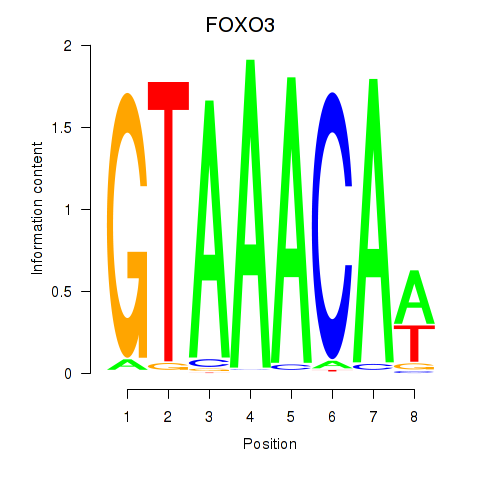
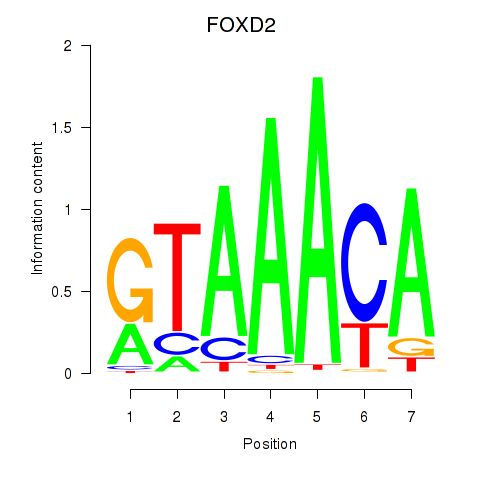
Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for FOXO3_FOXD2
Z-value: 0.95
Transcription factors associated with FOXO3_FOXD2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXO3
|
ENSG00000118689.10 | forkhead box O3 |
|
FOXD2
|
ENSG00000186564.5 | forkhead box D2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXD2 | hg19_v2_chr1_+_47901689_47901689 | -0.08 | 2.2e-01 | Click! |
| FOXO3 | hg19_v2_chr6_+_108882069_108882087, hg19_v2_chr6_+_108881012_108881038 | 0.01 | 8.5e-01 | Click! |
Activity profile of FOXO3_FOXD2 motif
Sorted Z-values of FOXO3_FOXD2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_92539614 | 34.87 |
ENST00000256015.3
|
BTG1
|
B-cell translocation gene 1, anti-proliferative |
| chr6_-_42016385 | 26.43 |
ENST00000502771.1
ENST00000508143.1 ENST00000514588.1 ENST00000510503.1 ENST00000415497.2 ENST00000372988.4 |
CCND3
|
cyclin D3 |
| chr7_-_99698338 | 23.75 |
ENST00000354230.3
ENST00000425308.1 |
MCM7
|
minichromosome maintenance complex component 7 |
| chr13_-_46716969 | 23.13 |
ENST00000435666.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr2_-_158345462 | 20.80 |
ENST00000439355.1
ENST00000540637.1 |
CYTIP
|
cytohesin 1 interacting protein |
| chr2_-_136875712 | 20.49 |
ENST00000241393.3
|
CXCR4
|
chemokine (C-X-C motif) receptor 4 |
| chr12_-_71551652 | 20.48 |
ENST00000546561.1
|
TSPAN8
|
tetraspanin 8 |
| chr5_+_169064245 | 17.46 |
ENST00000256935.8
|
DOCK2
|
dedicator of cytokinesis 2 |
| chr12_-_31479045 | 17.20 |
ENST00000539409.1
ENST00000395766.1 |
FAM60A
|
family with sequence similarity 60, member A |
| chr8_+_31497271 | 16.66 |
ENST00000520407.1
|
NRG1
|
neuregulin 1 |
| chr2_-_152146385 | 16.61 |
ENST00000414946.1
ENST00000243346.5 |
NMI
|
N-myc (and STAT) interactor |
| chr7_+_106809406 | 16.13 |
ENST00000468410.1
ENST00000478930.1 ENST00000464009.1 ENST00000222574.4 |
HBP1
|
HMG-box transcription factor 1 |
| chr13_-_46756351 | 16.01 |
ENST00000323076.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr14_-_23288930 | 14.56 |
ENST00000554517.1
ENST00000285850.7 ENST00000397529.2 ENST00000555702.1 |
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr3_-_49066811 | 13.87 |
ENST00000442157.1
ENST00000326739.4 |
IMPDH2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2 |
| chr4_-_174256276 | 13.81 |
ENST00000296503.5
|
HMGB2
|
high mobility group box 2 |
| chr5_-_98262240 | 13.59 |
ENST00000284049.3
|
CHD1
|
chromodomain helicase DNA binding protein 1 |
| chr10_+_99079008 | 12.78 |
ENST00000371021.3
|
FRAT1
|
frequently rearranged in advanced T-cell lymphomas |
| chr10_-_43904608 | 12.72 |
ENST00000337970.3
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F |
| chr7_+_129906660 | 12.40 |
ENST00000222481.4
|
CPA2
|
carboxypeptidase A2 (pancreatic) |
| chr10_-_43904235 | 12.13 |
ENST00000356053.3
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F |
| chr6_+_135502466 | 11.96 |
ENST00000367814.4
|
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr6_+_135502408 | 11.75 |
ENST00000341911.5
ENST00000442647.2 ENST00000316528.8 |
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr17_-_29641104 | 11.57 |
ENST00000577894.1
ENST00000330927.4 |
EVI2B
|
ecotropic viral integration site 2B |
| chr12_-_772901 | 10.49 |
ENST00000305108.4
|
NINJ2
|
ninjurin 2 |
| chr11_+_10476851 | 10.40 |
ENST00000396553.2
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chrX_+_9431324 | 10.30 |
ENST00000407597.2
ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X
|
transducin (beta)-like 1X-linked |
| chr3_-_141868293 | 10.15 |
ENST00000317104.7
ENST00000494358.1 |
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr6_-_31550192 | 10.10 |
ENST00000429299.2
ENST00000446745.2 |
LTB
|
lymphotoxin beta (TNF superfamily, member 3) |
| chr10_-_99094458 | 10.06 |
ENST00000371019.2
|
FRAT2
|
frequently rearranged in advanced T-cell lymphomas 2 |
| chr19_+_12902289 | 9.90 |
ENST00000302754.4
|
JUNB
|
jun B proto-oncogene |
| chr13_-_31038370 | 9.80 |
ENST00000399489.1
ENST00000339872.4 |
HMGB1
|
high mobility group box 1 |
| chr12_-_46384334 | 9.72 |
ENST00000369367.3
ENST00000266589.6 ENST00000395453.2 ENST00000395454.2 |
SCAF11
|
SR-related CTD-associated factor 11 |
| chr6_+_89791507 | 9.66 |
ENST00000354922.3
|
PNRC1
|
proline-rich nuclear receptor coactivator 1 |
| chr5_-_137674000 | 9.64 |
ENST00000510119.1
ENST00000513970.1 |
CDC25C
|
cell division cycle 25C |
| chr1_+_117297007 | 9.21 |
ENST00000369478.3
ENST00000369477.1 |
CD2
|
CD2 molecule |
| chr19_-_39826639 | 9.06 |
ENST00000602185.1
ENST00000598034.1 ENST00000601387.1 ENST00000595636.1 ENST00000253054.8 ENST00000594700.1 ENST00000597595.1 |
GMFG
|
glia maturation factor, gamma |
| chr17_+_65373531 | 8.90 |
ENST00000580974.1
|
PITPNC1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr2_+_97481974 | 8.88 |
ENST00000377060.3
ENST00000305510.3 |
CNNM3
|
cyclin M3 |
| chr3_-_107777208 | 8.79 |
ENST00000398258.3
|
CD47
|
CD47 molecule |
| chr12_-_71551868 | 8.78 |
ENST00000247829.3
|
TSPAN8
|
tetraspanin 8 |
| chr17_+_65374075 | 8.63 |
ENST00000581322.1
|
PITPNC1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr16_+_69958887 | 8.57 |
ENST00000568684.1
|
WWP2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr1_+_145438469 | 8.51 |
ENST00000369317.4
|
TXNIP
|
thioredoxin interacting protein |
| chr5_-_131132658 | 8.46 |
ENST00000514667.1
ENST00000511848.1 ENST00000510461.1 |
CTC-432M15.3
FNIP1
|
Folliculin-interacting protein 1 folliculin interacting protein 1 |
| chr8_-_8318847 | 8.45 |
ENST00000521218.1
|
CTA-398F10.2
|
CTA-398F10.2 |
| chr6_+_135502501 | 8.40 |
ENST00000527615.1
ENST00000420123.2 ENST00000525369.1 ENST00000528774.1 ENST00000534121.1 ENST00000534044.1 ENST00000533624.1 |
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr13_-_41240717 | 8.39 |
ENST00000379561.5
|
FOXO1
|
forkhead box O1 |
| chr12_+_25205666 | 8.39 |
ENST00000547044.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr11_+_117049910 | 8.36 |
ENST00000431081.2
ENST00000524842.1 |
SIDT2
|
SID1 transmembrane family, member 2 |
| chr5_+_95998746 | 8.27 |
ENST00000508608.1
|
CAST
|
calpastatin |
| chr11_+_71938925 | 8.16 |
ENST00000538751.1
|
INPPL1
|
inositol polyphosphate phosphatase-like 1 |
| chr2_+_58655461 | 8.09 |
ENST00000429095.1
ENST00000429664.1 ENST00000452840.1 |
AC007092.1
|
long intergenic non-protein coding RNA 1122 |
| chr8_+_128748466 | 7.98 |
ENST00000524013.1
ENST00000520751.1 |
MYC
|
v-myc avian myelocytomatosis viral oncogene homolog |
| chr8_+_128748308 | 7.96 |
ENST00000377970.2
|
MYC
|
v-myc avian myelocytomatosis viral oncogene homolog |
| chr17_-_61777090 | 7.90 |
ENST00000578061.1
|
LIMD2
|
LIM domain containing 2 |
| chr17_-_29641084 | 7.87 |
ENST00000544462.1
|
EVI2B
|
ecotropic viral integration site 2B |
| chr17_-_38721711 | 7.86 |
ENST00000578085.1
ENST00000246657.2 |
CCR7
|
chemokine (C-C motif) receptor 7 |
| chr7_-_115670792 | 7.78 |
ENST00000265440.7
ENST00000393485.1 |
TFEC
|
transcription factor EC |
| chr1_+_150122034 | 7.66 |
ENST00000025469.6
ENST00000369124.4 |
PLEKHO1
|
pleckstrin homology domain containing, family O member 1 |
| chr8_+_26150628 | 7.62 |
ENST00000523925.1
ENST00000315985.7 |
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha |
| chr2_+_109223595 | 7.61 |
ENST00000410093.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr7_-_115608304 | 7.59 |
ENST00000457268.1
|
TFEC
|
transcription factor EC |
| chr4_-_105416039 | 7.53 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr2_+_27435179 | 7.50 |
ENST00000606999.1
ENST00000405489.3 |
ATRAID
|
all-trans retinoic acid-induced differentiation factor |
| chr11_-_72504637 | 7.30 |
ENST00000536377.1
ENST00000359373.5 |
STARD10
ARAP1
|
StAR-related lipid transfer (START) domain containing 10 ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr8_-_28243934 | 7.29 |
ENST00000521185.1
ENST00000520290.1 ENST00000344423.5 |
ZNF395
|
zinc finger protein 395 |
| chr15_-_40600026 | 7.27 |
ENST00000456256.2
ENST00000557821.1 |
PLCB2
|
phospholipase C, beta 2 |
| chr8_+_126442563 | 7.20 |
ENST00000311922.3
|
TRIB1
|
tribbles pseudokinase 1 |
| chr6_-_89927151 | 7.09 |
ENST00000454853.2
|
GABRR1
|
gamma-aminobutyric acid (GABA) A receptor, rho 1 |
| chr16_+_84801852 | 6.98 |
ENST00000569925.1
ENST00000567526.1 |
USP10
|
ubiquitin specific peptidase 10 |
| chr2_-_165424973 | 6.91 |
ENST00000543549.1
|
GRB14
|
growth factor receptor-bound protein 14 |
| chr18_+_57567180 | 6.83 |
ENST00000316660.6
ENST00000269518.9 |
PMAIP1
|
phorbol-12-myristate-13-acetate-induced protein 1 |
| chr19_+_13134772 | 6.78 |
ENST00000587760.1
ENST00000585575.1 |
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr5_+_176853702 | 6.75 |
ENST00000507633.1
ENST00000393576.3 ENST00000355958.5 ENST00000528793.1 ENST00000512684.1 |
GRK6
|
G protein-coupled receptor kinase 6 |
| chr11_-_72070206 | 6.69 |
ENST00000544382.1
|
CLPB
|
ClpB caseinolytic peptidase B homolog (E. coli) |
| chr1_+_104159999 | 6.68 |
ENST00000414303.2
ENST00000423678.1 |
AMY2A
|
amylase, alpha 2A (pancreatic) |
| chr3_+_136649311 | 6.53 |
ENST00000469404.1
ENST00000467911.1 |
NCK1
|
NCK adaptor protein 1 |
| chr7_-_140624499 | 6.50 |
ENST00000288602.6
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B |
| chr5_+_176853669 | 6.49 |
ENST00000355472.5
|
GRK6
|
G protein-coupled receptor kinase 6 |
| chr8_+_11666649 | 6.48 |
ENST00000528643.1
ENST00000525777.1 |
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr10_-_3827417 | 6.43 |
ENST00000497571.1
ENST00000542957.1 |
KLF6
|
Kruppel-like factor 6 |
| chr2_+_162087577 | 6.40 |
ENST00000439442.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr6_+_6588902 | 6.34 |
ENST00000230568.4
|
LY86
|
lymphocyte antigen 86 |
| chr14_-_58893832 | 6.31 |
ENST00000556007.2
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr1_+_111682058 | 6.31 |
ENST00000545121.1
|
CEPT1
|
choline/ethanolamine phosphotransferase 1 |
| chr20_+_36373032 | 6.18 |
ENST00000373473.1
|
CTNNBL1
|
catenin, beta like 1 |
| chr7_-_115670804 | 6.13 |
ENST00000320239.7
|
TFEC
|
transcription factor EC |
| chr1_-_160231451 | 6.01 |
ENST00000495887.1
|
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chr5_-_169725231 | 5.99 |
ENST00000046794.5
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
| chr17_-_36358166 | 5.98 |
ENST00000537432.1
|
TBC1D3
|
TBC1 domain family, member 3 |
| chr14_-_58894223 | 5.88 |
ENST00000555593.1
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr15_-_60884706 | 5.85 |
ENST00000449337.2
|
RORA
|
RAR-related orphan receptor A |
| chr14_+_24584508 | 5.80 |
ENST00000559354.1
ENST00000560459.1 ENST00000559593.1 ENST00000396941.4 ENST00000396936.1 |
DCAF11
|
DDB1 and CUL4 associated factor 11 |
| chr5_-_150460539 | 5.80 |
ENST00000520931.1
ENST00000520695.1 ENST00000521591.1 ENST00000518977.1 |
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr1_-_207095324 | 5.74 |
ENST00000530505.1
ENST00000367091.3 ENST00000442471.2 |
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr2_+_27434860 | 5.67 |
ENST00000380171.3
|
ATRAID
|
all-trans retinoic acid-induced differentiation factor |
| chr18_+_3449695 | 5.62 |
ENST00000343820.5
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chrX_-_15619076 | 5.52 |
ENST00000252519.3
|
ACE2
|
angiotensin I converting enzyme 2 |
| chr3_-_141868357 | 5.52 |
ENST00000489671.1
ENST00000475734.1 ENST00000467072.1 ENST00000499676.2 |
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr12_-_123187890 | 5.47 |
ENST00000328880.5
|
HCAR2
|
hydroxycarboxylic acid receptor 2 |
| chr4_-_103746683 | 5.46 |
ENST00000504211.1
ENST00000508476.1 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr6_+_32812568 | 5.33 |
ENST00000414474.1
|
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr14_+_32798462 | 5.31 |
ENST00000280979.4
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr13_-_99910673 | 5.18 |
ENST00000397473.2
ENST00000397470.2 |
GPR18
|
G protein-coupled receptor 18 |
| chr1_+_158815588 | 5.14 |
ENST00000438394.1
|
MNDA
|
myeloid cell nuclear differentiation antigen |
| chr14_+_56127989 | 5.13 |
ENST00000555573.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr17_-_42345487 | 5.12 |
ENST00000262418.6
|
SLC4A1
|
solute carrier family 4 (anion exchanger), member 1 (Diego blood group) |
| chr10_+_114710211 | 5.11 |
ENST00000349937.2
ENST00000369397.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr12_-_53343602 | 5.09 |
ENST00000546897.1
ENST00000552551.1 |
KRT8
|
keratin 8 |
| chr4_-_123542224 | 5.06 |
ENST00000264497.3
|
IL21
|
interleukin 21 |
| chr9_-_128246769 | 5.02 |
ENST00000444226.1
|
MAPKAP1
|
mitogen-activated protein kinase associated protein 1 |
| chr7_+_77469439 | 5.02 |
ENST00000450574.1
ENST00000416283.2 ENST00000248550.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr4_-_103746924 | 4.98 |
ENST00000505207.1
ENST00000502404.1 ENST00000507845.1 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr10_-_3827371 | 4.98 |
ENST00000469435.1
|
KLF6
|
Kruppel-like factor 6 |
| chr2_+_86947296 | 4.98 |
ENST00000283632.4
|
RMND5A
|
required for meiotic nuclear division 5 homolog A (S. cerevisiae) |
| chr2_+_68592305 | 4.97 |
ENST00000234313.7
|
PLEK
|
pleckstrin |
| chr4_-_165305086 | 4.96 |
ENST00000507270.1
ENST00000514618.1 ENST00000503008.1 |
MARCH1
|
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr5_-_135701164 | 4.96 |
ENST00000355180.3
ENST00000426057.2 ENST00000513104.1 |
TRPC7
|
transient receptor potential cation channel, subfamily C, member 7 |
| chr3_-_168865522 | 4.93 |
ENST00000464456.1
|
MECOM
|
MDS1 and EVI1 complex locus |
| chr2_-_160472952 | 4.93 |
ENST00000541068.2
ENST00000355831.2 ENST00000343439.5 ENST00000392782.1 |
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B |
| chr17_-_65241281 | 4.89 |
ENST00000358691.5
ENST00000580168.1 |
HELZ
|
helicase with zinc finger |
| chr3_-_57233966 | 4.86 |
ENST00000473921.1
ENST00000295934.3 |
HESX1
|
HESX homeobox 1 |
| chr14_-_65569057 | 4.84 |
ENST00000555419.1
ENST00000341653.2 |
MAX
|
MYC associated factor X |
| chrX_-_20236970 | 4.83 |
ENST00000379548.4
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr10_-_14050522 | 4.82 |
ENST00000342409.2
|
FRMD4A
|
FERM domain containing 4A |
| chr19_+_859425 | 4.78 |
ENST00000327726.6
|
CFD
|
complement factor D (adipsin) |
| chr12_-_76462713 | 4.76 |
ENST00000552056.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr1_-_161193349 | 4.75 |
ENST00000469730.2
ENST00000463273.1 ENST00000464492.1 ENST00000367990.3 ENST00000470459.2 ENST00000468465.1 ENST00000463812.1 |
APOA2
|
apolipoprotein A-II |
| chr12_+_21525818 | 4.70 |
ENST00000240652.3
ENST00000542023.1 ENST00000537593.1 |
IAPP
|
islet amyloid polypeptide |
| chr6_+_34204642 | 4.70 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chr16_+_30484021 | 4.67 |
ENST00000358164.5
|
ITGAL
|
integrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) |
| chr10_-_121296045 | 4.57 |
ENST00000392865.1
|
RGS10
|
regulator of G-protein signaling 10 |
| chr15_+_81589254 | 4.55 |
ENST00000394652.2
|
IL16
|
interleukin 16 |
| chr14_+_60715928 | 4.55 |
ENST00000395076.4
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
| chr14_-_65569244 | 4.48 |
ENST00000557277.1
ENST00000556892.1 |
MAX
|
MYC associated factor X |
| chr4_-_116034979 | 4.38 |
ENST00000264363.2
|
NDST4
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 4 |
| chr10_+_114709999 | 4.38 |
ENST00000355995.4
ENST00000545257.1 ENST00000543371.1 ENST00000536810.1 ENST00000355717.4 ENST00000538897.1 ENST00000534894.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr9_+_113431059 | 4.36 |
ENST00000416899.2
|
MUSK
|
muscle, skeletal, receptor tyrosine kinase |
| chr2_-_136678123 | 4.35 |
ENST00000422708.1
|
DARS
|
aspartyl-tRNA synthetase |
| chr3_+_172468472 | 4.34 |
ENST00000232458.5
ENST00000392692.3 |
ECT2
|
epithelial cell transforming sequence 2 oncogene |
| chr18_+_66465302 | 4.34 |
ENST00000360242.5
ENST00000358653.5 |
CCDC102B
|
coiled-coil domain containing 102B |
| chr10_+_104535994 | 4.34 |
ENST00000369889.4
|
WBP1L
|
WW domain binding protein 1-like |
| chr12_-_86650077 | 4.32 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr20_+_62795827 | 4.31 |
ENST00000328439.1
ENST00000536311.1 |
MYT1
|
myelin transcription factor 1 |
| chr14_-_65569186 | 4.31 |
ENST00000555932.1
ENST00000358664.4 ENST00000284165.6 ENST00000358402.4 ENST00000246163.2 ENST00000556979.1 ENST00000555667.1 ENST00000557746.1 ENST00000556443.1 |
MAX
|
MYC associated factor X |
| chr7_+_139528952 | 4.22 |
ENST00000416849.2
ENST00000436047.2 ENST00000414508.2 ENST00000448866.1 |
TBXAS1
|
thromboxane A synthase 1 (platelet) |
| chr8_-_117886955 | 4.18 |
ENST00000297338.2
|
RAD21
|
RAD21 homolog (S. pombe) |
| chr19_+_1205740 | 4.18 |
ENST00000326873.7
|
STK11
|
serine/threonine kinase 11 |
| chr16_+_30483962 | 4.16 |
ENST00000356798.6
|
ITGAL
|
integrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) |
| chr1_+_161136180 | 4.13 |
ENST00000352210.5
ENST00000367999.4 ENST00000544598.1 ENST00000535223.1 ENST00000432542.2 |
PPOX
|
protoporphyrinogen oxidase |
| chr11_-_11374904 | 4.12 |
ENST00000528848.2
|
CSNK2A3
|
casein kinase 2, alpha 3 polypeptide |
| chr20_-_62582475 | 4.10 |
ENST00000369908.5
|
UCKL1
|
uridine-cytidine kinase 1-like 1 |
| chrX_-_122756660 | 4.09 |
ENST00000441692.1
|
THOC2
|
THO complex 2 |
| chr1_-_111743285 | 4.09 |
ENST00000357640.4
|
DENND2D
|
DENN/MADD domain containing 2D |
| chr6_+_108882069 | 4.08 |
ENST00000406360.1
|
FOXO3
|
forkhead box O3 |
| chr8_+_99956662 | 4.07 |
ENST00000523368.1
ENST00000297565.4 ENST00000435298.2 |
OSR2
|
odd-skipped related transciption factor 2 |
| chr14_+_102276209 | 4.06 |
ENST00000445439.3
ENST00000334743.5 ENST00000557095.1 |
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr1_+_26798955 | 3.99 |
ENST00000361427.5
|
HMGN2
|
high mobility group nucleosomal binding domain 2 |
| chr19_-_42916499 | 3.98 |
ENST00000601189.1
ENST00000599211.1 |
LIPE
|
lipase, hormone-sensitive |
| chr12_+_25205568 | 3.98 |
ENST00000548766.1
ENST00000556887.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr1_-_27816641 | 3.96 |
ENST00000430629.2
|
WASF2
|
WAS protein family, member 2 |
| chr6_+_42584847 | 3.94 |
ENST00000372883.3
|
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr1_+_227127981 | 3.94 |
ENST00000366778.1
ENST00000366777.3 ENST00000458507.2 |
ADCK3
|
aarF domain containing kinase 3 |
| chr6_+_108977520 | 3.94 |
ENST00000540898.1
|
FOXO3
|
forkhead box O3 |
| chr19_+_16435625 | 3.92 |
ENST00000248071.5
ENST00000592003.1 |
KLF2
|
Kruppel-like factor 2 |
| chr17_+_57642886 | 3.92 |
ENST00000251241.4
ENST00000451169.2 ENST00000425628.3 ENST00000584385.1 ENST00000580030.1 |
DHX40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
| chr6_-_136847099 | 3.92 |
ENST00000438100.2
|
MAP7
|
microtubule-associated protein 7 |
| chr7_+_80231466 | 3.88 |
ENST00000309881.7
ENST00000534394.1 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr1_+_214161854 | 3.85 |
ENST00000435016.1
|
PROX1
|
prospero homeobox 1 |
| chr3_-_18466026 | 3.85 |
ENST00000417717.2
|
SATB1
|
SATB homeobox 1 |
| chr17_+_41561317 | 3.82 |
ENST00000540306.1
ENST00000262415.3 ENST00000605777.1 |
DHX8
|
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
| chr6_-_55739542 | 3.81 |
ENST00000446683.2
|
BMP5
|
bone morphogenetic protein 5 |
| chr2_+_198380763 | 3.80 |
ENST00000448447.2
ENST00000409360.1 |
MOB4
|
MOB family member 4, phocein |
| chr17_+_4618734 | 3.78 |
ENST00000571206.1
|
ARRB2
|
arrestin, beta 2 |
| chr7_-_76829125 | 3.77 |
ENST00000248598.5
|
FGL2
|
fibrinogen-like 2 |
| chr16_-_68033356 | 3.75 |
ENST00000393847.1
ENST00000573808.1 ENST00000572624.1 |
DPEP2
|
dipeptidase 2 |
| chr14_-_36988882 | 3.72 |
ENST00000498187.2
|
NKX2-1
|
NK2 homeobox 1 |
| chr7_-_16844611 | 3.71 |
ENST00000401412.1
ENST00000419304.2 |
AGR2
|
anterior gradient 2 |
| chr1_+_104293028 | 3.68 |
ENST00000370079.3
|
AMY1C
|
amylase, alpha 1C (salivary) |
| chr8_+_95653373 | 3.67 |
ENST00000358397.5
|
ESRP1
|
epithelial splicing regulatory protein 1 |
| chr10_+_70847852 | 3.66 |
ENST00000242465.3
|
SRGN
|
serglycin |
| chr18_+_28898052 | 3.66 |
ENST00000257192.4
|
DSG1
|
desmoglein 1 |
| chr2_+_161993412 | 3.65 |
ENST00000259075.2
ENST00000432002.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr16_-_86542455 | 3.64 |
ENST00000595886.1
ENST00000597578.1 ENST00000593604.1 |
FENDRR
|
FOXF1 adjacent non-coding developmental regulatory RNA |
| chr7_+_139529040 | 3.64 |
ENST00000455353.1
ENST00000458722.1 ENST00000411653.1 |
TBXAS1
|
thromboxane A synthase 1 (platelet) |
| chr19_-_12721616 | 3.64 |
ENST00000311437.6
|
ZNF490
|
zinc finger protein 490 |
| chr1_+_199996733 | 3.62 |
ENST00000236914.3
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr16_+_86612112 | 3.58 |
ENST00000320241.3
|
FOXL1
|
forkhead box L1 |
| chr11_-_34535332 | 3.54 |
ENST00000257832.2
ENST00000429939.2 |
ELF5
|
E74-like factor 5 (ets domain transcription factor) |
| chr4_-_143227088 | 3.52 |
ENST00000511838.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr8_+_24151553 | 3.51 |
ENST00000265769.4
ENST00000540823.1 ENST00000397649.3 |
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr15_+_81475047 | 3.49 |
ENST00000559388.1
|
IL16
|
interleukin 16 |
| chrX_-_106960285 | 3.46 |
ENST00000503515.1
ENST00000372397.2 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr10_-_48416849 | 3.45 |
ENST00000249598.1
|
GDF2
|
growth differentiation factor 2 |
| chr22_+_40342819 | 3.44 |
ENST00000407075.3
|
GRAP2
|
GRB2-related adaptor protein 2 |
| chr14_+_60716159 | 3.42 |
ENST00000325658.3
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
| chr5_-_150460914 | 3.40 |
ENST00000389378.2
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr1_+_74701062 | 3.37 |
ENST00000326637.3
|
TNNI3K
|
TNNI3 interacting kinase |
| chr4_+_147096837 | 3.37 |
ENST00000296581.5
ENST00000502781.1 |
LSM6
|
LSM6 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXO3_FOXD2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.7 | 32.1 | GO:1990922 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 5.5 | 16.6 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 3.7 | 7.4 | GO:0036022 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 3.1 | 9.2 | GO:0044501 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 2.9 | 20.5 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 2.8 | 28.5 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 2.8 | 19.6 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 2.7 | 16.4 | GO:0072131 | kidney mesenchyme morphogenesis(GO:0072131) metanephric mesenchyme morphogenesis(GO:0072133) |
| 2.7 | 35.4 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 2.7 | 8.0 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 2.7 | 26.7 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 2.6 | 7.9 | GO:2000525 | dendritic cell dendrite assembly(GO:0097026) regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 2.6 | 7.7 | GO:0035623 | regulation of pronephros size(GO:0035565) renal glucose absorption(GO:0035623) |
| 2.4 | 9.8 | GO:2000426 | plasmacytoid dendritic cell activation(GO:0002270) T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) regulation of restriction endodeoxyribonuclease activity(GO:0032072) negative regulation of apoptotic cell clearance(GO:2000426) |
| 2.4 | 39.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 2.4 | 14.6 | GO:1990822 | basic amino acid transmembrane transport(GO:1990822) |
| 2.4 | 7.2 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 2.3 | 13.9 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 2.2 | 6.5 | GO:1903676 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 2.1 | 10.4 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 1.9 | 11.6 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 1.9 | 3.8 | GO:1905069 | allantois development(GO:1905069) |
| 1.9 | 7.4 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 1.8 | 11.0 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 1.8 | 7.3 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 1.8 | 5.3 | GO:2001250 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 1.7 | 8.7 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 1.7 | 10.4 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 1.7 | 5.0 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 1.7 | 8.4 | GO:0035947 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) response to fluoride(GO:1902617) |
| 1.7 | 5.0 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 1.6 | 9.9 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 1.6 | 9.8 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 1.6 | 6.2 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 1.6 | 14.0 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 1.5 | 4.6 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 1.5 | 4.5 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) |
| 1.4 | 4.3 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 1.4 | 10.1 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 1.4 | 4.3 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 1.4 | 13.8 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 1.4 | 4.1 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 1.3 | 3.9 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 1.3 | 6.5 | GO:0006696 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 1.3 | 9.1 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 1.3 | 7.5 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 1.2 | 13.7 | GO:0033227 | dsRNA transport(GO:0033227) |
| 1.2 | 3.7 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 1.2 | 9.7 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 1.1 | 5.5 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 1.1 | 6.6 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 1.1 | 5.5 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 1.0 | 5.2 | GO:0021759 | globus pallidus development(GO:0021759) |
| 1.0 | 3.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 1.0 | 7.1 | GO:2000332 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 1.0 | 6.0 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 1.0 | 3.0 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 1.0 | 3.0 | GO:0045957 | regulation of complement activation, alternative pathway(GO:0030451) negative regulation of complement activation, alternative pathway(GO:0045957) |
| 1.0 | 3.8 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 1.0 | 6.7 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.9 | 2.8 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.9 | 2.8 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.9 | 1.8 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.9 | 7.3 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.9 | 9.0 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.9 | 22.8 | GO:1904886 | beta-catenin destruction complex disassembly(GO:1904886) |
| 0.9 | 4.4 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.9 | 2.6 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.8 | 2.5 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.8 | 5.0 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.8 | 5.0 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.8 | 8.3 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.8 | 3.2 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.8 | 2.4 | GO:0072560 | glandular epithelial cell maturation(GO:0002071) type B pancreatic cell maturation(GO:0072560) |
| 0.8 | 3.2 | GO:0050883 | negative regulation of sodium:proton antiporter activity(GO:0032416) gastric motility(GO:0035482) gastric emptying(GO:0035483) musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.8 | 0.8 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 0.8 | 3.8 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.8 | 8.3 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.7 | 3.0 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.7 | 11.9 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.7 | 0.7 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.7 | 3.6 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.7 | 2.9 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.7 | 2.8 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.7 | 6.3 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.7 | 4.2 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.7 | 4.1 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.7 | 4.1 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.7 | 2.7 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.7 | 13.9 | GO:0032740 | positive regulation of interleukin-17 production(GO:0032740) |
| 0.7 | 13.2 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.7 | 3.9 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.6 | 1.9 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.6 | 14.2 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.6 | 8.8 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.6 | 2.5 | GO:0072183 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of nephron tubule epithelial cell differentiation(GO:0072183) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) negative regulation of epithelial cell differentiation involved in kidney development(GO:2000697) |
| 0.6 | 1.3 | GO:0048867 | stem cell fate determination(GO:0048867) |
| 0.6 | 1.9 | GO:0019557 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.6 | 12.4 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.6 | 3.1 | GO:0007619 | courtship behavior(GO:0007619) |
| 0.6 | 26.4 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.6 | 11.7 | GO:0008228 | opsonization(GO:0008228) |
| 0.6 | 3.7 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.6 | 1.2 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.6 | 4.8 | GO:0035934 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 0.6 | 1.8 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) negative regulation of B cell differentiation(GO:0045578) regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.6 | 7.0 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.6 | 8.5 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.6 | 2.2 | GO:2000568 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.6 | 5.6 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.6 | 5.0 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.6 | 3.3 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.5 | 7.6 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.5 | 3.3 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.5 | 5.4 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.5 | 3.2 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.5 | 2.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.5 | 5.4 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) establishment or maintenance of cytoskeleton polarity(GO:0030952) |
| 0.5 | 1.6 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 0.5 | 1.6 | GO:1904316 | positive regulation of neutrophil degranulation(GO:0043315) cellular response to gravity(GO:0071258) positive regulation of neutrophil activation(GO:1902565) regulation of transcytosis(GO:1904298) positive regulation of transcytosis(GO:1904300) regulation of maternal process involved in parturition(GO:1904301) positive regulation of maternal process involved in parturition(GO:1904303) response to 2-O-acetyl-1-O-hexadecyl-sn-glycero-3-phosphocholine(GO:1904316) cellular response to 2-O-acetyl-1-O-hexadecyl-sn-glycero-3-phosphocholine(GO:1904317) |
| 0.5 | 4.8 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.5 | 2.6 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.5 | 6.3 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.5 | 4.7 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.5 | 1.6 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.5 | 2.0 | GO:0002159 | desmosome assembly(GO:0002159) endothelial cell-cell adhesion(GO:0071603) |
| 0.5 | 1.5 | GO:0019087 | transformation of host cell by virus(GO:0019087) |
| 0.5 | 21.3 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.5 | 3.9 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.5 | 5.5 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.5 | 1.8 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.5 | 1.4 | GO:0019836 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.4 | 2.2 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.4 | 7.9 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.4 | 3.0 | GO:0034350 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.4 | 3.5 | GO:0070235 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.4 | 0.9 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.4 | 1.7 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.4 | 1.3 | GO:0065001 | specification of axis polarity(GO:0065001) negative regulation of tooth mineralization(GO:0070171) |
| 0.4 | 2.6 | GO:1904721 | negative regulation of mRNA cleavage(GO:0031438) negative regulation of immunoglobulin secretion(GO:0051025) negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.4 | 1.7 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.4 | 7.2 | GO:0072659 | protein localization to plasma membrane(GO:0072659) |
| 0.4 | 8.0 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.4 | 1.3 | GO:0001777 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.4 | 2.1 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.4 | 3.2 | GO:0001878 | response to yeast(GO:0001878) |
| 0.4 | 2.4 | GO:0032875 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.4 | 3.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.4 | 5.8 | GO:0021702 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.4 | 3.1 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.4 | 0.8 | GO:0033078 | extrathymic T cell differentiation(GO:0033078) |
| 0.4 | 0.8 | GO:0002011 | morphogenesis of an epithelial sheet(GO:0002011) |
| 0.4 | 1.5 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.4 | 7.9 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.4 | 10.5 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.4 | 11.6 | GO:0050913 | sensory perception of bitter taste(GO:0050913) |
| 0.4 | 4.7 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.4 | 2.9 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.4 | 1.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.4 | 1.1 | GO:2000543 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) positive regulation of gastrulation(GO:2000543) |
| 0.4 | 2.8 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.3 | 1.7 | GO:0010976 | positive regulation of neuron projection development(GO:0010976) |
| 0.3 | 1.7 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.3 | 6.1 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.3 | 2.0 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.3 | 4.0 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.3 | 6.0 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.3 | 24.8 | GO:0071349 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.3 | 4.0 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.3 | 3.2 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.3 | 20.1 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.3 | 3.5 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.3 | 1.3 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.3 | 6.4 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.3 | 5.1 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.3 | 9.6 | GO:1902751 | positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.3 | 2.1 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.3 | 18.0 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.3 | 12.7 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.3 | 0.9 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.3 | 7.0 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.3 | 3.4 | GO:0051573 | negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.3 | 2.2 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.3 | 1.1 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) negative regulation of skeletal muscle cell proliferation(GO:0014859) regulation of skeletal muscle tissue growth(GO:0048631) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.3 | 1.9 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.3 | 1.0 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.3 | 4.3 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.2 | 1.7 | GO:0098501 | polynucleotide dephosphorylation(GO:0098501) |
| 0.2 | 2.5 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.2 | 1.2 | GO:1902775 | mitochondrial ribosome assembly(GO:0061668) mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.2 | 1.4 | GO:0070627 | ferrous iron import(GO:0070627) ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.2 | 0.9 | GO:0032431 | activation of phospholipase A2 activity(GO:0032431) |
| 0.2 | 17.8 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.2 | 1.4 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.2 | 15.1 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.2 | 5.9 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.2 | 1.1 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.2 | 7.6 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.2 | 5.1 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.2 | 0.7 | GO:0048690 | modulation by virus of host transcription(GO:0019056) regulation of sprouting of injured axon(GO:0048686) positive regulation of sprouting of injured axon(GO:0048687) regulation of axon extension involved in regeneration(GO:0048690) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.2 | 4.8 | GO:0043555 | regulation of translation in response to stress(GO:0043555) |
| 0.2 | 1.1 | GO:0044266 | multicellular organismal protein catabolic process(GO:0044254) protein digestion(GO:0044256) multicellular organismal macromolecule catabolic process(GO:0044266) |
| 0.2 | 0.9 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.2 | 2.8 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.2 | 2.9 | GO:0007617 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.2 | 3.6 | GO:0006516 | N-acetylglucosamine metabolic process(GO:0006044) glycoprotein catabolic process(GO:0006516) |
| 0.2 | 0.8 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.2 | 1.0 | GO:1904016 | response to Thyroglobulin triiodothyronine(GO:1904016) |
| 0.2 | 2.2 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.2 | 0.8 | GO:0048840 | otolith development(GO:0048840) |
| 0.2 | 1.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.2 | 1.7 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.2 | 1.3 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.2 | 1.5 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.2 | 2.3 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.2 | 0.7 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.2 | 4.6 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.2 | 7.1 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.2 | 1.0 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.2 | 2.6 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.2 | 2.4 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.2 | 1.2 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.2 | 2.2 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.2 | 2.8 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.2 | 2.7 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.2 | 2.0 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.2 | 3.2 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.2 | 3.4 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.2 | 3.4 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.2 | 0.6 | GO:0061743 | motor learning(GO:0061743) |
| 0.2 | 4.6 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.2 | 13.9 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.2 | 0.6 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.2 | 2.8 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.2 | 2.6 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.2 | 3.0 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.1 | 2.2 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 1.5 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.1 | 1.6 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 1.8 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.1 | 4.9 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.1 | 13.2 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.1 | 3.1 | GO:0042772 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.1 | 0.7 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.1 | 1.8 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 2.1 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) |
| 0.1 | 1.2 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.1 | 0.7 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.1 | 3.3 | GO:0072662 | protein targeting to peroxisome(GO:0006625) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.1 | 10.3 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.1 | 0.6 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.1 | 0.6 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 0.8 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.1 | 5.8 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 1.2 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 1.3 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.1 | 8.7 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.1 | 12.1 | GO:0007338 | single fertilization(GO:0007338) |
| 0.1 | 13.3 | GO:0009267 | cellular response to starvation(GO:0009267) |
| 0.1 | 5.7 | GO:0016445 | somatic diversification of immunoglobulins(GO:0016445) |
| 0.1 | 0.9 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.1 | 0.8 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.1 | 1.4 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.1 | 6.2 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 1.3 | GO:0035878 | nail development(GO:0035878) |
| 0.1 | 1.8 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 2.6 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.1 | 0.6 | GO:0002027 | regulation of heart rate(GO:0002027) |
| 0.1 | 6.7 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.1 | 1.2 | GO:0098779 | mitophagy in response to mitochondrial depolarization(GO:0098779) |
| 0.1 | 0.6 | GO:0045779 | negative regulation of bone resorption(GO:0045779) negative regulation of bone remodeling(GO:0046851) |
| 0.1 | 1.5 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.1 | 2.8 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 2.3 | GO:0010659 | cardiac muscle cell apoptotic process(GO:0010659) |
| 0.1 | 1.8 | GO:0070328 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.1 | 0.7 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 4.6 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.1 | 1.7 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.1 | 4.0 | GO:0031640 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) |
| 0.1 | 0.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 0.7 | GO:0009988 | cell-cell recognition(GO:0009988) |
| 0.1 | 0.2 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.1 | 5.0 | GO:0071347 | cellular response to interleukin-1(GO:0071347) |
| 0.1 | 0.7 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.1 | 0.7 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 8.4 | GO:0031424 | keratinization(GO:0031424) |
| 0.1 | 0.3 | GO:1902950 | regulation of dendritic spine maintenance(GO:1902950) positive regulation of dendritic spine maintenance(GO:1902952) regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.1 | 1.0 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 1.1 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 2.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.4 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 2.4 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.1 | 0.5 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 3.1 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.1 | 0.6 | GO:0046606 | negative regulation of centrosome cycle(GO:0046606) |
| 0.1 | 1.1 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.1 | 1.3 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.1 | 0.2 | GO:0097283 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.1 | 0.7 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 0.7 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.1 | 0.5 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.1 | 1.1 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.1 | 2.3 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 0.2 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 2.0 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.5 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.0 | 0.3 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 6.6 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 4.0 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.8 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) |
| 0.0 | 3.3 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 2.3 | GO:0032091 | negative regulation of protein binding(GO:0032091) |
| 0.0 | 0.5 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.0 | 0.2 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 1.5 | GO:1903844 | regulation of cellular response to transforming growth factor beta stimulus(GO:1903844) |
| 0.0 | 2.5 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.7 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 1.5 | GO:0034724 | DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 | 0.3 | GO:0032438 | melanosome organization(GO:0032438) pigment granule organization(GO:0048753) |
| 0.0 | 0.8 | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) |
| 0.0 | 0.5 | GO:0042113 | B cell activation(GO:0042113) |
| 0.0 | 0.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 4.0 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.5 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.8 | GO:0007264 | small GTPase mediated signal transduction(GO:0007264) |
| 0.0 | 1.2 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) |
| 0.0 | 0.8 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 1.8 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 1.9 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 1.0 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.7 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.4 | GO:0042481 | regulation of odontogenesis(GO:0042481) |
| 0.0 | 0.1 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:1902963 | positive regulation of beta-amyloid clearance(GO:1900223) regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.0 | 1.4 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 0.8 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.9 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 2.5 | GO:0008213 | protein methylation(GO:0006479) protein alkylation(GO:0008213) |
| 0.0 | 0.6 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 1.4 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 0.3 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.2 | GO:1901374 | acetate ester transport(GO:1901374) |
| 0.0 | 3.7 | GO:0043413 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 2.9 | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process(GO:0043161) |
| 0.0 | 0.1 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 0.0 | 0.4 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.9 | GO:0001892 | embryonic placenta development(GO:0001892) |
| 0.0 | 1.7 | GO:0030168 | platelet activation(GO:0030168) |
| 0.0 | 0.8 | GO:0030816 | positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) |
| 0.0 | 2.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 1.2 | GO:0098869 | cellular oxidant detoxification(GO:0098869) |
| 0.0 | 0.3 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.2 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 1.4 | GO:0042493 | response to drug(GO:0042493) |
| 0.0 | 0.2 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 1.0 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 15.0 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 2.6 | 7.7 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 2.0 | 10.2 | GO:0036398 | TCR signalosome(GO:0036398) |
| 2.0 | 39.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 1.9 | 11.6 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 1.8 | 23.8 | GO:0042555 | MCM complex(GO:0042555) |
| 1.8 | 5.4 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 1.7 | 5.0 | GO:0034657 | GID complex(GO:0034657) |
| 1.5 | 8.8 | GO:0034687 | integrin alphaL-beta2 complex(GO:0034687) |
| 1.4 | 7.0 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 1.2 | 9.9 | GO:0035976 | AP1 complex(GO:0035976) |
| 1.1 | 7.4 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 1.0 | 4.2 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 1.0 | 7.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 1.0 | 2.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.9 | 2.7 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.9 | 2.7 | GO:0031251 | PAN complex(GO:0031251) |
| 0.8 | 5.6 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.8 | 3.1 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.8 | 1.5 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.7 | 17.2 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.7 | 1.4 | GO:0032154 | cell division site(GO:0032153) cleavage furrow(GO:0032154) cell division site part(GO:0032155) cell surface furrow(GO:0097610) |
| 0.7 | 2.8 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.7 | 5.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.6 | 1.2 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.6 | 20.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.6 | 1.8 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.6 | 2.3 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.6 | 2.8 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.5 | 8.8 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.5 | 2.4 | GO:0032021 | NELF complex(GO:0032021) |
| 0.5 | 29.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.4 | 4.7 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.4 | 7.0 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.4 | 4.5 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.4 | 13.2 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.4 | 6.2 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.4 | 3.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.4 | 13.6 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.3 | 2.4 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.3 | 21.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.3 | 42.8 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.3 | 23.4 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.3 | 63.6 | GO:0101002 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.3 | 32.3 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.3 | 4.0 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.3 | 9.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.3 | 5.0 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.3 | 0.6 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.3 | 8.2 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.3 | 5.4 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.3 | 6.9 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.3 | 1.8 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.3 | 2.8 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.3 | 1.0 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.3 | 4.0 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.2 | 0.9 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.2 | 36.0 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.2 | 7.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.2 | 1.5 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.2 | 4.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.2 | 7.1 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.2 | 1.8 | GO:0032039 | integrator complex(GO:0032039) |
| 0.2 | 4.1 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.2 | 2.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.2 | 1.3 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.2 | 1.5 | GO:0001741 | XY body(GO:0001741) |
| 0.2 | 4.8 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.2 | 12.4 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.2 | 1.4 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.2 | 0.9 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.2 | 0.8 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.1 | 1.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 3.0 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 53.0 | GO:0005769 | early endosome(GO:0005769) |
| 0.1 | 1.6 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 1.3 | GO:0098642 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.1 | 3.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 2.7 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 19.5 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 6.7 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 2.1 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 2.6 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 10.8 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 1.2 | GO:0097197 | perinuclear endoplasmic reticulum(GO:0097038) tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 1.7 | GO:0044216 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.1 | 2.0 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 0.8 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 4.7 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 2.8 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 9.7 | GO:0005886 | plasma membrane(GO:0005886) |
| 0.1 | 8.2 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.1 | 4.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.4 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 37.2 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 2.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 1.9 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 5.7 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.1 | 2.8 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 4.1 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.1 | 1.5 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 2.0 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 10.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 1.5 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.1 | 20.4 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.1 | 3.1 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.1 | 0.9 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 2.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 0.4 | GO:0030018 | Z disc(GO:0030018) I band(GO:0031674) |
| 0.1 | 1.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 1.8 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 5.3 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 3.1 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.9 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 2.1 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 1.5 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 1.1 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.1 | 2.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 0.6 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 1.1 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 9.5 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.9 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 5.7 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 10.4 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 33.4 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 3.8 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.5 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 1.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.7 | GO:0005929 | cilium(GO:0005929) |
| 0.0 | 4.7 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 1.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.6 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 1.1 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 2.1 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 3.0 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.9 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 1.6 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 1.5 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 1.0 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 1.0 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.5 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 1.4 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.2 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 0.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.3 | GO:0045211 | postsynaptic membrane(GO:0045211) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.4 | 13.2 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 3.5 | 13.9 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 3.5 | 13.8 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 2.9 | 32.1 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 2.6 | 7.9 | GO:0036134 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 2.3 | 13.7 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 2.0 | 26.4 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 2.0 | 19.6 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 1.9 | 17.5 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 1.8 | 11.0 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 1.8 | 5.3 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 1.7 | 6.7 | GO:0016160 | amylase activity(GO:0016160) |
| 1.6 | 9.8 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 1.6 | 6.5 | GO:0004310 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 1.6 | 15.9 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 1.6 | 6.3 | GO:0004307 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 1.4 | 8.3 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 1.3 | 17.5 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 1.3 | 4.0 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 1.3 | 14.6 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 1.3 | 6.5 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 1.3 | 8.8 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 1.3 | 3.8 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 1.3 | 5.0 | GO:0035276 | ethanol binding(GO:0035276) |
| 1.2 | 3.7 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 1.2 | 4.8 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 1.2 | 10.4 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 1.1 | 9.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 1.1 | 4.5 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 1.1 | 5.4 | GO:0002046 | opsin binding(GO:0002046) |
| 1.1 | 4.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 1.0 | 6.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 1.0 | 4.0 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 1.0 | 3.0 | GO:0001861 | complement component C4b receptor activity(GO:0001861) complement component C3b receptor activity(GO:0004877) |
| 1.0 | 3.9 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 1.0 | 6.7 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.9 | 2.8 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.9 | 2.8 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.9 | 5.5 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.9 | 5.5 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.9 | 2.7 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.9 | 2.6 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.9 | 4.3 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.8 | 5.8 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.8 | 37.3 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.8 | 8.2 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.8 | 8.0 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.8 | 6.2 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.8 | 3.0 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.7 | 2.9 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.7 | 4.3 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.7 | 3.5 | GO:0034597 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.7 | 12.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.6 | 3.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.6 | 5.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.6 | 15.9 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.6 | 3.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.6 | 3.1 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.6 | 3.7 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.6 | 3.7 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.6 | 7.9 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.6 | 24.8 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.6 | 2.9 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.6 | 4.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.6 | 1.8 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.6 | 1.7 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.5 | 8.0 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.5 | 1.6 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.5 | 10.7 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.5 | 7.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.5 | 5.6 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.5 | 2.0 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.5 | 13.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.5 | 1.5 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.5 | 7.3 | GO:0031702 | angiotensin receptor binding(GO:0031701) type 1 angiotensin receptor binding(GO:0031702) |
| 0.5 | 2.4 | GO:0070728 | leucine binding(GO:0070728) |
| 0.5 | 3.9 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.5 | 27.3 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.5 | 4.7 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.5 | 1.9 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.5 | 3.2 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.4 | 2.6 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.4 | 1.7 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.4 | 2.9 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.4 | 6.2 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.4 | 10.0 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.4 | 0.8 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.4 | 16.1 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.4 | 9.7 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.4 | 3.5 | GO:0043426 | MRF binding(GO:0043426) |
| 0.4 | 3.8 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.4 | 1.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.4 | 2.2 | GO:0023030 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.4 | 2.2 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.4 | 7.0 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.4 | 4.8 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.4 | 1.4 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.4 | 0.7 | GO:0033612 | receptor serine/threonine kinase binding(GO:0033612) |
| 0.4 | 3.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.3 | 8.0 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.3 | 1.0 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.3 | 4.1 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.3 | 2.6 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.3 | 4.3 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.3 | 11.9 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.3 | 7.0 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.3 | 7.6 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.3 | 2.2 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.3 | 3.4 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.3 | 2.7 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.3 | 3.6 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.3 | 4.7 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.3 | 5.3 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.3 | 5.6 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.3 | 2.8 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.3 | 10.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.3 | 4.3 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.2 | 6.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.2 | 4.1 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.2 | 10.4 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.2 | 1.2 | GO:0033265 | choline binding(GO:0033265) |
| 0.2 | 3.0 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.2 | 2.0 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.2 | 2.0 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.2 | 9.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.2 | 1.7 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.2 | 2.6 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.2 | 2.7 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.2 | 1.0 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.2 | 0.8 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.2 | 13.8 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.2 | 15.3 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.2 | 0.6 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.2 | 0.8 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.2 | 1.4 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.2 | 12.2 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.2 | 7.1 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.2 | 4.5 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.2 | 0.6 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.2 | 2.7 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.2 | 14.6 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.2 | 3.1 | GO:0008656 | translation activator activity(GO:0008494) cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.2 | 2.7 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.2 | 4.8 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.2 | 6.4 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.2 | 0.9 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.2 | 5.3 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.2 | 1.9 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.2 | 1.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.2 | 0.5 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.2 | 11.2 | GO:0008026 | ATP-dependent RNA helicase activity(GO:0004004) ATP-dependent helicase activity(GO:0008026) RNA-dependent ATPase activity(GO:0008186) purine NTP-dependent helicase activity(GO:0070035) |
| 0.2 | 1.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.2 | 1.6 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.2 | 3.2 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.2 | 1.8 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.2 | 4.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.2 | 10.4 | GO:0002039 | p53 binding(GO:0002039) |
| 0.2 | 1.5 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.2 | 35.1 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.2 | 1.1 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.2 | 0.9 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.2 | 0.9 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.2 | 1.7 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.2 | 0.8 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.2 | 1.5 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.1 | 1.6 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 5.8 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.1 | 2.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.6 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 15.1 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 0.7 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 2.2 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 70.7 | GO:0003712 | transcription cofactor activity(GO:0003712) |
| 0.1 | 0.6 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 3.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 1.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 5.3 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.1 | 6.7 | GO:0004386 | helicase activity(GO:0004386) |
| 0.1 | 2.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 0.4 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.1 | 0.6 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 1.2 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 3.0 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 2.0 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 1.7 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 0.5 | GO:0070851 | growth factor receptor binding(GO:0070851) |
| 0.1 | 0.3 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 2.9 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 10.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 1.7 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.6 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 2.5 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 1.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 3.3 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 0.2 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.1 | 1.7 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.1 | 1.8 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 0.6 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 1.5 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 0.6 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.1 | 0.9 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 4.8 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 1.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 7.3 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 1.1 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 0.2 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.1 | 0.6 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.1 | 7.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 2.1 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 1.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.6 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 1.1 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 1.5 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 4.0 | GO:0016684 | peroxidase activity(GO:0004601) oxidoreductase activity, acting on peroxide as acceptor(GO:0016684) |
| 0.1 | 0.7 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 0.4 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 1.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 1.1 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 1.0 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 1.3 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 8.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 0.9 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 2.4 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 1.3 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.5 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 13.3 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 1.8 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 9.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.1 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.2 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.0 | 1.2 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.4 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 0.7 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.5 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.5 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.9 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.5 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.9 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 1.4 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.1 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.0 | 0.7 | GO:0032934 | sterol binding(GO:0032934) |
| 0.0 | 3.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 1.0 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 0.9 | GO:0005262 | calcium channel activity(GO:0005262) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 26.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 1.0 | 9.1 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.9 | 61.4 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.7 | 32.0 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.6 | 14.8 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.6 | 18.9 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.6 | 13.9 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.6 | 21.8 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.5 | 26.2 | PID MYC PATHWAY | C-MYC pathway |
| 0.5 | 20.5 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.4 | 17.9 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.4 | 11.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.4 | 16.6 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.4 | 17.5 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.3 | 21.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.3 | 8.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.3 | 11.9 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.3 | 7.9 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.3 | 18.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.3 | 2.5 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.2 | 25.6 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.2 | 18.5 | PID E2F PATHWAY | E2F transcription factor network |
| 0.2 | 6.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.2 | 6.8 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.2 | 1.7 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.2 | 6.1 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.2 | 7.2 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.2 | 2.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.2 | 4.3 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 2.9 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 1.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 1.2 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 1.7 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 4.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 2.9 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 9.8 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.1 | 3.2 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.1 | 8.0 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 2.9 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.1 | 5.1 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 4.6 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 7.6 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 0.9 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 2.6 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 1.7 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 5.7 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 2.0 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 3.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 4.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 0.5 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 2.7 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 0.8 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 29.6 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 3.6 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 3.9 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 1.5 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.1 | 6.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 3.2 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.1 | 1.0 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 1.5 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 2.8 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 0.6 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 4.2 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 0.8 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.1 | 0.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 1.1 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 3.3 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 1.8 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 2.0 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 2.1 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.5 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.7 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 20.5 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 1.5 | 23.8 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 1.3 | 32.1 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 1.2 | 23.6 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.9 | 10.4 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.8 | 21.9 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.8 | 13.9 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.8 | 19.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.7 | 34.7 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.7 | 53.6 | REACTOME TRANSCRIPTIONAL ACTIVITY OF SMAD2 SMAD3 SMAD4 HETEROTRIMER | Genes involved in Transcriptional activity of SMAD2/SMAD3:SMAD4 heterotrimer |
| 0.7 | 6.7 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.7 | 18.0 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.6 | 5.0 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.6 | 4.7 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.6 | 6.4 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.6 | 7.9 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.6 | 2.8 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.5 | 18.7 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.5 | 9.6 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.5 | 1.5 | REACTOME SHC RELATED EVENTS | Genes involved in SHC-related events |
| 0.5 | 10.0 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.5 | 8.9 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.5 | 9.7 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.4 | 5.0 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.4 | 10.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.4 | 4.1 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.4 | 21.3 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.4 | 8.0 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.4 | 5.9 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.4 | 7.4 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.4 | 39.4 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.4 | 10.4 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.4 | 2.9 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.4 | 3.9 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.4 | 6.7 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.3 | 13.3 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.3 | 11.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.3 | 23.2 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.3 | 4.1 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 0.3 | 5.0 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.2 | 5.8 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.2 | 2.6 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.2 | 3.8 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.2 | 3.0 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.2 | 6.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 4.5 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.2 | 1.4 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.2 | 5.0 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 6.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.2 | 2.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.2 | 4.0 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 1.6 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.2 | 1.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.2 | 2.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.2 | 25.9 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.2 | 3.5 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.2 | 7.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 5.1 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.1 | 2.7 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 8.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 2.8 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.1 | 5.5 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 11.9 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 2.8 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 2.6 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 4.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 1.4 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.1 | 0.9 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.1 | 6.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.8 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 0.6 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 2.0 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 0.9 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 13.1 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 12.0 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 1.5 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 3.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 3.0 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 2.5 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.1 | 1.5 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.1 | 2.2 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 1.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 1.0 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 2.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.1 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 2.2 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 3.1 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 2.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 2.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 0.5 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.1 | 2.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 1.5 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 0.6 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 3.4 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 0.8 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 1.0 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 3.1 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.1 | 2.0 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 3.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.9 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.1 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.6 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.8 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 2.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.8 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 1.7 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 1.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.2 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 1.6 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.4 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 3.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 1.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.4 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |