Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
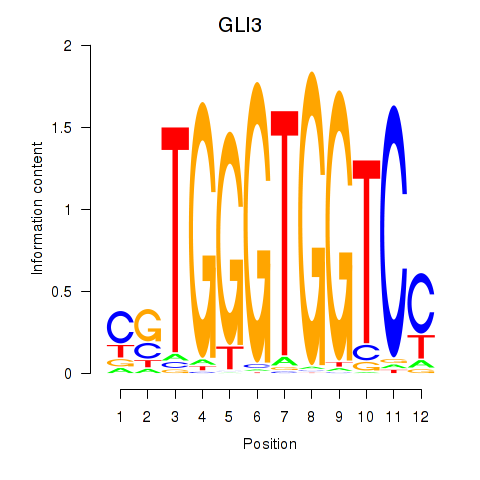
Results for GLI3
Z-value: 1.03
Transcription factors associated with GLI3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GLI3
|
ENSG00000106571.8 | GLI family zinc finger 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GLI3 | hg19_v2_chr7_-_42276612_42276782 | 0.34 | 1.8e-07 | Click! |
Activity profile of GLI3 motif
Sorted Z-values of GLI3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_153839149 | 24.09 |
ENST00000465093.1
ENST00000465817.1 |
ARHGEF26
|
Rho guanine nucleotide exchange factor (GEF) 26 |
| chr8_-_27462822 | 15.32 |
ENST00000522098.1
|
CLU
|
clusterin |
| chr22_+_23247030 | 13.72 |
ENST00000390324.2
|
IGLJ3
|
immunoglobulin lambda joining 3 |
| chr19_+_35521616 | 13.29 |
ENST00000595652.1
|
SCN1B
|
sodium channel, voltage-gated, type I, beta subunit |
| chr8_-_27468842 | 13.08 |
ENST00000523500.1
|
CLU
|
clusterin |
| chr19_+_45417921 | 12.88 |
ENST00000252491.4
ENST00000592885.1 ENST00000589781.1 |
APOC1
|
apolipoprotein C-I |
| chr11_-_111783919 | 12.13 |
ENST00000531198.1
ENST00000533879.1 |
CRYAB
|
crystallin, alpha B |
| chr19_+_45417812 | 12.07 |
ENST00000592535.1
|
APOC1
|
apolipoprotein C-I |
| chr5_-_42825983 | 11.40 |
ENST00000506577.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr11_-_111784005 | 11.39 |
ENST00000527899.1
|
CRYAB
|
crystallin, alpha B |
| chr3_+_45067659 | 11.31 |
ENST00000296130.4
|
CLEC3B
|
C-type lectin domain family 3, member B |
| chr1_+_6845384 | 11.10 |
ENST00000303635.7
|
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr19_+_45417504 | 10.97 |
ENST00000588750.1
ENST00000588802.1 |
APOC1
|
apolipoprotein C-I |
| chr19_+_45418067 | 10.56 |
ENST00000589078.1
ENST00000586638.1 |
APOC1
|
apolipoprotein C-I |
| chr4_-_57524061 | 10.38 |
ENST00000508121.1
|
HOPX
|
HOP homeobox |
| chr6_-_159420780 | 10.10 |
ENST00000449822.1
|
RSPH3
|
radial spoke 3 homolog (Chlamydomonas) |
| chr9_-_98079965 | 9.83 |
ENST00000289081.3
|
FANCC
|
Fanconi anemia, complementation group C |
| chr1_-_204380919 | 9.55 |
ENST00000367188.4
|
PPP1R15B
|
protein phosphatase 1, regulatory subunit 15B |
| chr5_-_149792295 | 9.38 |
ENST00000518797.1
ENST00000524315.1 ENST00000009530.7 ENST00000377795.3 |
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain |
| chr9_+_139871948 | 9.38 |
ENST00000224167.2
ENST00000457950.1 ENST00000371625.3 ENST00000371623.3 |
PTGDS
|
prostaglandin D2 synthase 21kDa (brain) |
| chr10_-_90712520 | 9.05 |
ENST00000224784.6
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chr3_-_15469006 | 8.93 |
ENST00000443029.1
ENST00000383790.3 ENST00000383789.5 |
METTL6
|
methyltransferase like 6 |
| chr17_+_4710391 | 8.65 |
ENST00000263088.6
ENST00000572940.1 |
PLD2
|
phospholipase D2 |
| chr19_-_17185848 | 8.55 |
ENST00000593360.1
|
HAUS8
|
HAUS augmin-like complex, subunit 8 |
| chr2_-_201936302 | 8.30 |
ENST00000453765.1
ENST00000452799.1 ENST00000446678.1 ENST00000418596.3 |
FAM126B
|
family with sequence similarity 126, member B |
| chr19_-_16653226 | 8.07 |
ENST00000198939.6
|
CHERP
|
calcium homeostasis endoplasmic reticulum protein |
| chr8_-_27468945 | 7.98 |
ENST00000405140.3
|
CLU
|
clusterin |
| chr14_+_29236269 | 7.97 |
ENST00000313071.4
|
FOXG1
|
forkhead box G1 |
| chr16_+_85061367 | 7.95 |
ENST00000538274.1
ENST00000258180.3 |
KIAA0513
|
KIAA0513 |
| chr2_+_17935383 | 7.88 |
ENST00000524465.1
ENST00000381254.2 ENST00000532257.1 |
GEN1
|
GEN1 Holliday junction 5' flap endonuclease |
| chr20_+_62185491 | 7.78 |
ENST00000370097.1
|
C20orf195
|
chromosome 20 open reading frame 195 |
| chr12_-_6798616 | 7.45 |
ENST00000355772.4
ENST00000417772.3 ENST00000396801.3 ENST00000396799.2 |
ZNF384
|
zinc finger protein 384 |
| chr19_+_2096868 | 7.40 |
ENST00000395296.1
ENST00000395301.3 |
IZUMO4
|
IZUMO family member 4 |
| chr14_+_94577074 | 7.39 |
ENST00000444961.1
ENST00000448882.1 ENST00000557098.1 ENST00000554800.1 ENST00000556544.1 ENST00000298902.5 ENST00000555819.1 ENST00000557634.1 ENST00000555744.1 |
IFI27
|
interferon, alpha-inducible protein 27 |
| chr4_+_166300084 | 7.27 |
ENST00000402744.4
|
CPE
|
carboxypeptidase E |
| chr2_+_17935119 | 7.22 |
ENST00000317402.7
|
GEN1
|
GEN1 Holliday junction 5' flap endonuclease |
| chr1_+_156123359 | 7.19 |
ENST00000368284.1
ENST00000368286.2 ENST00000438830.1 |
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr22_+_23241661 | 7.17 |
ENST00000390322.2
|
IGLJ2
|
immunoglobulin lambda joining 2 |
| chr8_+_21916710 | 7.15 |
ENST00000523266.1
ENST00000519907.1 |
DMTN
|
dematin actin binding protein |
| chr19_-_16653325 | 7.05 |
ENST00000546361.2
|
CHERP
|
calcium homeostasis endoplasmic reticulum protein |
| chr3_+_10068095 | 7.00 |
ENST00000287647.3
ENST00000383807.1 ENST00000383806.1 ENST00000419585.1 |
FANCD2
|
Fanconi anemia, complementation group D2 |
| chr11_-_111783595 | 6.98 |
ENST00000528628.1
|
CRYAB
|
crystallin, alpha B |
| chr19_+_35521572 | 6.83 |
ENST00000262631.5
|
SCN1B
|
sodium channel, voltage-gated, type I, beta subunit |
| chr2_-_136875712 | 6.81 |
ENST00000241393.3
|
CXCR4
|
chemokine (C-X-C motif) receptor 4 |
| chr14_+_102027688 | 6.77 |
ENST00000510508.4
ENST00000359323.3 |
DIO3
|
deiodinase, iodothyronine, type III |
| chr11_-_12030629 | 6.70 |
ENST00000396505.2
|
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr8_+_21916680 | 6.68 |
ENST00000358242.3
ENST00000415253.1 |
DMTN
|
dematin actin binding protein |
| chr1_-_20834586 | 6.67 |
ENST00000264198.3
|
MUL1
|
mitochondrial E3 ubiquitin protein ligase 1 |
| chr15_+_81591757 | 6.56 |
ENST00000558332.1
|
IL16
|
interleukin 16 |
| chr7_+_150498783 | 6.53 |
ENST00000475536.1
ENST00000468689.1 |
TMEM176A
|
transmembrane protein 176A |
| chr12_-_6798523 | 6.48 |
ENST00000319770.3
|
ZNF384
|
zinc finger protein 384 |
| chr3_-_46506358 | 6.46 |
ENST00000417439.1
ENST00000431944.1 |
LTF
|
lactotransferrin |
| chr20_-_50808236 | 6.44 |
ENST00000361387.2
|
ZFP64
|
ZFP64 zinc finger protein |
| chr1_+_156123318 | 6.42 |
ENST00000368285.3
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr19_+_39897453 | 6.33 |
ENST00000597629.1
ENST00000248673.3 ENST00000594045.1 ENST00000594442.1 |
ZFP36
|
ZFP36 ring finger protein |
| chr3_+_15468862 | 6.28 |
ENST00000396842.2
|
EAF1
|
ELL associated factor 1 |
| chr1_+_36396313 | 6.21 |
ENST00000324350.5
|
AGO3
|
argonaute RISC catalytic component 3 |
| chr16_+_84209539 | 6.16 |
ENST00000569735.1
|
DNAAF1
|
dynein, axonemal, assembly factor 1 |
| chr12_-_6798410 | 6.16 |
ENST00000361959.3
ENST00000436774.2 ENST00000544482.1 |
ZNF384
|
zinc finger protein 384 |
| chr7_+_150498610 | 6.15 |
ENST00000461345.1
|
TMEM176A
|
transmembrane protein 176A |
| chr15_+_84116106 | 6.08 |
ENST00000535412.1
ENST00000324537.5 |
SH3GL3
|
SH3-domain GRB2-like 3 |
| chr19_+_48281842 | 6.05 |
ENST00000509570.2
|
SEPW1
|
selenoprotein W, 1 |
| chr12_-_45270151 | 6.02 |
ENST00000429094.2
|
NELL2
|
NEL-like 2 (chicken) |
| chr20_+_36149602 | 5.99 |
ENST00000062104.2
ENST00000346199.2 |
NNAT
|
neuronatin |
| chr12_-_13248705 | 5.92 |
ENST00000396310.2
|
GSG1
|
germ cell associated 1 |
| chr17_+_17942684 | 5.91 |
ENST00000376345.3
|
GID4
|
GID complex subunit 4 |
| chr12_-_45270077 | 5.88 |
ENST00000551601.1
ENST00000549027.1 ENST00000452445.2 |
NELL2
|
NEL-like 2 (chicken) |
| chr17_+_17942594 | 5.87 |
ENST00000268719.4
|
GID4
|
GID complex subunit 4 |
| chr12_-_122751002 | 5.85 |
ENST00000267199.4
|
VPS33A
|
vacuolar protein sorting 33 homolog A (S. cerevisiae) |
| chr21_-_45079341 | 5.84 |
ENST00000443485.1
ENST00000291560.2 |
HSF2BP
|
heat shock transcription factor 2 binding protein |
| chr19_-_46318561 | 5.69 |
ENST00000221538.3
|
RSPH6A
|
radial spoke head 6 homolog A (Chlamydomonas) |
| chr11_-_65640325 | 5.62 |
ENST00000307998.6
|
EFEMP2
|
EGF containing fibulin-like extracellular matrix protein 2 |
| chr19_-_46318486 | 5.57 |
ENST00000597055.1
|
RSPH6A
|
radial spoke head 6 homolog A (Chlamydomonas) |
| chr6_-_46703069 | 5.47 |
ENST00000538237.1
ENST00000274793.7 |
PLA2G7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr6_-_167040731 | 5.41 |
ENST00000265678.4
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chr19_+_35629702 | 5.39 |
ENST00000351325.4
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr1_-_21978312 | 5.38 |
ENST00000359708.4
ENST00000290101.4 |
RAP1GAP
|
RAP1 GTPase activating protein |
| chr1_+_11866270 | 5.38 |
ENST00000376497.3
ENST00000376487.3 ENST00000376496.3 |
CLCN6
|
chloride channel, voltage-sensitive 6 |
| chr21_-_46330545 | 5.35 |
ENST00000320216.6
ENST00000397852.1 |
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr1_-_11866034 | 5.23 |
ENST00000376590.3
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr3_-_127542051 | 5.22 |
ENST00000398104.1
|
MGLL
|
monoglyceride lipase |
| chr3_-_15140629 | 5.17 |
ENST00000507357.1
ENST00000449050.1 ENST00000253699.3 ENST00000435849.3 ENST00000476527.2 |
ZFYVE20
|
zinc finger, FYVE domain containing 20 |
| chr6_-_28367510 | 5.11 |
ENST00000361028.1
|
ZSCAN12
|
zinc finger and SCAN domain containing 12 |
| chr11_-_65640198 | 5.10 |
ENST00000528176.1
|
EFEMP2
|
EGF containing fibulin-like extracellular matrix protein 2 |
| chr19_+_44084696 | 5.09 |
ENST00000562255.1
ENST00000569031.2 |
PINLYP
|
phospholipase A2 inhibitor and LY6/PLAUR domain containing |
| chr15_+_59499031 | 5.03 |
ENST00000307144.4
|
LDHAL6B
|
lactate dehydrogenase A-like 6B |
| chr3_+_49591881 | 5.01 |
ENST00000296452.4
|
BSN
|
bassoon presynaptic cytomatrix protein |
| chr19_+_58790314 | 5.01 |
ENST00000196548.5
ENST00000608843.1 |
ZNF8
ZNF8
|
Zinc finger protein 8 zinc finger protein 8 |
| chr12_+_50355647 | 5.01 |
ENST00000293599.6
|
AQP5
|
aquaporin 5 |
| chr4_-_17812309 | 4.97 |
ENST00000382247.1
ENST00000536863.1 |
DCAF16
|
DDB1 and CUL4 associated factor 16 |
| chr1_-_11865982 | 4.96 |
ENST00000418034.1
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr19_+_1103936 | 4.95 |
ENST00000354171.8
ENST00000589115.1 |
GPX4
|
glutathione peroxidase 4 |
| chr9_-_123476612 | 4.92 |
ENST00000426959.1
|
MEGF9
|
multiple EGF-like-domains 9 |
| chr16_+_28834303 | 4.88 |
ENST00000340394.8
ENST00000325215.6 ENST00000395547.2 ENST00000336783.4 ENST00000382686.4 ENST00000564304.1 |
ATXN2L
|
ataxin 2-like |
| chrX_-_40506766 | 4.87 |
ENST00000378421.1
ENST00000440784.2 ENST00000327877.5 ENST00000378426.1 ENST00000378418.2 |
CXorf38
|
chromosome X open reading frame 38 |
| chr19_-_3869012 | 4.86 |
ENST00000592398.1
ENST00000262961.4 ENST00000439086.2 |
ZFR2
|
zinc finger RNA binding protein 2 |
| chr10_-_27444143 | 4.86 |
ENST00000477432.1
|
YME1L1
|
YME1-like 1 ATPase |
| chr9_-_131872928 | 4.86 |
ENST00000455830.2
ENST00000393384.3 ENST00000318080.2 |
CRAT
|
carnitine O-acetyltransferase |
| chr11_-_2160180 | 4.85 |
ENST00000381406.4
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr4_-_74847800 | 4.82 |
ENST00000296029.3
|
PF4
|
platelet factor 4 |
| chr4_+_72052964 | 4.74 |
ENST00000264485.5
ENST00000425175.1 |
SLC4A4
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
| chr12_+_10365404 | 4.74 |
ENST00000266458.5
ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr14_-_107114267 | 4.68 |
ENST00000454421.2
|
IGHV3-64
|
immunoglobulin heavy variable 3-64 |
| chr19_-_33793430 | 4.64 |
ENST00000498907.2
|
CEBPA
|
CCAAT/enhancer binding protein (C/EBP), alpha |
| chr9_+_134000948 | 4.61 |
ENST00000359428.5
ENST00000411637.2 ENST00000451030.1 |
NUP214
|
nucleoporin 214kDa |
| chr9_+_34458771 | 4.51 |
ENST00000437363.1
ENST00000242317.4 |
DNAI1
|
dynein, axonemal, intermediate chain 1 |
| chr1_+_9005917 | 4.49 |
ENST00000549778.1
ENST00000480186.3 ENST00000377443.2 ENST00000377436.3 ENST00000377442.2 |
CA6
|
carbonic anhydrase VI |
| chr19_-_4454081 | 4.48 |
ENST00000591919.1
|
UBXN6
|
UBX domain protein 6 |
| chr16_+_67381289 | 4.45 |
ENST00000435835.3
|
LRRC36
|
leucine rich repeat containing 36 |
| chr7_-_994302 | 4.45 |
ENST00000265846.5
|
ADAP1
|
ArfGAP with dual PH domains 1 |
| chr17_+_42219267 | 4.43 |
ENST00000319977.4
ENST00000585683.1 |
C17orf53
|
chromosome 17 open reading frame 53 |
| chr19_+_782755 | 4.41 |
ENST00000606242.1
ENST00000586061.1 |
AC006273.5
|
AC006273.5 |
| chr4_-_5894777 | 4.41 |
ENST00000324989.7
|
CRMP1
|
collapsin response mediator protein 1 |
| chr7_-_105029812 | 4.41 |
ENST00000482897.1
|
SRPK2
|
SRSF protein kinase 2 |
| chr19_+_48281803 | 4.40 |
ENST00000601048.1
|
SEPW1
|
selenoprotein W, 1 |
| chr5_-_180237445 | 4.39 |
ENST00000393340.3
|
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr16_+_20817761 | 4.37 |
ENST00000568046.1
ENST00000261377.6 |
AC004381.6
|
Putative RNA exonuclease NEF-sp |
| chr16_+_67381263 | 4.34 |
ENST00000541146.1
ENST00000563189.1 ENST00000290940.7 |
LRRC36
|
leucine rich repeat containing 36 |
| chr19_+_49999631 | 4.32 |
ENST00000270625.2
ENST00000596873.1 ENST00000594493.1 ENST00000599561.1 |
RPS11
|
ribosomal protein S11 |
| chr8_+_133931648 | 4.28 |
ENST00000519178.1
ENST00000542445.1 |
TG
|
thyroglobulin |
| chr19_+_4969116 | 4.27 |
ENST00000588337.1
ENST00000159111.4 ENST00000381759.4 |
KDM4B
|
lysine (K)-specific demethylase 4B |
| chr6_+_163148973 | 4.27 |
ENST00000366888.2
|
PACRG
|
PARK2 co-regulated |
| chr1_+_19923454 | 4.25 |
ENST00000602662.1
ENST00000602293.1 ENST00000322753.6 |
MINOS1-NBL1
MINOS1
|
MINOS1-NBL1 readthrough mitochondrial inner membrane organizing system 1 |
| chr12_-_13248562 | 4.17 |
ENST00000457134.2
ENST00000537302.1 |
GSG1
|
germ cell associated 1 |
| chr17_+_7531281 | 4.16 |
ENST00000575729.1
ENST00000340624.5 |
SHBG
|
sex hormone-binding globulin |
| chr12_-_10542617 | 4.15 |
ENST00000240618.6
|
KLRK1
|
killer cell lectin-like receptor subfamily K, member 1 |
| chr1_+_11866207 | 4.13 |
ENST00000312413.6
ENST00000346436.6 |
CLCN6
|
chloride channel, voltage-sensitive 6 |
| chr19_+_5455421 | 4.12 |
ENST00000222033.4
|
ZNRF4
|
zinc and ring finger 4 |
| chr2_+_220379052 | 4.12 |
ENST00000347842.3
ENST00000358078.4 |
ASIC4
|
acid-sensing (proton-gated) ion channel family member 4 |
| chr17_+_42219383 | 4.11 |
ENST00000245382.6
|
C17orf53
|
chromosome 17 open reading frame 53 |
| chr16_+_57702099 | 4.10 |
ENST00000333493.4
ENST00000327655.6 |
GPR97
|
G protein-coupled receptor 97 |
| chr11_-_116708302 | 4.10 |
ENST00000375320.1
ENST00000359492.2 ENST00000375329.2 ENST00000375323.1 |
APOA1
|
apolipoprotein A-I |
| chr10_+_134145614 | 4.10 |
ENST00000368615.3
ENST00000392638.2 ENST00000344079.5 ENST00000356571.4 ENST00000368614.3 |
LRRC27
|
leucine rich repeat containing 27 |
| chrX_-_48056199 | 4.09 |
ENST00000311798.1
ENST00000347757.1 |
SSX5
|
synovial sarcoma, X breakpoint 5 |
| chr17_-_73851285 | 4.08 |
ENST00000589642.1
ENST00000593002.1 ENST00000590221.1 ENST00000344296.4 ENST00000587374.1 ENST00000585462.1 ENST00000433525.2 ENST00000254806.3 |
WBP2
|
WW domain binding protein 2 |
| chr4_+_76932326 | 4.06 |
ENST00000513353.1
ENST00000341029.5 |
ART3
|
ADP-ribosyltransferase 3 |
| chr12_-_52887034 | 4.05 |
ENST00000330722.6
|
KRT6A
|
keratin 6A |
| chr19_+_36208877 | 4.02 |
ENST00000420124.1
ENST00000222270.7 ENST00000341701.1 |
KMT2B
|
Histone-lysine N-methyltransferase 2B |
| chr14_-_60097524 | 3.98 |
ENST00000342503.4
|
RTN1
|
reticulon 1 |
| chr17_-_1420182 | 3.98 |
ENST00000421807.2
|
INPP5K
|
inositol polyphosphate-5-phosphatase K |
| chr1_-_31845914 | 3.98 |
ENST00000373713.2
|
FABP3
|
fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) |
| chr12_-_13248732 | 3.96 |
ENST00000396302.3
|
GSG1
|
germ cell associated 1 |
| chr5_+_80597419 | 3.96 |
ENST00000254037.2
ENST00000407610.3 ENST00000380199.5 |
ZCCHC9
|
zinc finger, CCHC domain containing 9 |
| chr20_+_44098346 | 3.95 |
ENST00000372676.3
|
WFDC2
|
WAP four-disulfide core domain 2 |
| chr14_-_94856987 | 3.95 |
ENST00000449399.3
ENST00000404814.4 |
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr9_+_90112590 | 3.92 |
ENST00000472284.1
|
DAPK1
|
death-associated protein kinase 1 |
| chr6_-_46703430 | 3.91 |
ENST00000537365.1
|
PLA2G7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr19_-_19754354 | 3.90 |
ENST00000587238.1
|
GMIP
|
GEM interacting protein |
| chr11_+_111783450 | 3.88 |
ENST00000537382.1
|
HSPB2
|
Homo sapiens heat shock 27kDa protein 2 (HSPB2), mRNA. |
| chr21_-_46340884 | 3.88 |
ENST00000302347.5
ENST00000517819.1 |
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr14_-_94857004 | 3.87 |
ENST00000557492.1
ENST00000448921.1 ENST00000437397.1 ENST00000355814.4 ENST00000393088.4 |
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr19_-_12886327 | 3.87 |
ENST00000397668.3
ENST00000587178.1 ENST00000264827.5 |
HOOK2
|
hook microtubule-tethering protein 2 |
| chr14_-_106330072 | 3.87 |
ENST00000488476.1
|
IGHJ5
|
immunoglobulin heavy joining 5 |
| chr14_-_94856951 | 3.86 |
ENST00000553327.1
ENST00000556955.1 ENST00000557118.1 ENST00000440909.1 |
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr17_+_8339189 | 3.85 |
ENST00000585098.1
ENST00000380025.4 ENST00000402554.3 ENST00000584866.1 ENST00000582490.1 |
NDEL1
|
nudE neurodevelopment protein 1-like 1 |
| chr7_-_150974494 | 3.84 |
ENST00000392811.2
|
SMARCD3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chr20_+_42143136 | 3.83 |
ENST00000373134.1
|
L3MBTL1
|
l(3)mbt-like 1 (Drosophila) |
| chr1_-_229569834 | 3.80 |
ENST00000366684.3
ENST00000366683.2 |
ACTA1
|
actin, alpha 1, skeletal muscle |
| chr3_-_169899504 | 3.77 |
ENST00000474275.1
ENST00000484931.1 ENST00000494943.1 ENST00000497658.1 ENST00000465896.1 ENST00000475729.1 ENST00000495893.2 ENST00000481639.1 ENST00000467570.1 ENST00000466189.1 |
PHC3
|
polyhomeotic homolog 3 (Drosophila) |
| chr3_-_53080047 | 3.77 |
ENST00000482396.1
ENST00000358080.2 ENST00000296295.6 ENST00000394752.3 |
SFMBT1
|
Scm-like with four mbt domains 1 |
| chr16_-_55867146 | 3.77 |
ENST00000422046.2
|
CES1
|
carboxylesterase 1 |
| chr14_-_21492113 | 3.75 |
ENST00000554094.1
|
NDRG2
|
NDRG family member 2 |
| chr3_+_187871060 | 3.74 |
ENST00000448637.1
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr6_-_52859968 | 3.74 |
ENST00000370959.1
|
GSTA4
|
glutathione S-transferase alpha 4 |
| chr11_+_1889880 | 3.73 |
ENST00000405957.2
|
LSP1
|
lymphocyte-specific protein 1 |
| chr11_-_2170786 | 3.72 |
ENST00000300632.5
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr20_+_44098385 | 3.72 |
ENST00000217425.5
ENST00000339946.3 |
WFDC2
|
WAP four-disulfide core domain 2 |
| chr3_-_127542021 | 3.72 |
ENST00000434178.2
|
MGLL
|
monoglyceride lipase |
| chr5_-_95297534 | 3.72 |
ENST00000513343.1
ENST00000431061.2 |
ELL2
|
elongation factor, RNA polymerase II, 2 |
| chr15_+_63569731 | 3.71 |
ENST00000261879.5
|
APH1B
|
APH1B gamma secretase subunit |
| chr9_+_131218336 | 3.68 |
ENST00000372814.3
|
ODF2
|
outer dense fiber of sperm tails 2 |
| chr1_+_27114589 | 3.67 |
ENST00000431541.1
ENST00000449950.2 ENST00000374145.1 |
PIGV
|
phosphatidylinositol glycan anchor biosynthesis, class V |
| chr19_+_35630022 | 3.67 |
ENST00000589209.1
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr5_-_79551838 | 3.65 |
ENST00000509193.1
ENST00000512972.2 |
SERINC5
|
serine incorporator 5 |
| chr1_-_157108266 | 3.64 |
ENST00000326786.4
|
ETV3
|
ets variant 3 |
| chr16_-_89556942 | 3.63 |
ENST00000301030.4
|
ANKRD11
|
ankyrin repeat domain 11 |
| chr20_+_49348081 | 3.62 |
ENST00000371610.2
|
PARD6B
|
par-6 family cell polarity regulator beta |
| chr19_+_18208603 | 3.61 |
ENST00000262811.6
|
MAST3
|
microtubule associated serine/threonine kinase 3 |
| chr6_+_89790459 | 3.61 |
ENST00000369472.1
|
PNRC1
|
proline-rich nuclear receptor coactivator 1 |
| chr6_-_41747595 | 3.61 |
ENST00000373018.3
|
FRS3
|
fibroblast growth factor receptor substrate 3 |
| chr16_-_850723 | 3.56 |
ENST00000248150.4
|
GNG13
|
guanine nucleotide binding protein (G protein), gamma 13 |
| chr2_+_25015968 | 3.55 |
ENST00000380834.2
ENST00000473706.1 |
CENPO
|
centromere protein O |
| chr20_-_62258394 | 3.53 |
ENST00000370077.1
|
GMEB2
|
glucocorticoid modulatory element binding protein 2 |
| chr14_+_105155925 | 3.52 |
ENST00000330634.7
ENST00000398337.4 ENST00000392634.4 |
INF2
|
inverted formin, FH2 and WH2 domain containing |
| chr20_+_44637526 | 3.51 |
ENST00000372330.3
|
MMP9
|
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
| chrX_-_7895755 | 3.46 |
ENST00000444736.1
ENST00000537427.1 ENST00000442940.1 |
PNPLA4
|
patatin-like phospholipase domain containing 4 |
| chr12_-_54978086 | 3.46 |
ENST00000553113.1
|
PPP1R1A
|
protein phosphatase 1, regulatory (inhibitor) subunit 1A |
| chr7_-_108166505 | 3.45 |
ENST00000426128.2
ENST00000427008.1 ENST00000388728.5 ENST00000257694.8 ENST00000422087.1 ENST00000453144.1 ENST00000436062.1 |
PNPLA8
|
patatin-like phospholipase domain containing 8 |
| chr9_+_87285539 | 3.45 |
ENST00000359847.3
|
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr12_-_54582655 | 3.44 |
ENST00000504338.1
ENST00000514685.1 ENST00000504797.1 ENST00000513838.1 ENST00000505128.1 ENST00000337581.3 ENST00000503306.1 ENST00000243112.5 ENST00000514196.1 ENST00000506169.1 ENST00000507904.1 ENST00000508394.2 |
SMUG1
|
single-strand-selective monofunctional uracil-DNA glycosylase 1 |
| chr19_+_708910 | 3.44 |
ENST00000264560.7
|
PALM
|
paralemmin |
| chr14_-_106322288 | 3.43 |
ENST00000390559.2
|
IGHM
|
immunoglobulin heavy constant mu |
| chr9_+_90112767 | 3.42 |
ENST00000408954.3
|
DAPK1
|
death-associated protein kinase 1 |
| chr21_-_46348694 | 3.41 |
ENST00000355153.4
ENST00000397850.2 |
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr9_-_113800317 | 3.41 |
ENST00000374431.3
|
LPAR1
|
lysophosphatidic acid receptor 1 |
| chr8_-_98290087 | 3.40 |
ENST00000322128.3
|
TSPYL5
|
TSPY-like 5 |
| chr11_-_115630900 | 3.40 |
ENST00000537070.1
ENST00000499809.1 ENST00000514294.2 ENST00000535683.1 |
LINC00900
|
long intergenic non-protein coding RNA 900 |
| chr1_+_92414928 | 3.40 |
ENST00000362005.3
ENST00000370389.2 ENST00000399546.2 ENST00000423434.1 ENST00000394530.3 ENST00000440509.1 |
BRDT
|
bromodomain, testis-specific |
| chrX_+_52780318 | 3.39 |
ENST00000375515.3
ENST00000276049.6 |
SSX2B
|
synovial sarcoma, X breakpoint 2B |
| chr11_-_117103208 | 3.37 |
ENST00000320934.3
ENST00000530269.1 |
PCSK7
|
proprotein convertase subtilisin/kexin type 7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of GLI3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.6 | 46.5 | GO:0010900 | negative regulation of phosphatidylcholine catabolic process(GO:0010900) |
| 6.7 | 20.1 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 6.1 | 36.4 | GO:1902847 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 5.0 | 15.1 | GO:0071140 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 2.6 | 10.5 | GO:2001151 | regulation of renal water transport(GO:2001151) positive regulation of renal water transport(GO:2001153) |
| 2.4 | 7.1 | GO:1901624 | negative regulation of lymphocyte chemotaxis(GO:1901624) |
| 2.3 | 9.3 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 2.3 | 13.8 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 2.2 | 6.5 | GO:1900159 | iron assimilation(GO:0033212) iron assimilation by chelation and transport(GO:0033214) positive regulation of bone mineralization involved in bone maturation(GO:1900159) negative regulation of tumor necrosis factor (ligand) superfamily member 11 production(GO:2000308) |
| 2.1 | 12.8 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 2.1 | 2.1 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 2.1 | 2.1 | GO:0032416 | negative regulation of sodium:proton antiporter activity(GO:0032416) |
| 2.0 | 10.2 | GO:0033274 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 2.0 | 23.8 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 1.9 | 9.4 | GO:0002905 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 1.8 | 5.4 | GO:0071810 | circadian temperature homeostasis(GO:0060086) regulation of fever generation by regulation of prostaglandin secretion(GO:0071810) positive regulation of fever generation by positive regulation of prostaglandin secretion(GO:0071812) positive regulation of ERK1 and ERK2 cascade via TNFSF11-mediated signaling(GO:0071848) regulation of fever generation by prostaglandin secretion(GO:0100009) |
| 1.8 | 5.4 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 1.8 | 8.9 | GO:0045918 | negative regulation of cytolysis(GO:0045918) |
| 1.7 | 13.9 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 1.7 | 6.9 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 1.7 | 30.5 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 1.7 | 6.7 | GO:1990262 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 1.7 | 13.3 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 1.7 | 6.6 | GO:1904566 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 1.7 | 5.0 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 1.6 | 1.6 | GO:0050684 | regulation of mRNA processing(GO:0050684) |
| 1.6 | 4.9 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 1.6 | 4.8 | GO:0051037 | histone displacement(GO:0001207) regulation of transcription involved in meiotic cell cycle(GO:0051037) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 1.6 | 9.6 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 1.6 | 10.9 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 1.5 | 4.4 | GO:1904530 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 1.5 | 7.3 | GO:0030070 | insulin processing(GO:0030070) |
| 1.3 | 6.7 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 1.3 | 4.0 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 1.3 | 8.9 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 1.2 | 6.2 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing by siRNA(GO:0090625) mRNA cleavage involved in gene silencing(GO:0098795) |
| 1.2 | 7.0 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 1.1 | 3.3 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 1.1 | 13.1 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 1.1 | 9.6 | GO:1903898 | negative regulation of PERK-mediated unfolded protein response(GO:1903898) negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 1.0 | 6.2 | GO:0010732 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 1.0 | 3.1 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 1.0 | 3.1 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) negative regulation of DNA catabolic process(GO:1903625) regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 1.0 | 14.4 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 1.0 | 4.1 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) |
| 1.0 | 14.1 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 1.0 | 5.0 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 1.0 | 4.0 | GO:0042335 | cuticle development(GO:0042335) |
| 1.0 | 3.0 | GO:1904328 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 1.0 | 6.8 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 1.0 | 4.9 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 1.0 | 8.6 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.9 | 6.5 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.9 | 3.7 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.9 | 4.4 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.8 | 4.2 | GO:0099551 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.8 | 2.5 | GO:0030451 | regulation of complement activation, alternative pathway(GO:0030451) negative regulation of complement activation, alternative pathway(GO:0045957) |
| 0.8 | 3.2 | GO:0034959 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.8 | 2.4 | GO:1904404 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 0.8 | 2.4 | GO:0009447 | putrescine catabolic process(GO:0009447) |
| 0.8 | 2.4 | GO:2000824 | negative regulation of androgen receptor activity(GO:2000824) |
| 0.8 | 0.8 | GO:0046164 | alcohol catabolic process(GO:0046164) |
| 0.8 | 3.9 | GO:0086047 | membrane depolarization during Purkinje myocyte cell action potential(GO:0086047) |
| 0.8 | 3.8 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.8 | 3.0 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.7 | 2.2 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) establishment of meiotic spindle localization(GO:0051295) |
| 0.7 | 3.6 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.7 | 12.8 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.7 | 2.0 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.7 | 4.0 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.7 | 3.3 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.6 | 3.2 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.6 | 4.5 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.6 | 7.0 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.6 | 2.5 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.6 | 12.6 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.6 | 6.0 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.6 | 2.4 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.6 | 4.1 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.6 | 2.3 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.6 | 9.4 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.5 | 2.2 | GO:0032053 | ciliary basal body organization(GO:0032053) positive regulation of protein localization to cilium(GO:1903566) |
| 0.5 | 3.3 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.5 | 1.6 | GO:1902996 | regulation of neurofibrillary tangle assembly(GO:1902996) |
| 0.5 | 2.2 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.5 | 4.3 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.5 | 2.1 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.5 | 2.7 | GO:0045626 | negative regulation of T-helper 1 cell differentiation(GO:0045626) response to odorant(GO:1990834) |
| 0.5 | 4.3 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.5 | 2.1 | GO:1990418 | response to insulin-like growth factor stimulus(GO:1990418) |
| 0.5 | 1.6 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.5 | 3.6 | GO:0009597 | detection of virus(GO:0009597) |
| 0.5 | 10.4 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.5 | 2.6 | GO:1904098 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.5 | 2.6 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.5 | 1.5 | GO:2001301 | lipoxin biosynthetic process(GO:2001301) lipoxin A4 metabolic process(GO:2001302) lipoxin A4 biosynthetic process(GO:2001303) |
| 0.5 | 3.0 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.5 | 10.5 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.5 | 2.0 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.5 | 1.5 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.5 | 2.0 | GO:0071373 | cellular response to luteinizing hormone stimulus(GO:0071373) |
| 0.5 | 3.4 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.5 | 2.0 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.5 | 8.2 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.5 | 4.3 | GO:0097012 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.5 | 1.9 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.5 | 1.4 | GO:1900082 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.5 | 0.5 | GO:2000196 | positive regulation of female gonad development(GO:2000196) |
| 0.5 | 0.9 | GO:0039694 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.5 | 1.4 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.5 | 6.5 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.5 | 1.8 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.5 | 3.6 | GO:0015705 | iodide transport(GO:0015705) |
| 0.5 | 3.2 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.4 | 1.8 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.4 | 1.3 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.4 | 3.6 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
| 0.4 | 4.9 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.4 | 1.3 | GO:0016999 | antibiotic metabolic process(GO:0016999) cellular amide catabolic process(GO:0043605) |
| 0.4 | 1.8 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.4 | 1.7 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.4 | 3.4 | GO:0046457 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.4 | 3.9 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.4 | 12.7 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.4 | 6.4 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.4 | 2.5 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.4 | 3.8 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.4 | 4.6 | GO:0045945 | urea cycle(GO:0000050) positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.4 | 2.1 | GO:0035356 | cellular triglyceride homeostasis(GO:0035356) |
| 0.4 | 3.8 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.4 | 1.7 | GO:1903597 | negative regulation of gap junction assembly(GO:1903597) |
| 0.4 | 1.2 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 0.4 | 6.6 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.4 | 1.2 | GO:1902910 | regulation of monophenol monooxygenase activity(GO:0032771) positive regulation of monophenol monooxygenase activity(GO:0032773) negative regulation of catagen(GO:0051796) regulation of hair cycle by canonical Wnt signaling pathway(GO:0060901) positive regulation of melanosome transport(GO:1902910) |
| 0.4 | 0.8 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.4 | 1.9 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.4 | 1.9 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.4 | 3.8 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) |
| 0.4 | 1.1 | GO:0019474 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.4 | 2.2 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.4 | 2.5 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.4 | 1.4 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.4 | 1.1 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.4 | 7.8 | GO:0097502 | mannosylation(GO:0097502) |
| 0.4 | 1.4 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.3 | 1.0 | GO:0044705 | mating behavior(GO:0007617) reproductive behavior(GO:0019098) multi-organism reproductive behavior(GO:0044705) |
| 0.3 | 1.0 | GO:0030264 | nuclear fragmentation involved in apoptotic nuclear change(GO:0030264) positive regulation of cardiac vascular smooth muscle cell differentiation(GO:2000724) |
| 0.3 | 1.0 | GO:0070086 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) ubiquitin-dependent endocytosis(GO:0070086) regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.3 | 1.0 | GO:0090155 | glucosylceramide biosynthetic process(GO:0006679) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.3 | 2.9 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.3 | 4.2 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.3 | 2.9 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.3 | 0.3 | GO:0086100 | endothelin receptor signaling pathway(GO:0086100) |
| 0.3 | 2.6 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.3 | 0.6 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.3 | 4.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.3 | 5.1 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.3 | 5.7 | GO:0090494 | dopamine uptake involved in synaptic transmission(GO:0051583) catecholamine uptake involved in synaptic transmission(GO:0051934) catecholamine uptake(GO:0090493) dopamine uptake(GO:0090494) |
| 0.3 | 1.0 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.3 | 0.9 | GO:0003257 | positive regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003257) mesoderm migration involved in gastrulation(GO:0007509) positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.3 | 0.9 | GO:1903960 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) negative regulation of anion transmembrane transport(GO:1903960) |
| 0.3 | 0.9 | GO:0001554 | luteolysis(GO:0001554) |
| 0.3 | 4.0 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.3 | 6.0 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.3 | 1.5 | GO:0034351 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.3 | 0.9 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.3 | 1.2 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.3 | 0.9 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.3 | 2.8 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.3 | 2.2 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.3 | 2.8 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.3 | 10.2 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.3 | 6.1 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.3 | 2.5 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.3 | 4.4 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.3 | 6.5 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.3 | 3.3 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.3 | 1.6 | GO:0019732 | antifungal humoral response(GO:0019732) |
| 0.3 | 2.2 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.3 | 1.3 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.3 | 1.9 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.3 | 1.3 | GO:0051384 | response to glucocorticoid(GO:0051384) |
| 0.3 | 1.3 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.3 | 3.4 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.3 | 1.6 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.3 | 13.5 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.3 | 2.6 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.3 | 2.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.3 | 0.8 | GO:0007418 | ventral midline development(GO:0007418) smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.3 | 2.3 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.3 | 1.5 | GO:1904628 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.3 | 1.5 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.2 | 15.8 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.2 | 1.0 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.2 | 6.1 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.2 | 1.4 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 0.2 | 1.2 | GO:0044860 | protein localization to plasma membrane raft(GO:0044860) |
| 0.2 | 0.7 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 0.2 | 0.7 | GO:0009584 | detection of visible light(GO:0009584) |
| 0.2 | 4.6 | GO:0034776 | response to histamine(GO:0034776) cellular response to histamine(GO:0071420) |
| 0.2 | 1.8 | GO:1902525 | regulation of protein monoubiquitination(GO:1902525) |
| 0.2 | 1.8 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.2 | 0.7 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.2 | 0.9 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.2 | 1.8 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.2 | 0.4 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.2 | 4.6 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.2 | 0.7 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.2 | 17.1 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.2 | 5.3 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.2 | 3.7 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.2 | 0.6 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.2 | 1.5 | GO:1903433 | regulation of constitutive secretory pathway(GO:1903433) |
| 0.2 | 2.1 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.2 | 0.6 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.2 | 4.3 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.2 | 0.6 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.2 | 0.8 | GO:0032415 | regulation of sodium:proton antiporter activity(GO:0032415) |
| 0.2 | 8.7 | GO:0036465 | synaptic vesicle recycling(GO:0036465) |
| 0.2 | 2.8 | GO:0042711 | maternal behavior(GO:0042711) parental behavior(GO:0060746) |
| 0.2 | 2.0 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.2 | 1.2 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.2 | 3.7 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.2 | 5.2 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.2 | 3.7 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.2 | 5.7 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.2 | 1.1 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.2 | 1.9 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.2 | 0.9 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.2 | 1.9 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.2 | 0.6 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.2 | 1.1 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.2 | 3.6 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.2 | 1.8 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.2 | 1.8 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.2 | 1.4 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.2 | 1.6 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.2 | 2.8 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.2 | 0.7 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.2 | 4.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.2 | 1.5 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.2 | 0.7 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.2 | 0.2 | GO:0002625 | regulation of T cell antigen processing and presentation(GO:0002625) |
| 0.2 | 1.7 | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
| 0.2 | 1.7 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.2 | 1.5 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.2 | 0.7 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.2 | 0.5 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.2 | 16.3 | GO:0007286 | spermatid development(GO:0007286) |
| 0.2 | 5.3 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.2 | 6.5 | GO:0009214 | cyclic nucleotide catabolic process(GO:0009214) |
| 0.2 | 0.9 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 6.1 | GO:0003254 | regulation of membrane depolarization(GO:0003254) |
| 0.1 | 1.3 | GO:0090037 | positive regulation of protein kinase C signaling(GO:0090037) |
| 0.1 | 1.5 | GO:0015871 | choline transport(GO:0015871) |
| 0.1 | 2.9 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.1 | 3.1 | GO:0045821 | positive regulation of nucleotide catabolic process(GO:0030813) positive regulation of glycolytic process(GO:0045821) positive regulation of cofactor metabolic process(GO:0051194) positive regulation of coenzyme metabolic process(GO:0051197) |
| 0.1 | 1.6 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.1 | 0.6 | GO:0072396 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) |
| 0.1 | 8.6 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 2.4 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 1.3 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.1 | 1.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 | 0.6 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.1 | 2.1 | GO:1901642 | nucleoside transmembrane transport(GO:1901642) |
| 0.1 | 0.8 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.1 | 0.3 | GO:0072143 | renal interstitial fibroblast development(GO:0072141) mesangial cell development(GO:0072143) glomerular mesangial cell development(GO:0072144) |
| 0.1 | 1.7 | GO:0015671 | gas transport(GO:0015669) oxygen transport(GO:0015671) |
| 0.1 | 1.4 | GO:0060850 | regulation of transcription involved in cell fate commitment(GO:0060850) |
| 0.1 | 5.9 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 1.4 | GO:0010212 | response to ionizing radiation(GO:0010212) |
| 0.1 | 2.6 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 5.4 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.1 | 2.9 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.1 | 1.3 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 4.1 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 0.8 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.1 | 3.4 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.1 | 1.0 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.8 | GO:0045794 | negative regulation of cell volume(GO:0045794) |
| 0.1 | 1.8 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.1 | 2.7 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.1 | 1.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 1.2 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.1 | 0.4 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.1 | 3.0 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 3.7 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 0.5 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 2.7 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.1 | 2.8 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.1 | 0.6 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.1 | 2.6 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 | 1.9 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.1 | 5.6 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 2.6 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 0.3 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 | 1.4 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.1 | 0.1 | GO:1904386 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.1 | 1.1 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.1 | 0.2 | GO:0044340 | canonical Wnt signaling pathway involved in regulation of cell proliferation(GO:0044340) |
| 0.1 | 2.2 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.1 | 0.5 | GO:2001256 | negative regulation of mast cell activation involved in immune response(GO:0033007) negative regulation of mast cell degranulation(GO:0043305) regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 1.8 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.1 | 1.0 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.1 | 0.5 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.1 | 1.9 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.1 | 0.7 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.1 | 3.1 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.1 | 1.4 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.1 | 0.3 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.1 | 1.8 | GO:0010762 | regulation of fibroblast migration(GO:0010762) |
| 0.1 | 0.5 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 2.7 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.1 | 1.0 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.1 | 0.2 | GO:0035989 | tendon development(GO:0035989) negative regulation of gastrulation(GO:2000542) |
| 0.1 | 0.7 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.1 | 1.0 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.1 | 0.5 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.3 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.1 | 3.0 | GO:0051972 | regulation of telomerase activity(GO:0051972) |
| 0.1 | 0.6 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.1 | 3.3 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 1.0 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.1 | 1.5 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.1 | 3.5 | GO:0034113 | heterotypic cell-cell adhesion(GO:0034113) |
| 0.1 | 0.1 | GO:0071168 | protein localization to chromatin(GO:0071168) |
| 0.1 | 0.3 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 0.6 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.1 | 0.4 | GO:0030728 | ovulation(GO:0030728) |
| 0.1 | 0.8 | GO:0045023 | G0 to G1 transition(GO:0045023) |
| 0.1 | 0.3 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.1 | 0.3 | GO:0043931 | ossification involved in bone maturation(GO:0043931) |
| 0.1 | 1.9 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 0.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.5 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.1 | 0.1 | GO:0021940 | positive regulation of cerebellar granule cell precursor proliferation(GO:0021940) |
| 0.1 | 0.2 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.1 | 1.2 | GO:0006706 | steroid catabolic process(GO:0006706) |
| 0.1 | 0.4 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.1 | 0.4 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.1 | 0.9 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.1 | 1.9 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 4.0 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 1.0 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.1 | 3.7 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.1 | 2.1 | GO:0090280 | positive regulation of calcium ion import(GO:0090280) |
| 0.1 | 0.4 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 1.7 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.1 | 1.0 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.1 | 4.4 | GO:0005977 | glycogen metabolic process(GO:0005977) |
| 0.1 | 2.4 | GO:0003407 | neural retina development(GO:0003407) |
| 0.1 | 0.9 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 0.3 | GO:0010529 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 | 1.4 | GO:0031102 | neuron projection regeneration(GO:0031102) |
| 0.0 | 1.2 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 1.2 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.3 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.7 | GO:0008206 | bile acid metabolic process(GO:0008206) |
| 0.0 | 1.1 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 0.7 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.0 | 1.2 | GO:0051966 | regulation of synaptic transmission, glutamatergic(GO:0051966) |
| 0.0 | 0.2 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.0 | 2.2 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.0 | 0.5 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 1.2 | GO:0030866 | cortical cytoskeleton organization(GO:0030865) cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 1.4 | GO:1901186 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) positive regulation of ERBB signaling pathway(GO:1901186) |
| 0.0 | 0.3 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 3.2 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 2.3 | GO:0030509 | BMP signaling pathway(GO:0030509) |
| 0.0 | 0.3 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.5 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.2 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 1.5 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 1.9 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.0 | 0.5 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.3 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 1.1 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.0 | 0.2 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.3 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.1 | GO:0048541 | mucosal-associated lymphoid tissue development(GO:0048537) Peyer's patch development(GO:0048541) Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 2.0 | GO:0042102 | positive regulation of T cell proliferation(GO:0042102) |
| 0.0 | 0.2 | GO:0046855 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.8 | GO:0098780 | response to mitochondrial depolarisation(GO:0098780) |
| 0.0 | 0.9 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 1.1 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.9 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.4 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 1.4 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.6 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.9 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.3 | GO:0007625 | grooming behavior(GO:0007625) |
| 0.0 | 0.2 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.0 | 2.3 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 1.4 | GO:0035418 | protein localization to synapse(GO:0035418) |
| 0.0 | 1.5 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.1 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 1.4 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.4 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.6 | GO:0006476 | protein deacetylation(GO:0006476) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.8 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.3 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.3 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.2 | GO:0010990 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.0 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.0 | 0.3 | GO:0015698 | inorganic anion transport(GO:0015698) |
| 0.0 | 9.0 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 0.0 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.0 | 0.1 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.0 | 1.1 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.6 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.8 | GO:0001764 | neuron migration(GO:0001764) |
| 0.0 | 0.6 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.8 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 46.7 | GO:0042627 | chylomicron(GO:0042627) |
| 2.5 | 12.6 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 2.4 | 40.5 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 2.3 | 11.3 | GO:0001652 | granular component(GO:0001652) |
| 2.3 | 9.0 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 2.1 | 33.7 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 2.0 | 14.0 | GO:0001520 | outer dense fiber(GO:0001520) |
| 1.6 | 4.8 | GO:0097679 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) other organism cytoplasm(GO:0097679) |
| 1.5 | 4.6 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 1.5 | 10.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 1.3 | 9.4 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 1.3 | 3.9 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 1.3 | 6.3 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 1.2 | 20.3 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 1.2 | 20.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 1.0 | 3.9 | GO:0070695 | FHF complex(GO:0070695) |
| 1.0 | 8.6 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.9 | 4.7 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.9 | 2.8 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.9 | 3.7 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.9 | 6.5 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) phagocytic vesicle lumen(GO:0097013) |
| 0.9 | 12.5 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.9 | 7.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.8 | 4.9 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.8 | 3.2 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.8 | 11.0 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.7 | 3.0 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.7 | 5.9 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.7 | 2.2 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.7 | 4.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.7 | 2.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.7 | 2.0 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.6 | 2.5 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.6 | 3.0 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.6 | 9.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.5 | 3.6 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.5 | 1.4 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.5 | 9.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.4 | 2.5 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.4 | 7.7 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.4 | 19.7 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.4 | 1.5 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.4 | 5.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.4 | 3.0 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.4 | 1.5 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.4 | 16.7 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.3 | 1.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.3 | 3.7 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.3 | 3.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.3 | 0.6 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.3 | 3.2 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.3 | 0.9 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.3 | 1.8 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.3 | 3.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.3 | 3.3 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.3 | 3.8 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.3 | 2.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.3 | 11.7 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.3 | 2.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.3 | 3.1 | GO:0000800 | lateral element(GO:0000800) |
| 0.2 | 2.9 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.2 | 1.9 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.2 | 3.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.2 | 0.7 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.2 | 1.8 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.2 | 1.8 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.2 | 2.4 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.2 | 1.4 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.2 | 11.5 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.2 | 2.2 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.2 | 3.8 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.2 | 1.3 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.2 | 2.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 6.6 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.2 | 1.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.2 | 3.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 3.8 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.2 | 1.4 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.2 | 0.5 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.2 | 1.2 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.2 | 36.5 | GO:0001726 | ruffle(GO:0001726) |
| 0.2 | 3.6 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 5.5 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.1 | 5.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 1.0 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 0.6 | GO:0043293 | apoptosome(GO:0043293) |
| 0.1 | 2.1 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 0.4 | GO:0016938 | kinesin I complex(GO:0016938) |
| 0.1 | 2.9 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 1.4 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 1.8 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 2.3 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 0.5 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 0.8 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 3.9 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 2.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 4.7 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 3.7 | GO:0090568 | nuclear transcriptional repressor complex(GO:0090568) |
| 0.1 | 11.4 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 4.2 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 1.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 2.6 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 9.6 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 4.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 4.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 1.8 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 1.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 2.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 0.5 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.1 | 5.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 3.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 1.8 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 1.5 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 2.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 2.9 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 1.6 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.1 | 4.1 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 6.0 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.1 | 6.7 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 1.6 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 6.3 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 4.9 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.1 | 2.0 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 2.0 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 0.6 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 1.2 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.1 | 2.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 0.4 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 3.8 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 1.5 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.1 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 4.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 3.6 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 2.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 1.0 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 10.9 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 2.5 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.8 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 5.5 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.3 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 3.4 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 2.9 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.5 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.6 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 1.8 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 1.7 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 1.7 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 3.0 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 3.8 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.0 | 2.7 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 2.1 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 13.1 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 0.6 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.2 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 2.7 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 2.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 5.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 2.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 4.6 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.9 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.0 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.0 | 21.1 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 6.4 | 50.8 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 2.3 | 9.4 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 2.2 | 9.0 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 2.0 | 10.2 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 1.9 | 7.7 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 1.9 | 15.1 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 1.9 | 5.7 | GO:0005330 | dopamine:sodium symporter activity(GO:0005330) |
| 1.8 | 9.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 1.8 | 36.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 1.8 | 12.6 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 1.8 | 10.5 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 1.7 | 6.9 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 1.6 | 6.3 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 1.5 | 3.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 1.5 | 9.2 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 1.4 | 12.8 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 1.3 | 4.0 | GO:0070538 | oleic acid binding(GO:0070538) |
| 1.3 | 3.8 | GO:0032093 | SAM domain binding(GO:0032093) |
| 1.2 | 4.9 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 1.1 | 14.7 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 1.1 | 6.8 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 1.1 | 3.3 | GO:0004961 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 1.1 | 3.2 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 1.0 | 3.0 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 1.0 | 2.9 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 1.0 | 2.9 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 1.0 | 4.8 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.9 | 2.8 | GO:0004040 | amidase activity(GO:0004040) fucose binding(GO:0042806) |
| 0.9 | 0.9 | GO:0098847 | sequence-specific single stranded DNA binding(GO:0098847) |
| 0.9 | 3.7 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.9 | 27.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.9 | 3.6 | GO:0031716 | calcitonin receptor activity(GO:0004948) calcitonin receptor binding(GO:0031716) |
| 0.9 | 5.3 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.9 | 3.4 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.9 | 3.4 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.8 | 4.2 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.8 | 14.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.8 | 2.5 | GO:0004877 | complement component C4b receptor activity(GO:0001861) complement component C3b receptor activity(GO:0004877) |
| 0.8 | 4.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.8 | 8.9 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.8 | 2.4 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.8 | 3.2 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.8 | 7.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.8 | 3.2 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.8 | 3.8 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.7 | 4.5 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.7 | 2.2 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.7 | 2.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.7 | 2.2 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.7 | 5.0 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 0.7 | 2.1 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.7 | 2.0 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.7 | 2.6 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.6 | 2.6 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.6 | 2.5 | GO:0016160 | amylase activity(GO:0016160) |
| 0.6 | 2.5 | GO:0042019 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.6 | 4.9 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.6 | 4.9 | GO:0016406 | carnitine O-acyltransferase activity(GO:0016406) |
| 0.6 | 1.8 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.6 | 2.4 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.6 | 4.7 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.6 | 1.7 | GO:0004913 | interleukin-4 receptor activity(GO:0004913) |
| 0.6 | 2.2 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.5 | 4.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.5 | 6.4 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.5 | 7.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.5 | 4.2 | GO:0005497 | androgen binding(GO:0005497) |
| 0.5 | 8.3 | GO:0015250 | water channel activity(GO:0015250) |
| 0.5 | 5.7 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.5 | 1.5 | GO:0047977 | hepoxilin-epoxide hydrolase activity(GO:0047977) |
| 0.5 | 1.5 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.5 | 6.3 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.5 | 2.4 | GO:0052901 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 0.5 | 8.5 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.5 | 5.6 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.5 | 1.4 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.5 | 6.5 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.5 | 3.2 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.4 | 8.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.4 | 2.2 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.4 | 1.3 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.4 | 9.6 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.4 | 3.5 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.4 | 3.0 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.4 | 5.1 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.4 | 1.3 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.4 | 4.2 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.4 | 4.6 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.4 | 12.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.4 | 6.3 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.4 | 7.8 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.4 | 18.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.4 | 7.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.4 | 1.9 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.4 | 1.5 | GO:0030107 | HLA-A specific inhibitory MHC class I receptor activity(GO:0030107) |
| 0.4 | 10.6 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.4 | 19.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.4 | 1.8 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.3 | 1.0 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.3 | 3.8 | GO:0004568 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 0.3 | 1.4 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.3 | 0.7 | GO:0019976 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.3 | 4.7 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.3 | 33.1 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.3 | 1.0 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.3 | 6.0 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.3 | 1.0 | GO:0047322 | [hydroxymethylglutaryl-CoA reductase (NADPH)] kinase activity(GO:0047322) [acetyl-CoA carboxylase] kinase activity(GO:0050405) |
| 0.3 | 2.3 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.3 | 1.9 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.3 | 1.0 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.3 | 5.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.3 | 1.3 | GO:0030760 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.3 | 6.6 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.3 | 13.6 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.3 | 0.9 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) |
| 0.3 | 2.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.3 | 5.1 | GO:0031005 | filamin binding(GO:0031005) |
| 0.3 | 8.0 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.3 | 3.3 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.3 | 1.5 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.3 | 4.0 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.3 | 0.9 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.3 | 1.4 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.3 | 6.2 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.3 | 0.8 | GO:0019961 | interferon binding(GO:0019961) |
| 0.3 | 11.7 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.3 | 0.8 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.3 | 4.0 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.3 | 17.0 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.3 | 7.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.3 | 5.7 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.3 | 3.4 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.3 | 2.8 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.2 | 1.5 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.2 | 2.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.2 | 0.7 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.2 | 4.0 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.2 | 0.9 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.2 | 0.7 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.2 | 3.7 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.2 | 0.6 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.2 | 2.1 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.2 | 1.2 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.2 | 1.2 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.2 | 1.8 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.2 | 1.6 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.2 | 1.2 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.2 | 2.5 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.2 | 5.0 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.2 | 4.4 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.2 | 1.1 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.2 | 3.2 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.2 | 1.5 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.2 | 1.8 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.2 | 1.3 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.2 | 1.8 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.2 | 1.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.2 | 2.1 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.2 | 0.9 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.2 | 0.7 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.2 | 7.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.2 | 1.3 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.2 | 1.0 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 5.5 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.2 | 2.6 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.2 | 1.3 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.2 | 0.5 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.2 | 0.8 | GO:0097157 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) pre-mRNA intronic binding(GO:0097157) |
| 0.2 | 3.1 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.2 | 1.2 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.2 | 2.9 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.2 | 2.5 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.2 | 3.6 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 4.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 1.6 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 1.8 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 1.7 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 1.2 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 15.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.0 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.1 | 1.9 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 1.8 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.1 | 2.9 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 1.3 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.1 | 1.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 1.7 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 2.6 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.9 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.1 | 1.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 5.3 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.1 | 6.6 | GO:0004112 | cyclic-nucleotide phosphodiesterase activity(GO:0004112) |
| 0.1 | 0.7 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.1 | 1.5 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.5 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.1 | 3.5 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.1 | 2.8 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 12.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 2.0 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 1.5 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 18.3 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 0.6 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 1.6 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.1 | 2.1 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.1 | 0.3 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.1 | 2.2 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 0.5 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 0.6 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.1 | 0.6 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.1 | 1.6 | GO:0016918 | retinal binding(GO:0016918) |
| 0.1 | 4.1 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 2.3 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 2.1 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 0.6 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.1 | 3.6 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 2.5 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 0.4 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 1.6 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 3.3 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 2.4 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 9.4 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 0.9 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 2.3 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 1.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 2.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 7.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 15.1 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.1 | 2.1 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.1 | 1.1 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.1 | 0.3 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.1 | 0.2 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.1 | 0.6 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 1.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 1.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 5.2 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 1.1 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 0.6 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.1 | 0.6 | GO:0043028 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.1 | 1.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 4.1 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 0.7 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 5.0 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 2.1 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.5 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.3 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.3 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 1.4 | GO:0008066 | glutamate receptor activity(GO:0008066) |
| 0.0 | 0.5 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.9 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 8.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.0 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.8 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) |
| 0.0 | 1.2 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 2.3 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.7 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 3.7 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.0 | 2.2 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 2.6 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 0.3 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 6.8 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.3 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.6 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.2 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.9 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.6 | GO:0016917 | GABA receptor activity(GO:0016917) |
| 0.0 | 0.8 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.0 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.3 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 0.1 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.0 | 0.8 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.0 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 1.2 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 1.1 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 2.5 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 0.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.4 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.2 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 1.0 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.0 | 0.1 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.1 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.1 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.4 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.2 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.1 | GO:0048487 | beta-tubulin binding(GO:0048487) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 6.6 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.7 | 1.3 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.5 | 3.7 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.4 | 22.3 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.4 | 9.0 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.3 | 6.3 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.3 | 47.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.3 | 11.8 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.3 | 0.9 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.3 | 0.8 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.3 | 9.2 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.3 | 13.4 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.2 | 2.4 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.2 | 4.5 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.2 | 4.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.2 | 0.8 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.2 | 3.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.2 | 9.9 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.2 | 1.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.2 | 4.9 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.2 | 5.9 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.2 | 8.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.2 | 6.7 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.2 | 0.6 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.2 | 4.8 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.2 | 6.6 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 2.9 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.1 | 4.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 6.8 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 4.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 4.8 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.1 | 3.1 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 3.7 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 1.2 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 8.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 2.4 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 20.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.1 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 3.7 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 2.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 2.6 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 1.8 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 4.0 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.1 | 5.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 2.7 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 2.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 0.9 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 0.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 3.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 5.9 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 1.6 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 0.7 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.1 | 2.3 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 3.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 0.3 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.1 | 2.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 1.1 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 2.5 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.1 | 2.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 2.3 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 12.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 0.9 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 3.0 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 0.5 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 1.3 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.8 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.0 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.3 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.2 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 5.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.2 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.5 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 3.6 | REACTOME DOWNSTREAM SIGNALING OF ACTIVATED FGFR | Genes involved in Downstream signaling of activated FGFR |
| 1.6 | 6.3 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 1.4 | 6.8 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 1.1 | 2.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.9 | 0.9 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.9 | 12.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.8 | 27.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.8 | 1.6 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.6 | 17.0 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.5 | 2.2 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.4 | 12.0 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.4 | 5.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.4 | 9.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.4 | 5.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.4 | 10.0 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.4 | 5.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.3 | 9.0 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.3 | 7.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.3 | 5.1 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.3 | 3.4 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.3 | 0.9 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.3 | 6.8 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.3 | 39.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.2 | 10.9 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.2 | 3.7 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.2 | 4.4 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.2 | 4.2 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.2 | 21.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.2 | 7.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 3.6 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.2 | 6.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 3.6 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.2 | 6.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.2 | 3.4 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.2 | 5.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 10.2 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.2 | 2.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.2 | 1.0 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.2 | 4.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 4.6 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.2 | 3.6 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 4.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 2.2 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 0.9 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 5.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 5.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 3.2 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 5.2 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.1 | 7.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 2.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.1 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.1 | 6.7 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 2.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 0.9 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 2.0 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.1 | 1.8 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.1 | 8.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 2.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.6 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.1 | 1.0 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 4.6 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.1 | 1.2 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 5.9 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 8.9 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 9.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 1.6 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 2.0 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.1 | 2.9 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 4.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 1.8 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.1 | 2.7 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.1 | 0.4 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 2.2 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 2.1 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 2.3 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 1.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 0.7 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 1.5 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 1.9 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 6.1 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.6 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 4.3 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 2.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.0 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.6 | REACTOME SIGNALING BY FGFR1 MUTANTS | Genes involved in Signaling by FGFR1 mutants |
| 0.0 | 0.9 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 2.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.7 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.8 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.3 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.1 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.0 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |