26
|
ribosomal protein S26 |
| chr7_-_115670792 | 3.47 |
ENST00000265440.7
ENST00000393485.1 |
TFEC
|
transcription factor EC |
| chr17_+_41561317 | 3.46 |
ENST00000540306.1
ENST00000262415.3 ENST00000605777.1 |
DHX8
|
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
| chr3_-_49066811 | 3.46 |
ENST00000442157.1
ENST00000326739.4 |
IMPDH2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2 |
| chr13_-_49975632 | 3.45 |
ENST00000457041.1
ENST00000355854.4 |
CAB39L
|
calcium binding protein 39-like |
| chr3_+_98451093 | 3.44 |
ENST00000483910.1
ENST00000460774.1 |
ST3GAL6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr6_-_131949305 | 3.43 |
ENST00000368053.4
ENST00000354577.4 ENST00000403834.3 ENST00000540546.1 ENST00000368068.3 ENST00000368060.3 |
MED23
|
mediator complex subunit 23 |
| chr12_+_53836339 | 3.40 |
ENST00000549135.1
|
PRR13
|
proline rich 13 |
| chr16_+_81812863 | 3.38 |
ENST00000359376.3
|
PLCG2
|
phospholipase C, gamma 2 (phosphatidylinositol-specific) |
| chr2_+_143635067 | 3.37 |
ENST00000264170.4
|
KYNU
|
kynureninase |
| chr19_-_18433910 | 3.36 |
ENST00000594828.3
ENST00000593829.1 |
LSM4
|
LSM4 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr14_+_56078695 | 3.33 |
ENST00000416613.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr17_-_26662440 | 3.29 |
ENST00000578122.1
|
IFT20
|
intraflagellar transport 20 homolog (Chlamydomonas) |
| chr16_-_21663919 | 3.28 |
ENST00000569602.1
|
IGSF6
|
immunoglobulin superfamily, member 6 |
| chr14_-_95624227 | 3.26 |
ENST00000526495.1
|
DICER1
|
dicer 1, ribonuclease type III |
| chr6_+_74171301 | 3.24 |
ENST00000415954.2
ENST00000498286.1 ENST00000370305.1 ENST00000370300.4 |
MTO1
|
mitochondrial tRNA translation optimization 1 |
| chr10_+_111967345 | 3.22 |
ENST00000332674.5
ENST00000453116.1 |
MXI1
|
MAX interactor 1, dimerization protein |
| chr7_-_139876812 | 3.20 |
ENST00000397560.2
|
JHDM1D
|
lysine (K)-specific demethylase 7A |
| chr5_-_150460539 | 3.12 |
ENST00000520931.1
ENST00000520695.1 ENST00000521591.1 ENST00000518977.1 |
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr12_+_32832134 | 3.07 |
ENST00000452533.2
|
DNM1L
|
dynamin 1-like |
| chr4_-_153332886 | 3.04 |
ENST00000603841.1
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr5_+_156607829 | 3.01 |
ENST00000422843.3
|
ITK
|
IL2-inducible T-cell kinase |
| chr14_-_102605983 | 3.00 |
ENST00000334701.7
|
HSP90AA1
|
heat shock protein 90kDa alpha (cytosolic), class A member 1 |
| chr7_+_77469439 | 3.00 |
ENST00000450574.1
ENST00000416283.2 ENST00000248550.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr7_-_76829125 | 2.99 |
ENST00000248598.5
|
FGL2
|
fibrinogen-like 2 |
| chrX_+_40440146 | 2.96 |
ENST00000535539.1
ENST00000378438.4 ENST00000436783.1 ENST00000544975.1 ENST00000535777.1 ENST00000447485.1 ENST00000423649.1 |
ATP6AP2
|
ATPase, H+ transporting, lysosomal accessory protein 2 |
| chr21_+_34602377 | 2.96 |
ENST00000342101.3
ENST00000413881.1 ENST00000443073.1 |
IFNAR2
|
interferon (alpha, beta and omega) receptor 2 |
| chr8_+_26247878 | 2.95 |
ENST00000518611.1
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr13_+_41885341 | 2.95 |
ENST00000379406.3
ENST00000379367.3 ENST00000403412.3 |
NAA16
|
N(alpha)-acetyltransferase 16, NatA auxiliary subunit |
| chr1_-_1342617 | 2.95 |
ENST00000482352.1
ENST00000344843.7 |
MRPL20
|
mitochondrial ribosomal protein L20 |
| chr16_+_56969284 | 2.94 |
ENST00000568358.1
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr6_-_42185583 | 2.93 |
ENST00000053468.3
|
MRPS10
|
mitochondrial ribosomal protein S10 |
| chr5_+_96212185 | 2.88 |
ENST00000379904.4
|
ERAP2
|
endoplasmic reticulum aminopeptidase 2 |
| chr11_-_9286921 | 2.88 |
ENST00000328194.3
|
DENND5A
|
DENN/MADD domain containing 5A |
| chr7_+_80275953 | 2.84 |
ENST00000538969.1
ENST00000544133.1 ENST00000433696.2 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr3_-_15382875 | 2.79 |
ENST00000408919.3
|
SH3BP5
|
SH3-domain binding protein 5 (BTK-associated) |
| chr3_-_150920979 | 2.77 |
ENST00000309180.5
ENST00000480322.1 |
GPR171
|
G protein-coupled receptor 171 |
| chr11_+_67374323 | 2.77 |
ENST00000322776.6
ENST00000532303.1 ENST00000532244.1 ENST00000528328.1 ENST00000529927.1 ENST00000532343.1 ENST00000415352.2 ENST00000533075.1 ENST00000529867.1 ENST00000530638.1 |
NDUFV1
|
NADH dehydrogenase (ubiquinone) flavoprotein 1, 51kDa |
| chr20_+_48552908 | 2.77 |
ENST00000244061.2
|
RNF114
|
ring finger protein 114 |
| chr2_-_27886460 | 2.75 |
ENST00000404798.2
ENST00000405491.1 ENST00000464789.2 ENST00000406540.1 |
SUPT7L
|
suppressor of Ty 7 (S. cerevisiae)-like |
| chr18_+_21033239 | 2.74 |
ENST00000581585.1
ENST00000577501.1 |
RIOK3
|
RIO kinase 3 |
| chr1_+_161123536 | 2.73 |
ENST00000368003.5
|
UFC1
|
ubiquitin-fold modifier conjugating enzyme 1 |
| chrX_+_77154935 | 2.67 |
ENST00000481445.1
|
COX7B
|
cytochrome c oxidase subunit VIIb |
| chr1_+_52082751 | 2.63 |
ENST00000447887.1
ENST00000435686.2 ENST00000428468.1 ENST00000453295.1 |
OSBPL9
|
oxysterol binding protein-like 9 |
| chr17_-_29641084 | 2.63 |
ENST00000544462.1
|
EVI2B
|
ecotropic viral integration site 2B |
| chr1_-_169555779 | 2.61 |
ENST00000367797.3
ENST00000367796.3 |
F5
|
coagulation factor V (proaccelerin, labile factor) |
| chr7_+_149570049 | 2.59 |
ENST00000421974.2
ENST00000456496.2 |
ATP6V0E2
|
ATPase, H+ transporting V0 subunit e2 |
| chr15_+_58430368 | 2.59 |
ENST00000558772.1
ENST00000219919.4 |
AQP9
|
aquaporin 9 |
| chr4_+_79567314 | 2.56 |
ENST00000503539.1
ENST00000504675.1 |
RP11-792D21.2
|
long intergenic non-protein coding RNA 1094 |
| chr3_-_105588231 | 2.53 |
ENST00000545639.1
ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr16_+_28858004 | 2.53 |
ENST00000322610.8
|
SH2B1
|
SH2B adaptor protein 1 |
| chr14_+_102276132 | 2.51 |
ENST00000350249.3
ENST00000557621.1 ENST00000556946.1 |
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr10_-_99094458 | 2.50 |
ENST00000371019.2
|
FRAT2
|
frequently rearranged in advanced T-cell lymphomas 2 |
| chr14_+_57046530 | 2.49 |
ENST00000536419.1
ENST00000538838.1 |
TMEM260
|
transmembrane protein 260 |
| chr19_+_36630454 | 2.48 |
ENST00000246533.3
|
CAPNS1
|
calpain, small subunit 1 |
| chr11_-_47447767 | 2.47 |
ENST00000530651.1
ENST00000524447.2 ENST00000531051.2 ENST00000526993.1 ENST00000602866.1 |
PSMC3
|
proteasome (prosome, macropain) 26S subunit, ATPase, 3 |
| chr21_-_33975547 | 2.41 |
ENST00000431599.1
|
C21orf59
|
chromosome 21 open reading frame 59 |
| chr2_+_109204743 | 2.39 |
ENST00000332345.6
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr20_+_36322408 | 2.38 |
ENST00000361383.6
ENST00000447935.1 ENST00000405275.2 |
CTNNBL1
|
catenin, beta like 1 |
| chr6_-_131949200 | 2.37 |
ENST00000539158.1
ENST00000368058.1 |
MED23
|
mediator complex subunit 23 |
| chr15_+_58430567 | 2.36 |
ENST00000536493.1
|
AQP9
|
aquaporin 9 |
| chr1_+_87797351 | 2.36 |
ENST00000370542.1
|
LMO4
|
LIM domain only 4 |
| chr7_+_80275752 | 2.35 |
ENST00000419819.2
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr15_+_42697018 | 2.35 |
ENST00000397204.4
|
CAPN3
|
calpain 3, (p94) |
| chr7_+_80275621 | 2.35 |
ENST00000426978.1
ENST00000432207.1 |
CD36
|
CD36 molecule (thrombospondin receptor) |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXA2_FOXJ3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.3 | 25.0 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 4.6 | 32.3 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 3.1 | 15.5 | GO:0048539 | immunoglobulin biosynthetic process(GO:0002378) bone marrow development(GO:0048539) |
| 3.0 | 11.9 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 2.5 | 10.0 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 2.3 | 11.5 | GO:0006696 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 1.9 | 9.5 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 1.9 | 18.9 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 1.9 | 5.6 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 1.8 | 7.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 1.7 | 22.3 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 1.7 | 5.1 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) mitochondrial membrane fission(GO:0090149) |
| 1.7 | 8.4 | GO:0051585 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) regulation of peroxidase activity(GO:2000468) positive regulation of peroxidase activity(GO:2000470) |
| 1.5 | 4.6 | GO:0097052 | tryptophan catabolic process to acetyl-CoA(GO:0019442) L-kynurenine metabolic process(GO:0097052) |
| 1.5 | 4.6 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 1.5 | 9.0 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 1.5 | 10.2 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 1.4 | 13.0 | GO:0071724 | toll-like receptor TLR6:TLR2 signaling pathway(GO:0038124) response to diacyl bacterial lipopeptide(GO:0071724) cellular response to diacyl bacterial lipopeptide(GO:0071726) |
| 1.4 | 15.6 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 1.4 | 4.2 | GO:2000909 | regulation of cholesterol import(GO:0060620) negative regulation of cholesterol import(GO:0060621) regulation of sterol import(GO:2000909) negative regulation of sterol import(GO:2000910) |
| 1.4 | 8.3 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 1.3 | 8.0 | GO:0044211 | CTP salvage(GO:0044211) |
| 1.3 | 7.7 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 1.2 | 4.8 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 1.2 | 1.2 | GO:1902227 | negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) |
| 1.1 | 10.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 1.1 | 3.4 | GO:0002316 | follicular B cell differentiation(GO:0002316) activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 1.1 | 6.7 | GO:0030885 | regulation of myeloid dendritic cell activation(GO:0030885) positive regulation of interferon-gamma secretion(GO:1902715) |
| 1.1 | 3.3 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 1.1 | 4.3 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 1.1 | 14.0 | GO:0051451 | myoblast migration(GO:0051451) |
| 1.0 | 5.2 | GO:0097338 | response to clozapine(GO:0097338) |
| 1.0 | 3.1 | GO:0044501 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 1.0 | 10.1 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 1.0 | 4.9 | GO:0015722 | canalicular bile acid transport(GO:0015722) pyrimidine nucleobase transport(GO:0015855) urea transmembrane transport(GO:0071918) purine nucleobase transmembrane transport(GO:1904823) |
| 1.0 | 3.9 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.9 | 7.4 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.9 | 3.5 | GO:0072308 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of nephron tubule epithelial cell differentiation(GO:0072183) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) negative regulation of epithelial cell differentiation involved in kidney development(GO:2000697) |
| 0.9 | 14.7 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.8 | 18.6 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.8 | 5.9 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.8 | 2.3 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.8 | 2.3 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.7 | 3.0 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.7 | 7.1 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.7 | 2.1 | GO:1903676 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.7 | 2.1 | GO:0051685 | maintenance of ER location(GO:0051685) |
| 0.7 | 2.0 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.6 | 11.2 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.6 | 4.9 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.6 | 9.9 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.6 | 3.0 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.6 | 9.5 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.6 | 1.8 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.6 | 4.1 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.6 | 3.5 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.6 | 2.3 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.5 | 6.4 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.5 | 1.6 | GO:0021764 | amygdala development(GO:0021764) |
| 0.5 | 2.6 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.5 | 3.0 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.5 | 3.0 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.5 | 1.9 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.5 | 4.7 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.5 | 3.8 | GO:0001878 | response to yeast(GO:0001878) |
| 0.5 | 16.8 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.4 | 3.6 | GO:0070235 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.4 | 8.3 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.4 | 8.7 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.4 | 5.2 | GO:0042407 | cristae formation(GO:0042407) |
| 0.4 | 5.6 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.4 | 7.7 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.4 | 20.2 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.4 | 3.6 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.4 | 20.7 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.4 | 2.7 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.4 | 8.5 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.4 | 3.8 | GO:0043382 | positive regulation of memory T cell differentiation(GO:0043382) |
| 0.4 | 1.9 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.4 | 3.0 | GO:0048069 | positive regulation of transforming growth factor beta1 production(GO:0032914) eye pigmentation(GO:0048069) |
| 0.4 | 10.9 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.4 | 3.2 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.4 | 3.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.3 | 3.0 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.3 | 13.6 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.3 | 1.9 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.3 | 9.6 | GO:0042994 | cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.3 | 2.3 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.3 | 13.9 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.3 | 8.0 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.3 | 3.7 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.3 | 4.9 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.3 | 2.5 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.2 | 1.7 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.2 | 2.0 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.2 | 6.0 | GO:0006068 | ethanol catabolic process(GO:0006068) sulfation(GO:0051923) |
| 0.2 | 3.5 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.2 | 1.6 | GO:0002860 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.2 | 2.8 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.2 | 8.3 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.2 | 4.7 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.2 | 5.4 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.2 | 2.9 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.2 | 5.7 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.2 | 5.7 | GO:0097503 | sialylation(GO:0097503) |
| 0.2 | 2.4 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.2 | 2.9 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.2 | 7.1 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.2 | 2.3 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.2 | 0.6 | GO:0019557 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.2 | 1.4 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.2 | 4.5 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.2 | 4.8 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.2 | 1.0 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.2 | 10.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.2 | 7.6 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.2 | 7.9 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.2 | 4.6 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
| 0.1 | 1.0 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.1 | 12.2 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 0.9 | GO:0030805 | regulation of cyclic nucleotide catabolic process(GO:0030805) regulation of cAMP catabolic process(GO:0030820) regulation of purine nucleotide catabolic process(GO:0033121) regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.1 | 2.5 | GO:1904886 | beta-catenin destruction complex disassembly(GO:1904886) |
| 0.1 | 0.8 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.1 | 1.9 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.1 | 0.7 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.1 | 2.8 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.1 | 8.2 | GO:0016445 | somatic diversification of immunoglobulins(GO:0016445) |
| 0.1 | 9.6 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.1 | 3.5 | GO:0035456 | response to interferon-beta(GO:0035456) |
| 0.1 | 1.4 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.1 | 14.1 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 4.2 | GO:0050434 | positive regulation of viral transcription(GO:0050434) |
| 0.1 | 2.6 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.1 | 2.9 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 0.4 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.1 | 8.9 | GO:0045333 | cellular respiration(GO:0045333) |
| 0.1 | 1.3 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.1 | 1.7 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.1 | 6.0 | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process(GO:0043161) |
| 0.1 | 2.9 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
| 0.1 | 1.2 | GO:0034035 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.1 | 7.1 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.1 | 5.3 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.1 | 3.3 | GO:0045661 | regulation of myoblast differentiation(GO:0045661) |
| 0.1 | 1.0 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.1 | 1.5 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.1 | 3.2 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.1 | 1.7 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.1 | 8.1 | GO:0002504 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.1 | 0.3 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.1 | 6.9 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.2 | GO:0036114 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.0 | 2.6 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.5 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.0 | 2.5 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 1.3 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.2 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 2.0 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 4.5 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 1.6 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 1.8 | GO:0071349 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.0 | 0.3 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 1.3 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.0 | 0.7 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.4 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.0 | 2.3 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.0 | 2.8 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.3 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 3.0 | GO:1990823 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.2 | GO:0008206 | bile acid metabolic process(GO:0008206) |
| 0.0 | 1.8 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.4 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.7 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 5.6 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 0.0 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 0.0 | 0.3 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 3.1 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 4.6 | GO:0010976 | positive regulation of neuron projection development(GO:0010976) |
| 0.0 | 0.2 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.1 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 1.0 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 1.6 | GO:0002433 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.0 | 1.9 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 2.1 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 14.0 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 2.6 | 10.2 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 2.4 | 7.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 1.6 | 7.9 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 1.6 | 4.7 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 1.5 | 11.9 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 1.4 | 10.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 1.4 | 8.5 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 1.4 | 16.8 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 1.3 | 3.8 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 1.2 | 3.5 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 1.1 | 19.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 1.0 | 20.9 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.9 | 9.4 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.8 | 13.6 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.7 | 5.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.6 | 1.8 | GO:0044393 | microspike(GO:0044393) |
| 0.6 | 17.9 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.6 | 2.3 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.5 | 4.9 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.5 | 4.2 | GO:0034715 | U7 snRNP(GO:0005683) pICln-Sm protein complex(GO:0034715) |
| 0.5 | 2.9 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.4 | 1.3 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.4 | 3.0 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.4 | 9.0 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.4 | 3.0 | GO:0031415 | NatA complex(GO:0031415) |
| 0.4 | 5.7 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.4 | 5.6 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.4 | 4.7 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.4 | 7.8 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.4 | 1.9 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.4 | 1.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.4 | 4.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.3 | 1.7 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.3 | 2.3 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.3 | 1.7 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.3 | 5.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.2 | 4.2 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.2 | 5.2 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.2 | 3.3 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.2 | 12.0 | GO:0031430 | M band(GO:0031430) |
| 0.2 | 3.0 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 2.0 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.2 | 3.6 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.2 | 5.0 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.2 | 16.3 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.2 | 2.6 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.2 | 2.7 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.2 | 1.9 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.2 | 10.1 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.2 | 10.3 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.2 | 0.7 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.2 | 5.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.2 | 4.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.2 | 4.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 10.0 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 0.9 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 1.8 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.1 | 5.9 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 41.7 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 2.9 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 3.0 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.1 | 2.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 2.0 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 2.9 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 15.9 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 16.2 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 4.5 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 5.3 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 3.8 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 1.6 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 9.2 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 5.9 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 13.3 | GO:1904813 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.1 | 2.9 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 0.4 | GO:0032982 | myosin filament(GO:0032982) |
| 0.1 | 1.0 | GO:0098791 | Golgi subcompartment(GO:0098791) |
| 0.1 | 2.1 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.1 | 0.3 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 5.4 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 9.6 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 7.0 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.1 | 3.3 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 11.3 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 23.7 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.1 | 2.1 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 1.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 1.8 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.1 | 3.5 | GO:0015935 | small ribosomal subunit(GO:0015935) cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 3.9 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 14.2 | GO:0005769 | early endosome(GO:0005769) |
| 0.1 | 5.4 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 3.7 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 8.6 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.4 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 3.7 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 8.8 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 6.8 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 1.0 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.2 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 5.2 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 1.5 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 1.0 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 1.6 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 1.1 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 10.3 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 2.7 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 7.6 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 2.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 5.3 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 0.3 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.9 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.5 | GO:0032991 | macromolecular complex(GO:0032991) |
| 0.0 | 0.4 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 9.6 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.5 | GO:0005905 | clathrin-coated pit(GO:0005905) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.8 | 32.3 | GO:0004913 | interleukin-4 receptor activity(GO:0004913) interleukin-7 receptor activity(GO:0004917) |
| 4.7 | 18.9 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 3.9 | 11.7 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 2.9 | 11.5 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 1.9 | 5.6 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 1.8 | 14.7 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 1.7 | 8.4 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 1.6 | 9.5 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 1.4 | 4.3 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 1.4 | 5.7 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 1.3 | 10.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 1.2 | 4.9 | GO:0015254 | glycerol channel activity(GO:0015254) urea channel activity(GO:0015265) |
| 1.2 | 4.9 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 1.2 | 4.6 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 1.1 | 8.0 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 1.1 | 4.2 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 1.0 | 4.2 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 1.0 | 11.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 1.0 | 5.0 | GO:0004905 | type I interferon receptor activity(GO:0004905) |
| 1.0 | 11.9 | GO:0089720 | caspase binding(GO:0089720) |
| 0.9 | 4.7 | GO:0002046 | opsin binding(GO:0002046) |
| 0.9 | 10.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.9 | 20.3 | GO:0055103 | ligase regulator activity(GO:0055103) |
| 0.9 | 4.4 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.9 | 3.5 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.8 | 3.3 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.8 | 2.3 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.7 | 5.9 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.7 | 4.9 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.7 | 2.0 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.6 | 4.9 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.6 | 4.8 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.5 | 6.4 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.5 | 1.5 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.5 | 13.6 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.5 | 5.2 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.5 | 10.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.4 | 1.7 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.4 | 4.7 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.4 | 2.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.4 | 1.7 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.4 | 4.2 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.4 | 18.6 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.4 | 4.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.4 | 3.6 | GO:0043426 | MRF binding(GO:0043426) |
| 0.4 | 1.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.4 | 9.0 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.4 | 8.9 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.3 | 3.8 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.3 | 5.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.3 | 3.5 | GO:0035325 | Toll-like receptor binding(GO:0035325) |
| 0.3 | 1.2 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.3 | 3.0 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.3 | 7.9 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.3 | 7.9 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.3 | 1.9 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.3 | 7.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.3 | 3.4 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.3 | 1.0 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.2 | 10.0 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.2 | 3.9 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.2 | 6.9 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.2 | 2.3 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.2 | 1.6 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.2 | 3.0 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.2 | 10.3 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.2 | 6.0 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.2 | 2.3 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.2 | 1.2 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.2 | 3.9 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.2 | 1.4 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.2 | 3.6 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.2 | 4.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.2 | 1.1 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.2 | 6.6 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.2 | 1.9 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.2 | 8.8 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.2 | 0.6 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.1 | 2.8 | GO:0016502 | G-protein coupled nucleotide receptor activity(GO:0001608) purinergic nucleotide receptor activity(GO:0001614) nucleotide receptor activity(GO:0016502) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.4 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.1 | 5.9 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.1 | 22.7 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 1.2 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 5.5 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 3.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 2.9 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.7 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.1 | 2.0 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 3.7 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 2.6 | GO:0044769 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) |
| 0.1 | 5.5 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.1 | 3.0 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.2 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 1.8 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 2.7 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 10.2 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 0.3 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.1 | 7.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 26.6 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 4.9 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 2.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 3.1 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 4.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 2.1 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 13.1 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 8.8 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 2.5 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 9.1 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.1 | 3.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 5.7 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.1 | 0.9 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 0.3 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.1 | 1.7 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.1 | 0.3 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 0.9 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 1.8 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 3.8 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.1 | 3.6 | GO:0070035 | ATP-dependent RNA helicase activity(GO:0004004) ATP-dependent helicase activity(GO:0008026) RNA-dependent ATPase activity(GO:0008186) purine NTP-dependent helicase activity(GO:0070035) |
| 0.1 | 3.2 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.1 | 3.6 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 4.8 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 2.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 1.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.3 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.1 | 1.1 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 2.9 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 28.5 | GO:0019900 | kinase binding(GO:0019900) |
| 0.1 | 1.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 6.5 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 2.5 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.5 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 7.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 2.4 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 0.3 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 2.3 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.9 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 0.8 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 1.7 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 1.3 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 1.6 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 4.8 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.1 | GO:0017076 | purine nucleotide binding(GO:0017076) |
| 0.0 | 0.4 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 5.9 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 7.4 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 0.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 4.3 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 2.0 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 5.0 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 4.4 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 30.5 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.4 | 17.8 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.3 | 19.2 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.3 | 3.5 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.3 | 23.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.3 | 9.7 | PID MYC PATHWAY | C-MYC pathway |
| 0.2 | 3.4 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.2 | 6.3 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.2 | 8.1 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.2 | 7.0 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.2 | 7.3 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.2 | 9.0 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.2 | 19.5 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.2 | 8.4 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.2 | 4.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.2 | 4.1 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 5.4 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 3.9 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 4.5 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.1 | 1.7 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 3.0 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 5.1 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 6.4 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.1 | 9.1 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 2.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 2.8 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 1.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 2.0 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 6.9 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 1.8 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 1.4 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 2.5 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 3.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 4.2 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 3.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.7 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.2 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 1.0 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 32.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 1.0 | 11.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.9 | 18.9 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.7 | 20.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.7 | 5.9 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.6 | 9.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.6 | 15.5 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.5 | 6.0 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.4 | 17.8 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.4 | 10.1 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.4 | 11.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.4 | 52.4 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.4 | 4.9 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.4 | 3.5 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.3 | 4.9 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.3 | 4.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.3 | 8.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.3 | 2.0 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.3 | 4.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.3 | 7.5 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.3 | 7.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.2 | 16.9 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.2 | 3.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.2 | 3.0 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.2 | 8.0 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.2 | 4.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.2 | 3.5 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.2 | 5.7 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.2 | 3.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.2 | 17.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.2 | 4.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 12.8 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.2 | 4.5 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.2 | 3.9 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.2 | 2.5 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.2 | 5.2 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.2 | 6.1 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.2 | 9.4 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.2 | 13.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 2.9 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.1 | 3.5 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 4.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 1.9 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 7.1 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 4.9 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 9.0 | REACTOME APOPTOTIC EXECUTION PHASE | Genes involved in Apoptotic execution phase |
| 0.1 | 3.0 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 1.3 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 3.1 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.1 | 1.8 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 1.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 3.2 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 4.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 2.0 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.1 | 5.3 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 4.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 1.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 2.0 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.9 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 13.5 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 8.5 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 3.2 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 3.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 4.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.6 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 2.3 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 2.8 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 1.3 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 2.2 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.2 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |