Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
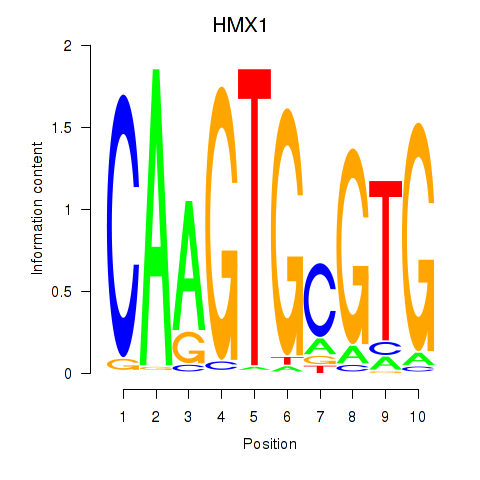
Results for HMX1
Z-value: 0.86
Transcription factors associated with HMX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HMX1
|
ENSG00000215612.5 | H6 family homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HMX1 | hg19_v2_chr4_-_8873531_8873543 | -0.52 | 2.1e-16 | Click! |
Activity profile of HMX1 motif
Sorted Z-values of HMX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_49066811 | 23.45 |
ENST00000442157.1
ENST00000326739.4 |
IMPDH2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2 |
| chr22_-_37640277 | 13.25 |
ENST00000401529.3
ENST00000249071.6 |
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr3_+_151986709 | 12.83 |
ENST00000495875.2
ENST00000493459.1 ENST00000324210.5 ENST00000459747.1 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr12_-_7077125 | 12.61 |
ENST00000545555.2
|
PHB2
|
prohibitin 2 |
| chrX_+_131157322 | 10.83 |
ENST00000481105.1
ENST00000354719.6 ENST00000394335.2 |
MST4
|
Serine/threonine-protein kinase MST4 |
| chrX_+_131157290 | 10.79 |
ENST00000394334.2
|
MST4
|
Serine/threonine-protein kinase MST4 |
| chr16_+_85645007 | 10.55 |
ENST00000405402.2
|
GSE1
|
Gse1 coiled-coil protein |
| chr13_+_37574678 | 10.37 |
ENST00000389704.3
|
EXOSC8
|
exosome component 8 |
| chr19_+_41768401 | 9.94 |
ENST00000352456.3
ENST00000595018.1 ENST00000597725.1 |
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chrX_-_153285251 | 9.91 |
ENST00000444230.1
ENST00000393682.1 ENST00000393687.2 ENST00000429936.2 ENST00000369974.2 |
IRAK1
|
interleukin-1 receptor-associated kinase 1 |
| chr1_-_153949751 | 9.35 |
ENST00000428469.1
|
JTB
|
jumping translocation breakpoint |
| chr22_-_37640456 | 9.26 |
ENST00000405484.1
ENST00000441619.1 ENST00000406508.1 |
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr16_+_88872176 | 9.24 |
ENST00000569140.1
|
CDT1
|
chromatin licensing and DNA replication factor 1 |
| chr4_-_71705027 | 9.10 |
ENST00000545193.1
|
GRSF1
|
G-rich RNA sequence binding factor 1 |
| chr6_-_31550192 | 9.06 |
ENST00000429299.2
ENST00000446745.2 |
LTB
|
lymphotoxin beta (TNF superfamily, member 3) |
| chr1_+_110163202 | 8.95 |
ENST00000531203.1
ENST00000256578.3 |
AMPD2
|
adenosine monophosphate deaminase 2 |
| chr22_-_39096661 | 8.82 |
ENST00000216039.5
|
JOSD1
|
Josephin domain containing 1 |
| chrX_-_153285395 | 8.42 |
ENST00000369980.3
|
IRAK1
|
interleukin-1 receptor-associated kinase 1 |
| chr5_-_81046841 | 8.34 |
ENST00000509013.2
ENST00000505980.1 ENST00000509053.1 |
SSBP2
|
single-stranded DNA binding protein 2 |
| chr6_-_7911042 | 8.23 |
ENST00000379757.4
|
TXNDC5
|
thioredoxin domain containing 5 (endoplasmic reticulum) |
| chr5_-_81046922 | 8.11 |
ENST00000514493.1
ENST00000320672.4 |
SSBP2
|
single-stranded DNA binding protein 2 |
| chr1_+_155278539 | 8.03 |
ENST00000447866.1
|
FDPS
|
farnesyl diphosphate synthase |
| chr13_+_76123883 | 7.95 |
ENST00000377595.3
|
UCHL3
|
ubiquitin carboxyl-terminal esterase L3 (ubiquitin thiolesterase) |
| chr1_-_25256368 | 7.77 |
ENST00000308873.6
|
RUNX3
|
runt-related transcription factor 3 |
| chrX_-_153599578 | 7.67 |
ENST00000360319.4
ENST00000344736.4 |
FLNA
|
filamin A, alpha |
| chr7_+_50344289 | 7.65 |
ENST00000413698.1
ENST00000359197.5 ENST00000331340.3 ENST00000357364.4 ENST00000343574.5 ENST00000349824.4 ENST00000346667.4 ENST00000440768.2 |
IKZF1
|
IKAROS family zinc finger 1 (Ikaros) |
| chr3_-_18466787 | 7.62 |
ENST00000338745.6
ENST00000450898.1 |
SATB1
|
SATB homeobox 1 |
| chr3_+_49711777 | 7.52 |
ENST00000442186.1
ENST00000438011.1 ENST00000457042.1 |
APEH
|
acylaminoacyl-peptide hydrolase |
| chr14_-_55369525 | 7.49 |
ENST00000543643.2
ENST00000536224.2 ENST00000395514.1 ENST00000491895.2 |
GCH1
|
GTP cyclohydrolase 1 |
| chr11_+_105948216 | 7.40 |
ENST00000278618.4
|
AASDHPPT
|
aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase |
| chr6_-_8102279 | 7.29 |
ENST00000488226.2
|
EEF1E1
|
eukaryotic translation elongation factor 1 epsilon 1 |
| chr1_+_155278625 | 7.22 |
ENST00000368356.4
ENST00000356657.6 |
FDPS
|
farnesyl diphosphate synthase |
| chr14_+_23776167 | 7.22 |
ENST00000554635.1
ENST00000557008.1 |
BCL2L2
BCL2L2-PABPN1
|
BCL2-like 2 BCL2L2-PABPN1 readthrough |
| chr1_-_54405773 | 7.15 |
ENST00000371376.1
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr14_+_23776024 | 6.80 |
ENST00000553781.1
ENST00000556100.1 ENST00000557236.1 ENST00000557579.1 |
BCL2L2-PABPN1
BCL2L2
|
BCL2L2-PABPN1 readthrough BCL2-like 2 |
| chr12_+_7060432 | 6.74 |
ENST00000318974.9
ENST00000456013.1 |
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr17_-_62097927 | 6.73 |
ENST00000578313.1
ENST00000584084.1 ENST00000579788.1 ENST00000579687.1 ENST00000578379.1 ENST00000578892.1 ENST00000412356.1 ENST00000418105.1 |
ICAM2
|
intercellular adhesion molecule 2 |
| chr2_-_130939115 | 6.70 |
ENST00000441135.1
ENST00000339679.7 ENST00000426662.2 ENST00000443958.2 ENST00000351288.6 ENST00000453750.1 ENST00000452225.2 |
SMPD4
|
sphingomyelin phosphodiesterase 4, neutral membrane (neutral sphingomyelinase-3) |
| chr22_-_29196030 | 6.68 |
ENST00000405219.3
|
XBP1
|
X-box binding protein 1 |
| chr5_-_81046904 | 6.53 |
ENST00000515395.1
|
SSBP2
|
single-stranded DNA binding protein 2 |
| chr6_-_8102714 | 6.46 |
ENST00000502429.1
ENST00000429723.2 ENST00000507463.1 ENST00000379715.5 |
EEF1E1
|
eukaryotic translation elongation factor 1 epsilon 1 |
| chr8_-_67525473 | 6.38 |
ENST00000522677.3
|
MYBL1
|
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
| chrX_+_69509927 | 6.33 |
ENST00000374403.3
|
KIF4A
|
kinesin family member 4A |
| chr17_+_40440481 | 6.27 |
ENST00000590726.2
ENST00000452307.2 ENST00000444283.1 ENST00000588868.1 |
STAT5A
|
signal transducer and activator of transcription 5A |
| chrX_-_20284958 | 6.25 |
ENST00000379565.3
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr16_-_1823114 | 6.23 |
ENST00000177742.3
ENST00000397375.2 |
MRPS34
|
mitochondrial ribosomal protein S34 |
| chr1_-_26232951 | 5.99 |
ENST00000426559.2
ENST00000455785.2 |
STMN1
|
stathmin 1 |
| chr16_-_20753114 | 5.97 |
ENST00000396083.2
|
THUMPD1
|
THUMP domain containing 1 |
| chr2_+_30454390 | 5.97 |
ENST00000395323.3
ENST00000406087.1 ENST00000404397.1 |
LBH
|
limb bud and heart development |
| chr11_-_67275542 | 5.88 |
ENST00000531506.1
|
CDK2AP2
|
cyclin-dependent kinase 2 associated protein 2 |
| chr12_-_49318715 | 5.73 |
ENST00000444214.2
|
FKBP11
|
FK506 binding protein 11, 19 kDa |
| chr20_-_62601218 | 5.59 |
ENST00000369888.1
|
ZNF512B
|
zinc finger protein 512B |
| chr19_-_13068012 | 5.52 |
ENST00000316939.1
|
GADD45GIP1
|
growth arrest and DNA-damage-inducible, gamma interacting protein 1 |
| chr3_-_52312636 | 5.46 |
ENST00000296490.3
|
WDR82
|
WD repeat domain 82 |
| chr8_+_145438870 | 5.36 |
ENST00000527931.1
|
FAM203B
|
family with sequence similarity 203, member B |
| chr11_-_65308082 | 5.26 |
ENST00000532661.1
|
LTBP3
|
latent transforming growth factor beta binding protein 3 |
| chr2_+_130939235 | 5.24 |
ENST00000425361.1
ENST00000457492.1 |
MZT2B
|
mitotic spindle organizing protein 2B |
| chr1_+_110163709 | 5.23 |
ENST00000369840.2
ENST00000527846.1 |
AMPD2
|
adenosine monophosphate deaminase 2 |
| chr7_-_150675372 | 5.23 |
ENST00000262186.5
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr5_+_10353780 | 5.16 |
ENST00000449913.2
ENST00000503788.1 ENST00000274140.5 |
MARCH6
|
membrane-associated ring finger (C3HC4) 6, E3 ubiquitin protein ligase |
| chr22_-_29196546 | 5.14 |
ENST00000403532.3
ENST00000216037.6 |
XBP1
|
X-box binding protein 1 |
| chr17_-_62097904 | 5.09 |
ENST00000583366.1
|
ICAM2
|
intercellular adhesion molecule 2 |
| chr16_+_28996364 | 5.06 |
ENST00000564277.1
|
LAT
|
linker for activation of T cells |
| chr1_+_43148625 | 5.03 |
ENST00000436427.1
|
YBX1
|
Y box binding protein 1 |
| chr1_-_28241226 | 5.00 |
ENST00000373912.3
ENST00000373909.3 |
RPA2
|
replication protein A2, 32kDa |
| chr1_+_154947148 | 4.95 |
ENST00000368436.1
ENST00000308987.5 |
CKS1B
|
CDC28 protein kinase regulatory subunit 1B |
| chr1_-_115259337 | 4.94 |
ENST00000369535.4
|
NRAS
|
neuroblastoma RAS viral (v-ras) oncogene homolog |
| chr10_-_99258135 | 4.87 |
ENST00000327238.10
ENST00000327277.7 ENST00000355839.6 ENST00000437002.1 ENST00000422685.1 |
MMS19
|
MMS19 nucleotide excision repair homolog (S. cerevisiae) |
| chr3_-_13461807 | 4.86 |
ENST00000254508.5
|
NUP210
|
nucleoporin 210kDa |
| chr6_+_33043703 | 4.84 |
ENST00000418931.2
ENST00000535465.1 |
HLA-DPB1
|
major histocompatibility complex, class II, DP beta 1 |
| chr9_-_127952187 | 4.81 |
ENST00000451402.1
ENST00000415905.1 |
PPP6C
|
protein phosphatase 6, catalytic subunit |
| chr9_-_127952032 | 4.80 |
ENST00000456642.1
ENST00000373546.3 ENST00000373547.4 |
PPP6C
|
protein phosphatase 6, catalytic subunit |
| chr1_-_207095324 | 4.73 |
ENST00000530505.1
ENST00000367091.3 ENST00000442471.2 |
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr3_+_20081515 | 4.72 |
ENST00000263754.4
|
KAT2B
|
K(lysine) acetyltransferase 2B |
| chr4_+_83956237 | 4.63 |
ENST00000264389.2
|
COPS4
|
COP9 signalosome subunit 4 |
| chr1_+_53480598 | 4.58 |
ENST00000430330.2
ENST00000408941.3 ENST00000478274.2 ENST00000484100.1 ENST00000435345.2 ENST00000488965.1 |
SCP2
|
sterol carrier protein 2 |
| chr1_+_154947126 | 4.53 |
ENST00000368439.1
|
CKS1B
|
CDC28 protein kinase regulatory subunit 1B |
| chr19_+_1407517 | 4.52 |
ENST00000336761.6
ENST00000233078.4 |
DAZAP1
|
DAZ associated protein 1 |
| chr11_-_62607036 | 4.48 |
ENST00000311713.7
ENST00000278856.4 |
WDR74
|
WD repeat domain 74 |
| chr12_+_53689309 | 4.40 |
ENST00000351500.3
ENST00000550846.1 ENST00000334478.4 ENST00000549759.1 |
PFDN5
|
prefoldin subunit 5 |
| chr17_-_47755436 | 4.40 |
ENST00000505581.1
ENST00000514121.1 ENST00000393328.2 ENST00000509079.1 ENST00000393331.3 ENST00000347630.2 ENST00000504102.1 |
SPOP
|
speckle-type POZ protein |
| chr4_+_83956312 | 4.29 |
ENST00000509317.1
ENST00000503682.1 ENST00000511653.1 |
COPS4
|
COP9 signalosome subunit 4 |
| chr14_+_31343747 | 4.27 |
ENST00000216361.4
ENST00000396618.3 ENST00000475087.1 |
COCH
|
cochlin |
| chr16_+_28996572 | 4.17 |
ENST00000360872.5
ENST00000566177.1 ENST00000354453.4 |
LAT
|
linker for activation of T cells |
| chr14_+_31343951 | 4.16 |
ENST00000556908.1
ENST00000555881.1 ENST00000460581.2 |
COCH
|
cochlin |
| chr20_+_30639991 | 4.15 |
ENST00000534862.1
ENST00000538448.1 ENST00000375862.2 |
HCK
|
hemopoietic cell kinase |
| chr2_-_24583168 | 4.11 |
ENST00000361999.3
|
ITSN2
|
intersectin 2 |
| chr12_+_4382917 | 4.10 |
ENST00000261254.3
|
CCND2
|
cyclin D2 |
| chr17_-_79876010 | 4.08 |
ENST00000328666.6
|
SIRT7
|
sirtuin 7 |
| chr22_+_38349670 | 4.07 |
ENST00000442738.2
ENST00000460648.1 ENST00000407936.1 ENST00000488684.1 ENST00000492213.1 ENST00000606538.1 ENST00000405557.1 |
POLR2F
|
polymerase (RNA) II (DNA directed) polypeptide F |
| chr3_-_67705006 | 3.98 |
ENST00000492795.1
ENST00000493112.1 ENST00000307227.5 |
SUCLG2
|
succinate-CoA ligase, GDP-forming, beta subunit |
| chr16_+_85646763 | 3.98 |
ENST00000411612.1
ENST00000253458.7 |
GSE1
|
Gse1 coiled-coil protein |
| chr14_-_21905395 | 3.92 |
ENST00000430710.3
ENST00000553283.1 |
CHD8
|
chromodomain helicase DNA binding protein 8 |
| chr16_+_28996416 | 3.82 |
ENST00000395456.2
ENST00000454369.2 |
LAT
|
linker for activation of T cells |
| chr3_+_47021168 | 3.79 |
ENST00000450053.3
ENST00000292309.5 ENST00000383740.2 |
NBEAL2
|
neurobeachin-like 2 |
| chr16_+_29472707 | 3.78 |
ENST00000565290.1
|
SULT1A4
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 4 |
| chr17_-_42298201 | 3.70 |
ENST00000527034.1
|
UBTF
|
upstream binding transcription factor, RNA polymerase I |
| chr9_-_136024721 | 3.67 |
ENST00000393160.3
|
RALGDS
|
ral guanine nucleotide dissociation stimulator |
| chr4_-_151936416 | 3.58 |
ENST00000510413.1
ENST00000507224.1 |
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr16_-_67190152 | 3.52 |
ENST00000486556.1
|
TRADD
|
TNFRSF1A-associated via death domain |
| chr11_-_417308 | 3.51 |
ENST00000397632.3
ENST00000382520.2 |
SIGIRR
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr6_+_163835669 | 3.51 |
ENST00000453779.2
ENST00000275262.7 ENST00000392127.2 ENST00000361752.3 |
QKI
|
QKI, KH domain containing, RNA binding |
| chr12_+_100867694 | 3.48 |
ENST00000392986.3
ENST00000549996.1 |
NR1H4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr14_+_72399114 | 3.45 |
ENST00000553525.1
ENST00000555571.1 |
RGS6
|
regulator of G-protein signaling 6 |
| chr1_-_51425902 | 3.44 |
ENST00000396153.2
|
FAF1
|
Fas (TNFRSF6) associated factor 1 |
| chr6_-_32157947 | 3.34 |
ENST00000375050.4
|
PBX2
|
pre-B-cell leukemia homeobox 2 |
| chr16_+_30212378 | 3.34 |
ENST00000569485.1
|
SULT1A3
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 |
| chr22_-_39268308 | 3.33 |
ENST00000407418.3
|
CBX6
|
chromobox homolog 6 |
| chr12_+_70133152 | 3.28 |
ENST00000550536.1
ENST00000362025.5 |
RAB3IP
|
RAB3A interacting protein |
| chr1_-_26232522 | 3.21 |
ENST00000399728.1
|
STMN1
|
stathmin 1 |
| chr11_-_128392085 | 3.20 |
ENST00000526145.2
ENST00000531611.1 ENST00000319397.6 ENST00000345075.4 ENST00000535549.1 |
ETS1
|
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
| chr16_+_30210552 | 3.20 |
ENST00000338971.5
|
SULT1A3
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 |
| chr17_-_41132010 | 3.19 |
ENST00000409103.1
ENST00000360221.4 |
PTGES3L-AARSD1
|
PTGES3L-AARSD1 readthrough |
| chr20_+_30640004 | 3.16 |
ENST00000520553.1
ENST00000518730.1 ENST00000375852.2 |
HCK
|
hemopoietic cell kinase |
| chrX_+_37545012 | 3.14 |
ENST00000378616.3
|
XK
|
X-linked Kx blood group (McLeod syndrome) |
| chr1_+_193091080 | 3.12 |
ENST00000367435.3
|
CDC73
|
cell division cycle 73 |
| chr3_-_195270162 | 3.12 |
ENST00000438848.1
ENST00000328432.3 |
PPP1R2
|
protein phosphatase 1, regulatory (inhibitor) subunit 2 |
| chr18_-_33709268 | 3.11 |
ENST00000269187.5
ENST00000590986.1 ENST00000440549.2 |
SLC39A6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr11_-_417388 | 3.08 |
ENST00000332725.3
|
SIGIRR
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr4_-_151936865 | 3.07 |
ENST00000535741.1
|
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr14_+_58666824 | 3.04 |
ENST00000254286.4
|
ACTR10
|
actin-related protein 10 homolog (S. cerevisiae) |
| chr1_-_32110467 | 2.98 |
ENST00000440872.2
ENST00000373703.4 |
PEF1
|
penta-EF-hand domain containing 1 |
| chr5_+_95998746 | 2.94 |
ENST00000508608.1
|
CAST
|
calpastatin |
| chr2_-_27545431 | 2.94 |
ENST00000233545.2
|
MPV17
|
MpV17 mitochondrial inner membrane protein |
| chr2_+_219081817 | 2.92 |
ENST00000315717.5
ENST00000420104.1 ENST00000295685.10 |
ARPC2
|
actin related protein 2/3 complex, subunit 2, 34kDa |
| chr14_-_25103388 | 2.91 |
ENST00000526004.1
ENST00000415355.3 |
GZMB
|
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
| chr22_+_47070490 | 2.78 |
ENST00000408031.1
|
GRAMD4
|
GRAM domain containing 4 |
| chr12_+_16035307 | 2.78 |
ENST00000538352.1
ENST00000025399.6 ENST00000419869.2 |
STRAP
|
serine/threonine kinase receptor associated protein |
| chr1_-_207095212 | 2.77 |
ENST00000420007.2
|
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr19_+_41313017 | 2.76 |
ENST00000595621.1
ENST00000595051.1 |
EGLN2
|
egl-9 family hypoxia-inducible factor 2 |
| chr1_+_39456895 | 2.72 |
ENST00000432648.3
ENST00000446189.2 ENST00000372984.4 |
AKIRIN1
|
akirin 1 |
| chr22_-_39268192 | 2.72 |
ENST00000216083.6
|
CBX6
|
chromobox homolog 6 |
| chr6_+_20403997 | 2.70 |
ENST00000535432.1
|
E2F3
|
E2F transcription factor 3 |
| chr4_-_109087872 | 2.68 |
ENST00000510624.1
|
LEF1
|
lymphoid enhancer-binding factor 1 |
| chr22_+_38349724 | 2.61 |
ENST00000470701.1
|
POLR2F
|
polymerase (RNA) II (DNA directed) polypeptide F |
| chr15_+_89346699 | 2.56 |
ENST00000558207.1
|
ACAN
|
aggrecan |
| chr5_+_156693159 | 2.56 |
ENST00000347377.6
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chrX_-_107019181 | 2.54 |
ENST00000315660.4
ENST00000372384.2 ENST00000502650.1 ENST00000506724.1 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr5_+_148206156 | 2.52 |
ENST00000305988.4
|
ADRB2
|
adrenoceptor beta 2, surface |
| chr1_+_110162448 | 2.52 |
ENST00000342115.4
ENST00000469039.2 ENST00000474459.1 ENST00000528667.1 |
AMPD2
|
adenosine monophosphate deaminase 2 |
| chr17_-_42298331 | 2.52 |
ENST00000343638.5
|
UBTF
|
upstream binding transcription factor, RNA polymerase I |
| chr1_+_110163682 | 2.51 |
ENST00000358729.4
|
AMPD2
|
adenosine monophosphate deaminase 2 |
| chr5_+_156693091 | 2.49 |
ENST00000318218.6
ENST00000442283.2 ENST00000522463.1 ENST00000521420.1 |
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr20_-_21494654 | 2.46 |
ENST00000377142.4
|
NKX2-2
|
NK2 homeobox 2 |
| chr1_-_167059830 | 2.45 |
ENST00000367868.3
|
GPA33
|
glycoprotein A33 (transmembrane) |
| chr7_+_44240520 | 2.44 |
ENST00000496112.1
ENST00000223369.2 |
YKT6
|
YKT6 v-SNARE homolog (S. cerevisiae) |
| chr7_+_872107 | 2.40 |
ENST00000405266.1
ENST00000401592.1 ENST00000403868.1 ENST00000425407.2 |
SUN1
|
Sad1 and UNC84 domain containing 1 |
| chr15_+_81489213 | 2.36 |
ENST00000559383.1
ENST00000394660.2 |
IL16
|
interleukin 16 |
| chr1_-_28241024 | 2.33 |
ENST00000313433.7
ENST00000444045.1 |
RPA2
|
replication protein A2, 32kDa |
| chr5_-_37371278 | 2.32 |
ENST00000231498.3
|
NUP155
|
nucleoporin 155kDa |
| chr5_-_37371163 | 2.27 |
ENST00000513532.1
|
NUP155
|
nucleoporin 155kDa |
| chr2_-_27545921 | 2.27 |
ENST00000402310.1
ENST00000405983.1 ENST00000403262.2 ENST00000428910.1 ENST00000402722.1 ENST00000399052.4 ENST00000380044.1 ENST00000405076.1 |
MPV17
|
MpV17 mitochondrial inner membrane protein |
| chr19_-_4124079 | 2.20 |
ENST00000394867.4
ENST00000262948.5 |
MAP2K2
|
mitogen-activated protein kinase kinase 2 |
| chr1_+_178995021 | 2.20 |
ENST00000263733.4
|
FAM20B
|
family with sequence similarity 20, member B |
| chr9_-_74525658 | 2.19 |
ENST00000333421.6
|
ABHD17B
|
abhydrolase domain containing 17B |
| chr3_-_107941230 | 2.17 |
ENST00000264538.3
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr1_-_51425772 | 2.16 |
ENST00000371778.4
|
FAF1
|
Fas (TNFRSF6) associated factor 1 |
| chr14_-_25103472 | 2.14 |
ENST00000216341.4
ENST00000382542.1 ENST00000382540.1 |
GZMB
|
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
| chr17_-_8151353 | 2.13 |
ENST00000315684.8
|
CTC1
|
CTS telomere maintenance complex component 1 |
| chrX_-_153744507 | 2.12 |
ENST00000442929.1
ENST00000426266.1 ENST00000359889.5 ENST00000369641.3 ENST00000447601.2 ENST00000434658.2 |
FAM3A
|
family with sequence similarity 3, member A |
| chr4_-_109087906 | 2.10 |
ENST00000515500.1
|
LEF1
|
lymphoid enhancer-binding factor 1 |
| chr11_+_65819802 | 2.10 |
ENST00000528302.1
ENST00000322535.6 ENST00000524627.1 ENST00000533595.1 ENST00000530322.1 |
SF3B2
|
splicing factor 3b, subunit 2, 145kDa |
| chr4_-_144940477 | 2.04 |
ENST00000513128.1
ENST00000429670.2 ENST00000502664.1 |
GYPB
|
glycophorin B (MNS blood group) |
| chr7_+_31009148 | 2.00 |
ENST00000409904.3
ENST00000409316.1 |
GHRHR
|
growth hormone releasing hormone receptor |
| chr17_-_55038375 | 1.93 |
ENST00000240316.4
|
COIL
|
coilin |
| chr9_+_116263778 | 1.88 |
ENST00000394646.3
|
RGS3
|
regulator of G-protein signaling 3 |
| chr9_-_140082983 | 1.87 |
ENST00000323927.2
|
ANAPC2
|
anaphase promoting complex subunit 2 |
| chr1_+_235490659 | 1.82 |
ENST00000488594.1
|
GGPS1
|
geranylgeranyl diphosphate synthase 1 |
| chr6_-_2245892 | 1.77 |
ENST00000380815.4
|
GMDS
|
GDP-mannose 4,6-dehydratase |
| chr12_+_4430371 | 1.76 |
ENST00000179259.4
|
C12orf5
|
chromosome 12 open reading frame 5 |
| chr12_+_100867486 | 1.76 |
ENST00000548884.1
|
NR1H4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr2_+_204801471 | 1.75 |
ENST00000316386.6
ENST00000435193.1 |
ICOS
|
inducible T-cell co-stimulator |
| chr9_+_140083099 | 1.73 |
ENST00000322310.5
|
SSNA1
|
Sjogren syndrome nuclear autoantigen 1 |
| chr2_+_46769798 | 1.70 |
ENST00000238738.4
|
RHOQ
|
ras homolog family member Q |
| chr9_+_116263639 | 1.66 |
ENST00000343817.5
|
RGS3
|
regulator of G-protein signaling 3 |
| chr4_-_144826682 | 1.63 |
ENST00000358615.4
ENST00000437468.2 |
GYPE
|
glycophorin E (MNS blood group) |
| chr11_-_78285804 | 1.60 |
ENST00000281038.5
ENST00000529571.1 |
NARS2
|
asparaginyl-tRNA synthetase 2, mitochondrial (putative) |
| chr17_-_41132410 | 1.60 |
ENST00000409446.3
ENST00000453594.1 ENST00000409399.1 ENST00000421990.2 |
PTGES3L
PTGES3L-AARSD1
|
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chrX_-_153744434 | 1.59 |
ENST00000369643.1
ENST00000393572.1 |
FAM3A
|
family with sequence similarity 3, member A |
| chr5_+_176784837 | 1.57 |
ENST00000408923.3
|
RGS14
|
regulator of G-protein signaling 14 |
| chr18_+_11981427 | 1.57 |
ENST00000269159.3
|
IMPA2
|
inositol(myo)-1(or 4)-monophosphatase 2 |
| chr22_+_37447771 | 1.56 |
ENST00000402077.3
ENST00000403888.3 ENST00000456470.1 |
KCTD17
|
potassium channel tetramerization domain containing 17 |
| chr2_-_24583583 | 1.52 |
ENST00000355123.4
|
ITSN2
|
intersectin 2 |
| chr16_+_3062457 | 1.49 |
ENST00000445369.2
|
CLDN9
|
claudin 9 |
| chr3_+_5020801 | 1.37 |
ENST00000256495.3
|
BHLHE40
|
basic helix-loop-helix family, member e40 |
| chr17_+_66508537 | 1.34 |
ENST00000392711.1
ENST00000585427.1 ENST00000589228.1 ENST00000536854.2 ENST00000588702.1 ENST00000589309.1 |
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr21_-_36259445 | 1.26 |
ENST00000399240.1
|
RUNX1
|
runt-related transcription factor 1 |
| chr9_-_26892765 | 1.24 |
ENST00000520187.1
ENST00000333916.5 |
CAAP1
|
caspase activity and apoptosis inhibitor 1 |
| chr8_+_38854418 | 1.21 |
ENST00000481513.1
ENST00000487273.2 |
ADAM9
|
ADAM metallopeptidase domain 9 |
| chr2_-_175462934 | 1.21 |
ENST00000392546.2
ENST00000436221.1 |
WIPF1
|
WAS/WASL interacting protein family, member 1 |
| chr6_-_89927151 | 1.17 |
ENST00000454853.2
|
GABRR1
|
gamma-aminobutyric acid (GABA) A receptor, rho 1 |
| chr15_+_89402148 | 1.17 |
ENST00000560601.1
|
ACAN
|
aggrecan |
| chr12_+_53399942 | 1.16 |
ENST00000262056.9
|
EIF4B
|
eukaryotic translation initiation factor 4B |
| chr17_+_79989500 | 1.16 |
ENST00000306897.4
|
RAC3
|
ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) |
| chr14_+_90422239 | 1.14 |
ENST00000393452.3
ENST00000554180.1 ENST00000393454.2 ENST00000553617.1 ENST00000335725.4 ENST00000357382.3 ENST00000556867.1 ENST00000553527.1 |
TDP1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr3_+_57994127 | 1.08 |
ENST00000490882.1
ENST00000295956.4 ENST00000358537.3 ENST00000429972.2 ENST00000348383.5 ENST00000357272.4 |
FLNB
|
filamin B, beta |
| chr19_-_10697895 | 1.04 |
ENST00000591240.1
ENST00000589684.1 ENST00000591676.1 ENST00000250244.6 ENST00000590923.1 |
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr19_+_41305406 | 1.04 |
ENST00000406058.2
ENST00000593726.1 |
EGLN2
|
egl-9 family hypoxia-inducible factor 2 |
| chr9_-_123555655 | 1.00 |
ENST00000340778.5
ENST00000453291.1 ENST00000608872.1 |
FBXW2
|
F-box and WD repeat domain containing 2 |
| chr14_-_99947168 | 0.96 |
ENST00000331768.5
|
SETD3
|
SET domain containing 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HMX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 11.8 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) regulation of lactation(GO:1903487) |
| 3.9 | 23.4 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 3.4 | 17.1 | GO:0045337 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) farnesyl diphosphate biosynthetic process(GO:0045337) |
| 3.1 | 9.2 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 2.6 | 10.4 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 2.5 | 7.5 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 2.4 | 7.3 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 2.4 | 11.9 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 1.9 | 21.3 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 1.9 | 19.2 | GO:0032264 | IMP salvage(GO:0032264) |
| 1.8 | 9.2 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 1.8 | 12.6 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 1.7 | 5.2 | GO:0001079 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 1.7 | 5.2 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 1.7 | 6.7 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 1.6 | 4.7 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 1.6 | 6.3 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 1.4 | 12.5 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 1.4 | 4.1 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 1.3 | 5.3 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 1.3 | 9.0 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 1.2 | 3.7 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 1.1 | 4.6 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 1.1 | 9.1 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 1.1 | 5.5 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 1.0 | 3.1 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 1.0 | 18.3 | GO:0034162 | toll-like receptor 9 signaling pathway(GO:0034162) |
| 1.0 | 4.9 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.9 | 6.6 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.9 | 6.5 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.9 | 4.6 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.9 | 13.8 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.8 | 2.5 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.8 | 2.5 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.8 | 4.0 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.7 | 7.4 | GO:0015939 | lysine metabolic process(GO:0006553) pantothenate metabolic process(GO:0015939) |
| 0.7 | 5.0 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.7 | 5.0 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.7 | 6.7 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.7 | 2.0 | GO:0046010 | positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) |
| 0.6 | 2.4 | GO:0044821 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.6 | 1.8 | GO:0042350 | GDP-L-fucose biosynthetic process(GO:0042350) 'de novo' GDP-L-fucose biosynthetic process(GO:0042351) |
| 0.6 | 8.9 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.5 | 7.1 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.5 | 2.2 | GO:0036289 | peptidyl-serine autophosphorylation(GO:0036289) |
| 0.5 | 7.6 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.5 | 1.6 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.5 | 6.2 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) |
| 0.4 | 3.6 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.4 | 3.9 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.4 | 1.7 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.4 | 2.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.4 | 3.0 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.4 | 6.3 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.4 | 1.2 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.4 | 3.2 | GO:1904996 | PML body organization(GO:0030578) positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.4 | 5.5 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.4 | 2.5 | GO:0070235 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.4 | 3.5 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.3 | 7.3 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.3 | 9.0 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.3 | 5.6 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.3 | 2.0 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.3 | 3.1 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.3 | 3.5 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.3 | 4.7 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.3 | 3.1 | GO:0001711 | endodermal cell fate commitment(GO:0001711) positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.3 | 2.7 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.3 | 0.8 | GO:1901253 | negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.3 | 1.6 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.2 | 4.1 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.2 | 0.7 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.2 | 1.7 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.2 | 1.7 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.2 | 6.3 | GO:0043555 | regulation of translation in response to stress(GO:0043555) |
| 0.2 | 3.8 | GO:0030220 | platelet formation(GO:0030220) |
| 0.2 | 13.0 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.2 | 2.9 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.2 | 4.5 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.2 | 6.0 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.2 | 1.9 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.2 | 2.8 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.2 | 2.4 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.2 | 8.8 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.2 | 7.5 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.2 | 6.7 | GO:0006363 | transcription elongation from RNA polymerase I promoter(GO:0006362) termination of RNA polymerase I transcription(GO:0006363) |
| 0.2 | 5.7 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.2 | 9.1 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.1 | 3.8 | GO:0050427 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.1 | 1.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 5.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 4.9 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.1 | 1.3 | GO:0021894 | regulation of neuron maturation(GO:0014041) cerebral cortex GABAergic interneuron development(GO:0021894) |
| 0.1 | 1.9 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.1 | 1.0 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 2.8 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.1 | 1.1 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.1 | 0.5 | GO:1901098 | regulation of autophagosome maturation(GO:1901096) positive regulation of autophagosome maturation(GO:1901098) |
| 0.1 | 1.8 | GO:0002517 | T cell tolerance induction(GO:0002517) |
| 0.1 | 1.5 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.1 | 0.4 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.1 | 1.7 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.1 | 3.1 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 | 6.2 | GO:0070125 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 0.1 | 12.8 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.1 | 1.4 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 11.8 | GO:0002223 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) |
| 0.1 | 0.5 | GO:0032006 | regulation of TOR signaling(GO:0032006) |
| 0.1 | 9.3 | GO:0008637 | apoptotic mitochondrial changes(GO:0008637) |
| 0.1 | 1.6 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.1 | 0.4 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.1 | 4.8 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.1 | 4.5 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.1 | 6.6 | GO:0030218 | erythrocyte differentiation(GO:0030218) |
| 0.1 | 1.1 | GO:0003334 | keratinocyte development(GO:0003334) cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.1 | 8.4 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.1 | 4.7 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 7.7 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.7 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 1.2 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.4 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.3 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 8.4 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 8.9 | GO:0000398 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 5.5 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.4 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.4 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 1.4 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.8 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.0 | 0.0 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 18.3 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 2.6 | 7.7 | GO:0031523 | Myb complex(GO:0031523) |
| 1.1 | 4.6 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 1.0 | 5.2 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 1.0 | 2.9 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.9 | 10.4 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.9 | 5.2 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) |
| 0.8 | 2.4 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.8 | 4.9 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.7 | 9.0 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.7 | 4.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.7 | 4.0 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.7 | 13.8 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.6 | 4.4 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.6 | 5.0 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.6 | 6.7 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.6 | 5.2 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.5 | 7.7 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.5 | 5.5 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.5 | 4.7 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.5 | 11.8 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.5 | 7.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.5 | 22.0 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.4 | 4.8 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.4 | 9.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.4 | 5.6 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.4 | 6.7 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.3 | 3.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.3 | 25.6 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.3 | 22.5 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.3 | 7.6 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.3 | 2.8 | GO:0032797 | SMN complex(GO:0032797) |
| 0.2 | 3.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 6.2 | GO:0005840 | ribosome(GO:0005840) |
| 0.2 | 4.8 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 3.0 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.2 | 12.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.2 | 2.4 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.2 | 0.7 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.2 | 8.5 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.2 | 3.5 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.2 | 5.2 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.2 | 2.1 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.2 | 3.9 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 7.0 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 4.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 20.9 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 8.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 3.0 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 2.7 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 9.8 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 0.4 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.1 | 3.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 1.2 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.1 | 9.2 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.1 | 5.5 | GO:0005761 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.1 | 7.6 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.1 | 0.5 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 1.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 11.3 | GO:0044452 | nucleolar part(GO:0044452) |
| 0.1 | 4.9 | GO:0018995 | host(GO:0018995) host cell(GO:0043657) |
| 0.1 | 2.5 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.1 | 7.8 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 3.8 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 1.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 7.5 | GO:0101002 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.0 | 0.8 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 4.7 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.7 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 1.2 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 5.5 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 1.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.5 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 0.8 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 1.5 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 1.6 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.9 | 23.4 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 4.3 | 12.8 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 3.4 | 17.1 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 2.6 | 18.3 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 2.1 | 19.2 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 1.9 | 7.7 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 1.7 | 5.2 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 1.7 | 16.9 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 1.5 | 4.6 | GO:0070538 | oleic acid binding(GO:0070538) |
| 1.4 | 4.1 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 1.4 | 9.5 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 1.2 | 7.5 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 1.1 | 6.5 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 1.0 | 4.0 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.9 | 4.7 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.9 | 6.2 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.9 | 5.2 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.9 | 9.5 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.8 | 2.5 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.7 | 6.7 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.6 | 3.5 | GO:0070513 | death domain binding(GO:0070513) |
| 0.6 | 6.4 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.6 | 1.7 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.5 | 2.0 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.5 | 2.9 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.4 | 3.6 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.4 | 9.0 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.4 | 8.7 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.4 | 3.9 | GO:0043426 | MRF binding(GO:0043426) |
| 0.4 | 5.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.4 | 2.0 | GO:0052832 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.4 | 8.2 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.4 | 3.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.4 | 6.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.4 | 6.7 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.4 | 7.4 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.3 | 7.5 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.3 | 17.5 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.3 | 10.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.3 | 1.1 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.3 | 13.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.3 | 4.5 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.2 | 28.0 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.2 | 3.8 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.2 | 6.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.2 | 5.7 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.2 | 5.2 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.2 | 3.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 2.4 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.2 | 7.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.2 | 0.8 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.1 | 5.6 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 4.8 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 2.4 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 8.8 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.1 | 0.4 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 2.4 | GO:0008603 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 9.6 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 2.4 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 12.2 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 3.2 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 3.1 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 1.7 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 11.8 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.1 | 8.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 1.6 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 3.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 2.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 4.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 2.0 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 5.6 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.1 | 1.6 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 2.0 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 2.0 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 14.4 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 8.9 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.4 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.4 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 1.6 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 1.2 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 6.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.8 | GO:0050661 | NADP binding(GO:0050661) |
| 0.0 | 1.5 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 4.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.7 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 13.0 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.6 | 18.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.5 | 30.0 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.4 | 6.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.3 | 6.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.3 | 4.9 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.3 | 8.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.2 | 4.8 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.2 | 11.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 21.6 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.2 | 7.7 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.2 | 4.1 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.2 | 14.8 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.2 | 5.0 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.2 | 2.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.2 | 12.6 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.2 | 9.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.2 | 5.9 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 11.1 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 7.3 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 3.5 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 8.0 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 5.0 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 5.0 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 5.6 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.1 | 8.5 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 8.9 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 4.1 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 7.2 | PID P73PATHWAY | p73 transcription factor network |
| 0.1 | 2.8 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 2.5 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 4.8 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 7.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 2.4 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 11.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.8 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.0 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 17.8 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 1.4 | 23.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 1.2 | 10.4 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.8 | 11.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.8 | 19.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.6 | 11.9 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.6 | 7.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.6 | 17.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.5 | 7.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.5 | 7.1 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.4 | 18.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.4 | 13.0 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.4 | 4.7 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.4 | 13.8 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.4 | 11.6 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.4 | 10.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.3 | 8.2 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.3 | 7.5 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.3 | 6.7 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.3 | 25.0 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.3 | 12.6 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.2 | 9.5 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.2 | 6.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.2 | 4.6 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.2 | 6.2 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.2 | 10.9 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.2 | 3.1 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.2 | 4.0 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.2 | 3.7 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 6.7 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 11.8 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 4.3 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.1 | 3.5 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 5.0 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 17.1 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 4.4 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.1 | 3.6 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 2.4 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.1 | 6.6 | REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE | Genes involved in MyD88:Mal cascade initiated on plasma membrane |
| 0.1 | 2.8 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 1.9 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.1 | 9.1 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 2.0 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 2.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.4 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.1 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 1.2 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.7 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 1.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 1.7 | REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | Genes involved in Recruitment of mitotic centrosome proteins and complexes |
| 0.0 | 0.4 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 1.1 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |