Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
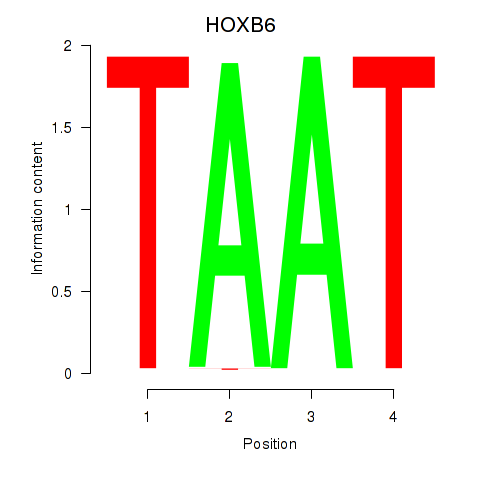
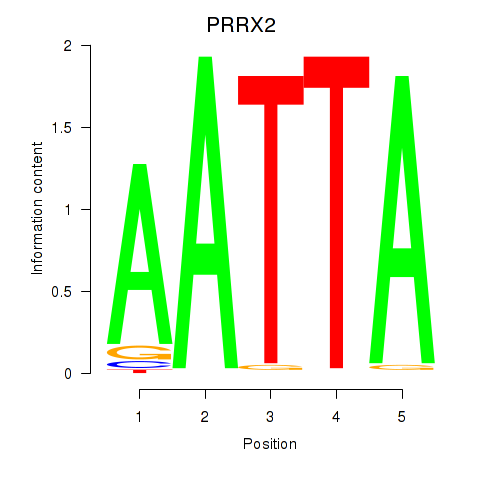
Results for HOXB6_PRRX2
Z-value: 2.13
Transcription factors associated with HOXB6_PRRX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXB6
|
ENSG00000108511.8 | homeobox B6 |
|
PRRX2
|
ENSG00000167157.9 | paired related homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PRRX2 | hg19_v2_chr9_+_132427883_132427951 | 0.49 | 2.0e-14 | Click! |
| HOXB6 | hg19_v2_chr17_-_46682321_46682362 | -0.06 | 3.6e-01 | Click! |
Activity profile of HOXB6_PRRX2 motif
Sorted Z-values of HOXB6_PRRX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_16761007 | 140.21 |
ENST00000354662.1
ENST00000441439.2 |
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr4_-_176733897 | 133.83 |
ENST00000393658.2
|
GPM6A
|
glycoprotein M6A |
| chr2_-_175711133 | 74.69 |
ENST00000409597.1
ENST00000413882.1 |
CHN1
|
chimerin 1 |
| chr12_-_16759711 | 64.75 |
ENST00000447609.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr16_+_7382745 | 62.91 |
ENST00000436368.2
ENST00000311745.5 ENST00000355637.4 ENST00000340209.4 |
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr2_-_50201327 | 58.09 |
ENST00000412315.1
|
NRXN1
|
neurexin 1 |
| chr18_-_53089723 | 56.11 |
ENST00000561992.1
ENST00000562512.2 |
TCF4
|
transcription factor 4 |
| chr1_+_50569575 | 52.82 |
ENST00000371827.1
|
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr5_-_88179302 | 52.26 |
ENST00000504921.2
|
MEF2C
|
myocyte enhancer factor 2C |
| chr13_-_67802549 | 47.29 |
ENST00000328454.5
ENST00000377865.2 |
PCDH9
|
protocadherin 9 |
| chr1_+_50574585 | 46.23 |
ENST00000371824.1
ENST00000371823.4 |
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr18_-_52989217 | 46.01 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr3_+_111717600 | 42.93 |
ENST00000273368.4
|
TAGLN3
|
transgelin 3 |
| chr8_-_18744528 | 41.55 |
ENST00000523619.1
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr8_+_80523321 | 40.64 |
ENST00000518111.1
|
STMN2
|
stathmin-like 2 |
| chr13_-_36429763 | 40.48 |
ENST00000379893.1
|
DCLK1
|
doublecortin-like kinase 1 |
| chr4_+_41258786 | 39.77 |
ENST00000503431.1
ENST00000284440.4 ENST00000508768.1 ENST00000512788.1 |
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
| chr2_-_224467093 | 38.80 |
ENST00000305409.2
|
SCG2
|
secretogranin II |
| chr3_+_111717511 | 37.27 |
ENST00000478951.1
ENST00000393917.2 |
TAGLN3
|
transgelin 3 |
| chr7_-_31380502 | 37.04 |
ENST00000297142.3
|
NEUROD6
|
neuronal differentiation 6 |
| chr12_-_16758304 | 36.21 |
ENST00000320122.6
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr12_-_91576561 | 35.81 |
ENST00000547568.2
ENST00000552962.1 |
DCN
|
decorin |
| chrX_-_13835147 | 35.69 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr3_+_111718036 | 35.35 |
ENST00000455401.2
|
TAGLN3
|
transgelin 3 |
| chr3_+_158787041 | 35.18 |
ENST00000471575.1
ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr5_-_24645078 | 33.67 |
ENST00000264463.4
|
CDH10
|
cadherin 10, type 2 (T2-cadherin) |
| chr12_-_16758059 | 32.75 |
ENST00000261169.6
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr12_+_79258444 | 32.61 |
ENST00000261205.4
|
SYT1
|
synaptotagmin I |
| chr3_+_111718173 | 32.11 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chr8_+_85618155 | 31.34 |
ENST00000523850.1
ENST00000521376.1 |
RALYL
|
RALY RNA binding protein-like |
| chr3_+_159557637 | 30.85 |
ENST00000445224.2
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr12_-_91576429 | 30.80 |
ENST00000552145.1
ENST00000546745.1 |
DCN
|
decorin |
| chr12_+_79258547 | 30.51 |
ENST00000457153.2
|
SYT1
|
synaptotagmin I |
| chr12_-_91576750 | 30.20 |
ENST00000228329.5
ENST00000303320.3 ENST00000052754.5 |
DCN
|
decorin |
| chr18_-_52989525 | 29.70 |
ENST00000457482.3
|
TCF4
|
transcription factor 4 |
| chr6_-_46293378 | 29.58 |
ENST00000330430.6
|
RCAN2
|
regulator of calcineurin 2 |
| chr3_+_115342349 | 28.54 |
ENST00000393780.3
|
GAP43
|
growth associated protein 43 |
| chr5_+_66300446 | 26.94 |
ENST00000261569.7
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr3_-_114477787 | 26.70 |
ENST00000464560.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr3_+_35721106 | 26.58 |
ENST00000474696.1
ENST00000412048.1 ENST00000396482.2 ENST00000432682.1 |
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr10_-_21786179 | 25.93 |
ENST00000377113.5
|
CASC10
|
cancer susceptibility candidate 10 |
| chrX_-_13835461 | 25.92 |
ENST00000316715.4
ENST00000356942.5 |
GPM6B
|
glycoprotein M6B |
| chr13_-_45010939 | 25.41 |
ENST00000261489.2
|
TSC22D1
|
TSC22 domain family, member 1 |
| chr3_-_114477962 | 25.04 |
ENST00000471418.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr11_+_131240373 | 24.56 |
ENST00000374791.3
ENST00000436745.1 |
NTM
|
neurotrimin |
| chr14_+_22977587 | 22.37 |
ENST00000390504.1
|
TRAJ33
|
T cell receptor alpha joining 33 |
| chr4_-_41884620 | 21.95 |
ENST00000504870.1
|
LINC00682
|
long intergenic non-protein coding RNA 682 |
| chr2_-_2334888 | 21.32 |
ENST00000428368.2
ENST00000399161.2 |
MYT1L
|
myelin transcription factor 1-like |
| chr16_-_29910853 | 21.27 |
ENST00000308713.5
|
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2 |
| chr8_-_102803163 | 21.10 |
ENST00000523645.1
ENST00000520346.1 ENST00000220931.6 ENST00000522448.1 ENST00000522951.1 ENST00000522252.1 ENST00000519098.1 |
NCALD
|
neurocalcin delta |
| chr12_-_91539918 | 20.99 |
ENST00000548218.1
|
DCN
|
decorin |
| chr11_-_129062093 | 20.90 |
ENST00000310343.9
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr1_-_156399184 | 20.42 |
ENST00000368243.1
ENST00000357975.4 ENST00000310027.5 ENST00000400991.2 |
C1orf61
|
chromosome 1 open reading frame 61 |
| chr1_+_65730385 | 20.39 |
ENST00000263441.7
ENST00000395325.3 |
DNAJC6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr9_+_87284622 | 20.32 |
ENST00000395882.1
|
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr11_-_111794446 | 20.24 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr12_-_91574142 | 20.24 |
ENST00000547937.1
|
DCN
|
decorin |
| chr2_+_183943464 | 20.14 |
ENST00000354221.4
|
DUSP19
|
dual specificity phosphatase 19 |
| chrX_+_135278908 | 19.70 |
ENST00000539015.1
ENST00000370683.1 |
FHL1
|
four and a half LIM domains 1 |
| chr8_-_120651020 | 19.58 |
ENST00000522826.1
ENST00000520066.1 ENST00000259486.6 ENST00000075322.6 |
ENPP2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr1_-_57045228 | 19.44 |
ENST00000371250.3
|
PPAP2B
|
phosphatidic acid phosphatase type 2B |
| chr12_+_79439405 | 19.37 |
ENST00000552744.1
|
SYT1
|
synaptotagmin I |
| chr17_-_78450398 | 19.26 |
ENST00000306773.4
|
NPTX1
|
neuronal pentraxin I |
| chrX_+_135251783 | 19.13 |
ENST00000394153.2
|
FHL1
|
four and a half LIM domains 1 |
| chr11_-_111782696 | 19.11 |
ENST00000227251.3
ENST00000526180.1 |
CRYAB
|
crystallin, alpha B |
| chr3_-_58613323 | 19.05 |
ENST00000474531.1
ENST00000465970.1 |
FAM107A
|
family with sequence similarity 107, member A |
| chrX_+_135279179 | 18.82 |
ENST00000370676.3
|
FHL1
|
four and a half LIM domains 1 |
| chr3_+_35722487 | 18.80 |
ENST00000441454.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr4_-_186877806 | 18.60 |
ENST00000355634.5
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr4_-_87028478 | 18.55 |
ENST00000515400.1
ENST00000395157.3 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr1_+_84630053 | 18.53 |
ENST00000394838.2
ENST00000370682.3 ENST00000432111.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr5_+_170736243 | 18.37 |
ENST00000296921.5
|
TLX3
|
T-cell leukemia homeobox 3 |
| chr6_+_45296048 | 18.24 |
ENST00000465038.2
ENST00000352853.5 ENST00000541979.1 ENST00000371438.1 |
RUNX2
|
runt-related transcription factor 2 |
| chr15_+_93443419 | 18.22 |
ENST00000557381.1
ENST00000420239.2 |
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr16_+_8806800 | 18.01 |
ENST00000561870.1
ENST00000396600.2 |
ABAT
|
4-aminobutyrate aminotransferase |
| chr8_+_104831472 | 17.95 |
ENST00000262231.10
ENST00000507740.1 |
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr18_-_21891460 | 17.59 |
ENST00000357041.4
|
OSBPL1A
|
oxysterol binding protein-like 1A |
| chr18_-_53070913 | 17.39 |
ENST00000568186.1
ENST00000564228.1 |
TCF4
|
transcription factor 4 |
| chr4_-_153274078 | 17.37 |
ENST00000263981.5
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr4_+_158141843 | 17.35 |
ENST00000509417.1
ENST00000296526.7 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr11_-_125366089 | 17.17 |
ENST00000366139.3
ENST00000278919.3 |
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr4_+_158141899 | 16.53 |
ENST00000264426.9
ENST00000506284.1 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr2_+_166428839 | 16.52 |
ENST00000342316.4
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3 |
| chr18_-_53177984 | 16.40 |
ENST00000543082.1
|
TCF4
|
transcription factor 4 |
| chr4_+_72204755 | 16.36 |
ENST00000512686.1
ENST00000340595.3 |
SLC4A4
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
| chr7_-_27183263 | 16.27 |
ENST00000222726.3
|
HOXA5
|
homeobox A5 |
| chrX_+_135252050 | 16.11 |
ENST00000449474.1
ENST00000345434.3 |
FHL1
|
four and a half LIM domains 1 |
| chr2_+_54785485 | 15.83 |
ENST00000333896.5
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1 |
| chr4_+_113970772 | 15.60 |
ENST00000504454.1
ENST00000394537.3 ENST00000357077.4 ENST00000264366.6 |
ANK2
|
ankyrin 2, neuronal |
| chr14_-_98444386 | 15.13 |
ENST00000556462.1
ENST00000556138.1 |
C14orf64
|
chromosome 14 open reading frame 64 |
| chr11_-_111782484 | 15.06 |
ENST00000533971.1
|
CRYAB
|
crystallin, alpha B |
| chr8_-_18541603 | 15.01 |
ENST00000428502.2
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr16_+_6533380 | 14.95 |
ENST00000552089.1
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr5_+_156712372 | 14.87 |
ENST00000541131.1
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr18_-_53069419 | 14.81 |
ENST00000570177.2
|
TCF4
|
transcription factor 4 |
| chr2_-_60780536 | 14.77 |
ENST00000538214.1
|
BCL11A
|
B-cell CLL/lymphoma 11A (zinc finger protein) |
| chrX_-_72434628 | 14.66 |
ENST00000536638.1
ENST00000373517.3 |
NAP1L2
|
nucleosome assembly protein 1-like 2 |
| chr15_+_34261089 | 14.56 |
ENST00000383263.5
|
CHRM5
|
cholinergic receptor, muscarinic 5 |
| chr18_-_53257027 | 14.49 |
ENST00000568740.1
ENST00000564403.2 ENST00000537578.1 |
TCF4
|
transcription factor 4 |
| chr1_+_50571949 | 14.39 |
ENST00000357083.4
|
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr2_+_102508955 | 14.34 |
ENST00000414004.2
|
FLJ20373
|
FLJ20373 |
| chr11_+_123396528 | 14.32 |
ENST00000322282.7
ENST00000529750.1 |
GRAMD1B
|
GRAM domain containing 1B |
| chr6_+_45296391 | 14.28 |
ENST00000371436.6
ENST00000576263.1 |
RUNX2
|
runt-related transcription factor 2 |
| chr8_+_104892639 | 14.20 |
ENST00000436393.2
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr4_+_158141806 | 14.16 |
ENST00000393815.2
|
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr5_+_161494521 | 14.08 |
ENST00000356592.3
|
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr9_+_103204553 | 13.94 |
ENST00000502978.1
ENST00000334943.6 |
MSANTD3-TMEFF1
TMEFF1
|
MSANTD3-TMEFF1 readthrough transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chr5_+_140810132 | 13.91 |
ENST00000252085.3
|
PCDHGA12
|
protocadherin gamma subfamily A, 12 |
| chr14_-_101036119 | 13.81 |
ENST00000355173.2
|
BEGAIN
|
brain-enriched guanylate kinase-associated |
| chr4_+_113739244 | 13.77 |
ENST00000503271.1
ENST00000503423.1 ENST00000506722.1 |
ANK2
|
ankyrin 2, neuronal |
| chr4_+_88754069 | 13.62 |
ENST00000395102.4
ENST00000497649.2 |
MEPE
|
matrix extracellular phosphoglycoprotein |
| chr22_-_36236623 | 13.61 |
ENST00000405409.2
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr3_-_114343039 | 13.52 |
ENST00000481632.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr7_-_37026108 | 13.36 |
ENST00000396045.3
|
ELMO1
|
engulfment and cell motility 1 |
| chr1_+_162602244 | 13.30 |
ENST00000367922.3
ENST00000367921.3 |
DDR2
|
discoidin domain receptor tyrosine kinase 2 |
| chr11_+_92085707 | 13.19 |
ENST00000525166.1
|
FAT3
|
FAT atypical cadherin 3 |
| chr17_+_76311791 | 13.17 |
ENST00000586321.1
|
AC061992.2
|
AC061992.2 |
| chr18_+_32290218 | 13.12 |
ENST00000348997.5
ENST00000588949.1 ENST00000597599.1 |
DTNA
|
dystrobrevin, alpha |
| chr17_+_41363854 | 13.11 |
ENST00000588693.1
ENST00000588659.1 ENST00000541594.1 ENST00000536052.1 ENST00000331615.3 |
TMEM106A
|
transmembrane protein 106A |
| chr12_-_50290839 | 13.10 |
ENST00000552863.1
|
FAIM2
|
Fas apoptotic inhibitory molecule 2 |
| chr2_+_166152283 | 13.08 |
ENST00000375427.2
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chr5_+_67586465 | 12.95 |
ENST00000336483.5
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr4_+_88896819 | 12.94 |
ENST00000237623.7
ENST00000395080.3 ENST00000508233.1 ENST00000360804.4 |
SPP1
|
secreted phosphoprotein 1 |
| chr11_-_84634217 | 12.83 |
ENST00000524982.1
|
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr2_+_166150541 | 12.69 |
ENST00000283256.6
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chr2_+_66662690 | 12.67 |
ENST00000488550.1
|
MEIS1
|
Meis homeobox 1 |
| chr12_+_54447637 | 12.45 |
ENST00000609810.1
ENST00000430889.2 |
HOXC4
HOXC4
|
homeobox C4 Homeobox protein Hox-C4 |
| chr1_+_155829286 | 12.31 |
ENST00000368324.4
|
SYT11
|
synaptotagmin XI |
| chr4_+_158142750 | 12.28 |
ENST00000505888.1
ENST00000449365.1 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr4_+_41614909 | 12.24 |
ENST00000509454.1
ENST00000396595.3 ENST00000381753.4 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr12_+_81110684 | 11.99 |
ENST00000228644.3
|
MYF5
|
myogenic factor 5 |
| chr2_+_210444748 | 11.94 |
ENST00000392194.1
|
MAP2
|
microtubule-associated protein 2 |
| chr6_-_6007200 | 11.91 |
ENST00000244766.2
|
NRN1
|
neuritin 1 |
| chr1_+_92632542 | 11.85 |
ENST00000409154.4
ENST00000370378.4 |
KIAA1107
|
KIAA1107 |
| chr2_+_210444142 | 11.85 |
ENST00000360351.4
ENST00000361559.4 |
MAP2
|
microtubule-associated protein 2 |
| chr2_+_162272605 | 11.84 |
ENST00000389554.3
|
TBR1
|
T-box, brain, 1 |
| chr9_-_124989804 | 11.78 |
ENST00000373755.2
ENST00000373754.2 |
LHX6
|
LIM homeobox 6 |
| chr5_+_59783941 | 11.77 |
ENST00000506884.1
ENST00000504876.2 |
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding) |
| chr5_+_161274940 | 11.53 |
ENST00000393943.4
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr4_+_88754113 | 11.49 |
ENST00000560249.1
ENST00000540395.1 ENST00000511670.1 ENST00000361056.3 |
MEPE
|
matrix extracellular phosphoglycoprotein |
| chr5_+_66254698 | 11.44 |
ENST00000405643.1
ENST00000407621.1 ENST00000432426.1 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr12_-_9268707 | 11.26 |
ENST00000318602.7
|
A2M
|
alpha-2-macroglobulin |
| chr3_-_127541194 | 11.24 |
ENST00000453507.2
|
MGLL
|
monoglyceride lipase |
| chr16_+_89334512 | 11.23 |
ENST00000602042.1
|
AC137932.1
|
AC137932.1 |
| chr8_-_27115931 | 11.18 |
ENST00000523048.1
|
STMN4
|
stathmin-like 4 |
| chr10_-_62149433 | 11.17 |
ENST00000280772.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr4_-_186877502 | 11.07 |
ENST00000431902.1
ENST00000284776.7 ENST00000415274.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr3_+_129247479 | 11.04 |
ENST00000296271.3
|
RHO
|
rhodopsin |
| chr16_-_21289627 | 10.99 |
ENST00000396023.2
ENST00000415987.2 |
CRYM
|
crystallin, mu |
| chr2_-_177502659 | 10.96 |
ENST00000295549.4
|
AC017048.3
|
long intergenic non-protein coding RNA 1116 |
| chr5_+_161274685 | 10.94 |
ENST00000428797.2
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr6_-_169654139 | 10.87 |
ENST00000366787.3
|
THBS2
|
thrombospondin 2 |
| chr5_+_156696362 | 10.78 |
ENST00000377576.3
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr5_+_161494770 | 10.62 |
ENST00000414552.2
ENST00000361925.4 |
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr9_-_124990680 | 10.61 |
ENST00000541397.2
ENST00000560485.1 |
LHX6
|
LIM homeobox 6 |
| chr2_-_183291741 | 10.56 |
ENST00000351439.5
ENST00000409365.1 |
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr5_-_11588907 | 10.44 |
ENST00000513598.1
ENST00000503622.1 |
CTNND2
|
catenin (cadherin-associated protein), delta 2 |
| chr10_-_49813090 | 10.39 |
ENST00000249601.4
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr9_+_2158443 | 10.23 |
ENST00000302401.3
ENST00000324954.5 ENST00000423555.1 ENST00000382186.1 |
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr2_+_171034646 | 10.21 |
ENST00000409044.3
ENST00000408978.4 |
MYO3B
|
myosin IIIB |
| chr18_-_53303123 | 10.19 |
ENST00000569357.1
ENST00000565124.1 ENST00000398339.1 |
TCF4
|
transcription factor 4 |
| chr5_-_1882858 | 10.14 |
ENST00000511126.1
ENST00000231357.2 |
IRX4
|
iroquois homeobox 4 |
| chr8_-_87755878 | 10.12 |
ENST00000320005.5
|
CNGB3
|
cyclic nucleotide gated channel beta 3 |
| chr2_+_66662510 | 10.09 |
ENST00000272369.9
ENST00000407092.2 |
MEIS1
|
Meis homeobox 1 |
| chr6_+_69942298 | 10.06 |
ENST00000238918.8
|
BAI3
|
brain-specific angiogenesis inhibitor 3 |
| chr17_-_26220366 | 10.02 |
ENST00000460380.2
ENST00000508862.1 ENST00000379102.3 ENST00000582441.1 |
LYRM9
RP1-66C13.4
|
LYR motif containing 9 Uncharacterized protein |
| chr13_+_96085847 | 10.01 |
ENST00000376873.3
|
CLDN10
|
claudin 10 |
| chr2_-_213403565 | 10.00 |
ENST00000342788.4
ENST00000436443.1 |
ERBB4
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 4 |
| chr5_+_140762268 | 9.98 |
ENST00000518325.1
|
PCDHGA7
|
protocadherin gamma subfamily A, 7 |
| chr5_+_161112563 | 9.96 |
ENST00000274545.5
|
GABRA6
|
gamma-aminobutyric acid (GABA) A receptor, alpha 6 |
| chr8_-_120685608 | 9.96 |
ENST00000427067.2
|
ENPP2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr11_-_123525289 | 9.86 |
ENST00000392770.2
ENST00000299333.3 ENST00000530277.1 |
SCN3B
|
sodium channel, voltage-gated, type III, beta subunit |
| chr3_+_69985792 | 9.80 |
ENST00000531774.1
|
MITF
|
microphthalmia-associated transcription factor |
| chr15_+_62853562 | 9.80 |
ENST00000561311.1
|
TLN2
|
talin 2 |
| chr15_+_74422585 | 9.79 |
ENST00000561740.1
ENST00000435464.1 ENST00000453268.2 |
ISLR2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr5_+_161275320 | 9.75 |
ENST00000437025.2
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr14_-_69261310 | 9.67 |
ENST00000336440.3
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr5_-_11589131 | 9.64 |
ENST00000511377.1
|
CTNND2
|
catenin (cadherin-associated protein), delta 2 |
| chr10_+_95517660 | 9.60 |
ENST00000371413.3
|
LGI1
|
leucine-rich, glioma inactivated 1 |
| chr5_-_115910091 | 9.60 |
ENST00000257414.8
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr4_-_99578776 | 9.58 |
ENST00000515287.1
|
TSPAN5
|
tetraspanin 5 |
| chr5_-_138210977 | 9.57 |
ENST00000274711.6
ENST00000521094.2 |
LRRTM2
|
leucine rich repeat transmembrane neuronal 2 |
| chr9_+_2158485 | 9.56 |
ENST00000417599.1
ENST00000382185.1 ENST00000382183.1 |
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr8_-_27115903 | 9.48 |
ENST00000350889.4
ENST00000519997.1 ENST00000519614.1 ENST00000522908.1 ENST00000265770.7 |
STMN4
|
stathmin-like 4 |
| chr18_-_53253112 | 9.42 |
ENST00000568673.1
ENST00000562847.1 ENST00000568147.1 |
TCF4
|
transcription factor 4 |
| chr17_-_29624343 | 9.32 |
ENST00000247271.4
|
OMG
|
oligodendrocyte myelin glycoprotein |
| chr5_+_174151536 | 9.25 |
ENST00000239243.6
ENST00000507785.1 |
MSX2
|
msh homeobox 2 |
| chr2_-_60780702 | 9.19 |
ENST00000359629.5
|
BCL11A
|
B-cell CLL/lymphoma 11A (zinc finger protein) |
| chr2_-_166060552 | 9.16 |
ENST00000283254.7
ENST00000453007.1 |
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit |
| chr15_+_43985084 | 9.04 |
ENST00000434505.1
ENST00000411750.1 |
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chrX_+_135251835 | 8.85 |
ENST00000456445.1
|
FHL1
|
four and a half LIM domains 1 |
| chr9_-_14308004 | 8.80 |
ENST00000493697.1
|
NFIB
|
nuclear factor I/B |
| chr4_-_99578789 | 8.69 |
ENST00000511651.1
ENST00000505184.1 |
TSPAN5
|
tetraspanin 5 |
| chr1_+_84630645 | 8.62 |
ENST00000394839.2
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr9_-_13165457 | 8.62 |
ENST00000542239.1
ENST00000538841.1 ENST00000433359.2 |
MPDZ
|
multiple PDZ domain protein |
| chr4_+_114214125 | 8.62 |
ENST00000509550.1
|
ANK2
|
ankyrin 2, neuronal |
| chr1_+_50575292 | 8.53 |
ENST00000371821.1
ENST00000371819.1 |
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr5_+_161112754 | 8.51 |
ENST00000523217.1
|
GABRA6
|
gamma-aminobutyric acid (GABA) A receptor, alpha 6 |
| chr11_-_111175739 | 8.48 |
ENST00000532918.1
|
COLCA1
|
colorectal cancer associated 1 |
| chr2_-_152589670 | 8.48 |
ENST00000604864.1
ENST00000603639.1 |
NEB
|
nebulin |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXB6_PRRX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 45.7 | 273.9 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 27.5 | 82.5 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 16.2 | 64.7 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 13.0 | 38.9 | GO:0007412 | axon target recognition(GO:0007412) |
| 12.7 | 38.0 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 12.2 | 146.6 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 10.2 | 40.6 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 9.6 | 28.7 | GO:1904800 | negative regulation of dendrite extension(GO:1903860) regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 8.2 | 65.3 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 7.6 | 7.6 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 6.2 | 61.7 | GO:0051610 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 5.4 | 27.1 | GO:0097338 | response to clozapine(GO:0097338) |
| 5.4 | 16.3 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) |
| 5.3 | 21.3 | GO:0072658 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 4.9 | 9.8 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 4.7 | 23.4 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 4.7 | 23.3 | GO:0099538 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 4.6 | 23.1 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 4.5 | 18.0 | GO:1904450 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 4.3 | 12.9 | GO:0035938 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 4.3 | 86.1 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 4.3 | 34.2 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 4.1 | 12.3 | GO:1905154 | negative regulation of membrane invagination(GO:1905154) |
| 3.8 | 15.1 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 3.7 | 7.5 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 3.7 | 37.0 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 3.6 | 3.6 | GO:0021781 | glial cell fate commitment(GO:0021781) |
| 3.4 | 20.1 | GO:0021764 | amygdala development(GO:0021764) |
| 3.3 | 10.0 | GO:2000366 | regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 3.3 | 13.2 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 3.2 | 9.7 | GO:0048382 | mesendoderm development(GO:0048382) |
| 3.2 | 25.7 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 3.1 | 12.4 | GO:0018032 | peptide amidation(GO:0001519) protein amidation(GO:0018032) peptide modification(GO:0031179) |
| 3.1 | 9.3 | GO:0051795 | positive regulation of catagen(GO:0051795) activation of meiosis(GO:0090427) |
| 3.1 | 39.9 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 3.0 | 121.2 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 3.0 | 54.4 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 3.0 | 3.0 | GO:1902285 | sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) |
| 2.9 | 14.7 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 2.9 | 29.1 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 2.9 | 40.4 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 2.9 | 17.2 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 2.8 | 56.6 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 2.8 | 2.8 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 2.7 | 13.6 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 2.6 | 7.8 | GO:0072276 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 2.5 | 15.1 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 2.4 | 31.6 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 2.4 | 24.2 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 2.4 | 16.8 | GO:0021740 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 2.3 | 4.7 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 2.3 | 18.5 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 2.3 | 25.4 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 2.3 | 20.4 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 2.2 | 6.7 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 2.2 | 59.4 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 2.1 | 4.3 | GO:0021779 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 2.1 | 8.5 | GO:0007525 | somatic muscle development(GO:0007525) |
| 2.1 | 6.2 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 2.0 | 30.5 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 2.0 | 4.0 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 2.0 | 23.7 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 1.9 | 7.7 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 1.9 | 7.5 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 1.9 | 5.6 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 1.8 | 40.3 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 1.8 | 5.4 | GO:0009214 | cAMP catabolic process(GO:0006198) cyclic nucleotide catabolic process(GO:0009214) |
| 1.8 | 3.6 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 1.8 | 5.4 | GO:0048867 | stem cell fate determination(GO:0048867) |
| 1.8 | 101.0 | GO:0003254 | regulation of membrane depolarization(GO:0003254) |
| 1.8 | 5.3 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 1.7 | 7.0 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 1.7 | 7.0 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 1.7 | 12.0 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 1.7 | 8.5 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 1.7 | 18.2 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 1.6 | 8.2 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 1.6 | 11.2 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 1.6 | 12.6 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 1.6 | 11.0 | GO:0090324 | negative regulation of oxidative phosphorylation(GO:0090324) |
| 1.5 | 25.7 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 1.5 | 25.1 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 1.5 | 13.1 | GO:0021681 | cerebellar granular layer development(GO:0021681) |
| 1.4 | 4.3 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 1.4 | 11.4 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 1.4 | 5.5 | GO:0036343 | psychomotor behavior(GO:0036343) motor behavior(GO:0061744) |
| 1.4 | 4.1 | GO:2000320 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 1.4 | 53.2 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 1.4 | 2.7 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 1.4 | 39.4 | GO:0008038 | neuron recognition(GO:0008038) |
| 1.3 | 2.7 | GO:0034505 | tooth mineralization(GO:0034505) |
| 1.3 | 13.3 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 1.3 | 14.4 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 1.3 | 2.6 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 1.3 | 11.6 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 1.3 | 3.9 | GO:0030035 | microspike assembly(GO:0030035) |
| 1.3 | 7.7 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 1.3 | 6.4 | GO:0048690 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 1.3 | 68.7 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 1.3 | 7.6 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 1.2 | 186.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 1.2 | 66.4 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 1.2 | 21.3 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 1.2 | 4.7 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 1.2 | 2.3 | GO:1990637 | response to prolactin(GO:1990637) |
| 1.2 | 13.9 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 1.1 | 3.4 | GO:0060325 | face morphogenesis(GO:0060325) |
| 1.1 | 6.7 | GO:0015811 | L-cystine transport(GO:0015811) |
| 1.1 | 11.2 | GO:0048262 | determination of dorsal/ventral asymmetry(GO:0048262) determination of dorsal identity(GO:0048263) |
| 1.1 | 12.2 | GO:0006554 | lysine catabolic process(GO:0006554) |
| 1.1 | 4.4 | GO:0036269 | swimming behavior(GO:0036269) |
| 1.1 | 4.4 | GO:0060023 | soft palate development(GO:0060023) |
| 1.1 | 14.2 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 1.1 | 5.5 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 1.1 | 4.3 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 1.1 | 3.3 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 1.1 | 1.1 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 1.1 | 3.2 | GO:0003099 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) baroreceptor response to increased systemic arterial blood pressure(GO:0001983) positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 1.1 | 7.4 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 1.1 | 4.2 | GO:1902159 | regulation of cyclic nucleotide-gated ion channel activity(GO:1902159) |
| 1.0 | 32.2 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 1.0 | 32.7 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 1.0 | 29.5 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 1.0 | 3.0 | GO:0090394 | negative regulation of excitatory postsynaptic potential(GO:0090394) |
| 1.0 | 4.9 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 1.0 | 6.8 | GO:0070166 | enamel mineralization(GO:0070166) |
| 1.0 | 9.7 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 1.0 | 1.0 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.9 | 1.9 | GO:0021612 | facial nerve structural organization(GO:0021612) |
| 0.9 | 4.6 | GO:0015824 | proline transport(GO:0015824) |
| 0.9 | 2.7 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.9 | 5.4 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.9 | 4.5 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.9 | 15.2 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.9 | 11.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.9 | 1.7 | GO:0050955 | thermoception(GO:0050955) |
| 0.8 | 15.8 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.8 | 3.3 | GO:0048807 | ectoderm and mesoderm interaction(GO:0007499) female genitalia morphogenesis(GO:0048807) positive regulation of somatic stem cell population maintenance(GO:1904674) |
| 0.8 | 0.8 | GO:0050787 | detoxification of mercury ion(GO:0050787) |
| 0.8 | 3.3 | GO:0061743 | motor learning(GO:0061743) |
| 0.8 | 4.8 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.8 | 6.9 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.8 | 5.3 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.8 | 3.0 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.8 | 3.0 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.8 | 3.0 | GO:0051136 | regulation of NK T cell differentiation(GO:0051136) |
| 0.7 | 6.0 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.7 | 5.2 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.7 | 5.9 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.7 | 24.3 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.7 | 5.1 | GO:0061325 | cell proliferation involved in outflow tract morphogenesis(GO:0061325) |
| 0.7 | 7.3 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.7 | 3.6 | GO:1901660 | calcium ion export(GO:1901660) calcium ion export from cell(GO:1990034) |
| 0.7 | 4.4 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.7 | 3.6 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.7 | 2.1 | GO:0043163 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.7 | 11.0 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.7 | 188.7 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
| 0.7 | 11.4 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.7 | 3.3 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.7 | 4.7 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.7 | 2.0 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.6 | 20.7 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.6 | 3.2 | GO:0003025 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) |
| 0.6 | 3.2 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.6 | 9.8 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.6 | 2.4 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.6 | 17.1 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.6 | 1.8 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.6 | 0.6 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.6 | 5.9 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.6 | 4.1 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.6 | 3.5 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.6 | 1.7 | GO:0010710 | regulation of collagen catabolic process(GO:0010710) negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.6 | 5.7 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.6 | 1.7 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.6 | 4.0 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.6 | 3.4 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.6 | 1.7 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.5 | 18.5 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.5 | 10.3 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.5 | 3.8 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.5 | 2.2 | GO:0002331 | pre-B cell differentiation(GO:0002329) pre-B cell allelic exclusion(GO:0002331) |
| 0.5 | 3.2 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.5 | 6.4 | GO:0010990 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.5 | 1.1 | GO:0042320 | regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) |
| 0.5 | 0.5 | GO:0035711 | T-helper 1 cell activation(GO:0035711) |
| 0.5 | 5.8 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.5 | 28.3 | GO:0051899 | membrane depolarization(GO:0051899) |
| 0.5 | 1.0 | GO:0021798 | neuroblast differentiation(GO:0014016) forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.5 | 25.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.5 | 4.1 | GO:0002870 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.5 | 15.8 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.5 | 3.0 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.5 | 5.4 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.5 | 5.4 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
| 0.5 | 2.0 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.5 | 4.4 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.5 | 1.5 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.5 | 2.9 | GO:1990418 | response to insulin-like growth factor stimulus(GO:1990418) |
| 0.5 | 1.0 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.5 | 10.6 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.5 | 1.9 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.5 | 2.8 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.5 | 3.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.5 | 27.0 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.5 | 3.2 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.5 | 3.6 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.4 | 1.3 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.4 | 2.7 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.4 | 4.4 | GO:0015865 | purine nucleotide transport(GO:0015865) purine ribonucleotide transport(GO:0015868) |
| 0.4 | 3.0 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 0.4 | 8.3 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.4 | 0.4 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.4 | 1.7 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.4 | 15.6 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.4 | 4.6 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.4 | 4.5 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.4 | 7.3 | GO:0015874 | norepinephrine transport(GO:0015874) |
| 0.4 | 1.5 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.4 | 1.5 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.4 | 0.8 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.4 | 3.0 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.4 | 3.0 | GO:1900165 | negative regulation of interleukin-6 secretion(GO:1900165) |
| 0.4 | 2.2 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.4 | 12.1 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.4 | 0.7 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.4 | 9.0 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.4 | 5.0 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.4 | 6.0 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.4 | 4.2 | GO:0035878 | nail development(GO:0035878) |
| 0.3 | 2.8 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.3 | 20.3 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.3 | 1.7 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.3 | 2.0 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.3 | 1.0 | GO:0070781 | arginine biosynthetic process via ornithine(GO:0042450) response to biotin(GO:0070781) |
| 0.3 | 2.3 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.3 | 24.8 | GO:0050806 | positive regulation of synaptic transmission(GO:0050806) |
| 0.3 | 4.5 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.3 | 6.0 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.3 | 3.8 | GO:1903671 | negative regulation of sprouting angiogenesis(GO:1903671) |
| 0.3 | 13.5 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.3 | 1.5 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.3 | 3.3 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.3 | 13.7 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.3 | 5.0 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.3 | 2.3 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.3 | 4.0 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.3 | 2.8 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.3 | 14.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.3 | 2.8 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.3 | 7.6 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.3 | 2.0 | GO:0072501 | cellular divalent inorganic anion homeostasis(GO:0072501) |
| 0.3 | 1.4 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.3 | 5.3 | GO:0050954 | sensory perception of sound(GO:0007605) sensory perception of mechanical stimulus(GO:0050954) |
| 0.3 | 3.1 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.3 | 0.8 | GO:0043416 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.2 | 2.2 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.2 | 2.0 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.2 | 6.1 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.2 | 0.7 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.2 | 1.7 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.2 | 1.3 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.2 | 1.7 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.2 | 26.9 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.2 | 4.4 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.2 | 0.8 | GO:0021997 | response to chlorate(GO:0010157) neural plate axis specification(GO:0021997) |
| 0.2 | 9.4 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.2 | 2.9 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.2 | 1.9 | GO:0034379 | very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.2 | 0.8 | GO:0086042 | cardiac muscle cell-cardiac muscle cell adhesion(GO:0086042) bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.2 | 0.7 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.2 | 3.7 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.2 | 1.3 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.2 | 2.0 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.2 | 2.2 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.2 | 0.5 | GO:0000965 | mitochondrial RNA 3'-end processing(GO:0000965) |
| 0.2 | 1.1 | GO:0048840 | otolith development(GO:0048840) |
| 0.2 | 0.3 | GO:0042441 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.2 | 0.3 | GO:0086053 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 0.2 | 4.2 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.2 | 0.5 | GO:0010578 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.2 | 2.3 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.2 | 4.4 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.2 | 1.6 | GO:2001054 | negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.2 | 9.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.2 | 2.5 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.2 | 0.8 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.2 | 0.9 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.2 | 0.9 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.2 | 1.5 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.2 | 4.4 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.1 | 1.6 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.1 | 0.9 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.1 | 1.6 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.1 | 0.4 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.1 | 1.4 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.1 | 0.4 | GO:0035947 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) |
| 0.1 | 5.1 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.1 | 3.3 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 6.3 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.1 | 0.3 | GO:1901876 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.1 | 0.8 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 3.5 | GO:0048536 | spleen development(GO:0048536) |
| 0.1 | 0.6 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.1 | 11.7 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.1 | 15.0 | GO:0006813 | potassium ion transport(GO:0006813) |
| 0.1 | 2.3 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 0.4 | GO:0042421 | norepinephrine biosynthetic process(GO:0042421) |
| 0.1 | 1.8 | GO:0061053 | somite development(GO:0061053) |
| 0.1 | 6.9 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 2.9 | GO:1902895 | positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.1 | 1.4 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) |
| 0.1 | 0.9 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.1 | 1.6 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.1 | 1.3 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 13.2 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 1.0 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.1 | 1.7 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.1 | 0.7 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.1 | 4.4 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.1 | 2.1 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 1.4 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.1 | 2.9 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 2.6 | GO:0009408 | response to heat(GO:0009408) |
| 0.1 | 0.5 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 1.8 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 0.3 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.1 | 0.4 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 0.4 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 1.8 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.1 | 2.1 | GO:0006986 | response to unfolded protein(GO:0006986) response to topologically incorrect protein(GO:0035966) |
| 0.1 | 2.6 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 0.7 | GO:0032232 | negative regulation of actin filament bundle assembly(GO:0032232) negative regulation of stress fiber assembly(GO:0051497) |
| 0.1 | 0.6 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.1 | 7.4 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.1 | 1.4 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 1.4 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.1 | 1.0 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 0.3 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 1.9 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 1.5 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.6 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 1.6 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 1.8 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 3.1 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 1.1 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 0.7 | GO:0048469 | cell maturation(GO:0048469) |
| 0.0 | 0.6 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 1.5 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 1.3 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.2 | GO:0045605 | negative regulation of epidermal cell differentiation(GO:0045605) |
| 0.0 | 2.6 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 2.3 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.8 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.3 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 2.0 | GO:0015698 | inorganic anion transport(GO:0015698) |
| 0.0 | 0.9 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.5 | GO:0050906 | detection of stimulus involved in sensory perception(GO:0050906) |
| 0.0 | 0.1 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.0 | 0.6 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.3 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 16.5 | 82.5 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 12.1 | 60.3 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 10.3 | 144.8 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 6.1 | 133.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 5.7 | 28.5 | GO:0032584 | growth cone membrane(GO:0032584) |
| 4.6 | 27.6 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 4.5 | 18.0 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 3.9 | 11.8 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 3.9 | 11.7 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 3.8 | 15.2 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 3.7 | 62.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 3.4 | 54.4 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 3.3 | 23.4 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 3.3 | 115.6 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 2.9 | 11.4 | GO:0031673 | H zone(GO:0031673) |
| 2.5 | 12.3 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 2.4 | 38.8 | GO:0031045 | dense core granule(GO:0031045) |
| 2.2 | 15.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 2.1 | 6.4 | GO:0072534 | perineuronal net(GO:0072534) |
| 2.0 | 17.6 | GO:0016013 | syntrophin complex(GO:0016013) |
| 1.9 | 26.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 1.9 | 16.8 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 1.7 | 51.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 1.5 | 21.1 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 1.4 | 27.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 1.2 | 12.1 | GO:0045180 | basal cortex(GO:0045180) |
| 1.2 | 49.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 1.1 | 13.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 1.1 | 4.4 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 1.1 | 5.4 | GO:0044308 | axonal spine(GO:0044308) |
| 1.0 | 10.5 | GO:0071953 | elastic fiber(GO:0071953) |
| 1.0 | 88.5 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 1.0 | 68.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.9 | 0.9 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.8 | 218.9 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.8 | 186.9 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.8 | 8.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.7 | 4.5 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.7 | 11.1 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.7 | 157.0 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.7 | 5.5 | GO:0030478 | actin cap(GO:0030478) |
| 0.7 | 6.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.7 | 3.4 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.7 | 4.0 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.6 | 3.2 | GO:0060198 | clathrin-sculpted vesicle(GO:0060198) clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.6 | 9.9 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.6 | 19.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.6 | 1.7 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.6 | 5.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.6 | 6.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.5 | 4.3 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.5 | 2.5 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.5 | 4.4 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.5 | 2.8 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.5 | 5.9 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.5 | 5.0 | GO:0005922 | connexon complex(GO:0005922) |
| 0.5 | 3.6 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.5 | 5.5 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.4 | 54.2 | GO:0030426 | growth cone(GO:0030426) |
| 0.4 | 43.8 | GO:0043204 | perikaryon(GO:0043204) |
| 0.4 | 0.8 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.4 | 50.9 | GO:0030018 | Z disc(GO:0030018) |
| 0.4 | 58.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.3 | 1.7 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.3 | 2.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.3 | 4.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.3 | 4.9 | GO:0031430 | M band(GO:0031430) |
| 0.3 | 56.0 | GO:0098794 | postsynapse(GO:0098794) |
| 0.3 | 1.7 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.3 | 6.2 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.3 | 1.8 | GO:0036019 | endolysosome(GO:0036019) |
| 0.3 | 7.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.2 | 8.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.2 | 3.6 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) |
| 0.2 | 1.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.2 | 12.5 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.2 | 5.7 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.2 | 44.7 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.2 | 4.4 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.2 | 3.0 | GO:0032059 | bleb(GO:0032059) |
| 0.2 | 12.7 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.2 | 1.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.2 | 5.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.2 | 2.8 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.2 | 10.2 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.2 | 13.3 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.2 | 9.4 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.2 | 1.5 | GO:0005921 | gap junction(GO:0005921) |
| 0.2 | 3.0 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.2 | 7.8 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.2 | 0.8 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 4.3 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.2 | 0.5 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.1 | 3.3 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 0.4 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 6.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 2.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 1.7 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.1 | 5.7 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 39.6 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 17.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 1.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 7.4 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 20.4 | GO:0030424 | axon(GO:0030424) |
| 0.1 | 8.9 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.1 | 1.1 | GO:0031674 | I band(GO:0031674) |
| 0.1 | 7.0 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.1 | 1.0 | GO:0032994 | protein-lipid complex(GO:0032994) plasma lipoprotein particle(GO:0034358) lipoprotein particle(GO:1990777) |
| 0.1 | 3.0 | GO:0005657 | replication fork(GO:0005657) |
| 0.1 | 2.1 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 1.6 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 0.3 | GO:0097060 | synaptic membrane(GO:0097060) |
| 0.1 | 18.2 | GO:0045202 | synapse(GO:0045202) |
| 0.1 | 1.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 11.6 | GO:0005770 | late endosome(GO:0005770) |
| 0.1 | 3.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 3.2 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.1 | 4.7 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 9.3 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 11.9 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.1 | 1.5 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 189.5 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.1 | 0.6 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 1.5 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 8.6 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.5 | GO:0098798 | mitochondrial protein complex(GO:0098798) |
| 0.0 | 0.6 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 1.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.4 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 2.2 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.4 | GO:0098791 | Golgi subcompartment(GO:0098791) |
| 0.0 | 0.4 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 1.9 | GO:0005929 | cilium(GO:0005929) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 17.0 | 220.6 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 16.5 | 82.5 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 9.9 | 39.8 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 7.4 | 29.5 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 6.4 | 32.2 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 6.0 | 71.4 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 5.4 | 32.7 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 5.0 | 25.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 4.7 | 23.3 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 4.5 | 18.0 | GO:0047298 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 4.4 | 57.4 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 4.1 | 65.3 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 3.8 | 11.3 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 3.2 | 107.8 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 3.1 | 68.3 | GO:0035198 | miRNA binding(GO:0035198) |
| 3.1 | 12.4 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 3.1 | 18.5 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 2.9 | 22.8 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 2.7 | 8.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 2.7 | 11.0 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 2.7 | 13.6 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 2.7 | 27.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 2.7 | 13.3 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 2.6 | 62.3 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 2.6 | 7.8 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 2.6 | 7.7 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 2.4 | 7.3 | GO:0005330 | dopamine:sodium symporter activity(GO:0005330) |
| 2.4 | 49.8 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 2.2 | 6.6 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 2.2 | 159.2 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 2.2 | 56.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 2.0 | 13.7 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 1.9 | 15.2 | GO:0043559 | insulin binding(GO:0043559) |
| 1.8 | 3.6 | GO:0004945 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 1.8 | 7.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 1.7 | 5.2 | GO:0005549 | odorant binding(GO:0005549) |
| 1.7 | 27.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 1.7 | 51.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 1.6 | 8.2 | GO:0004359 | glutaminase activity(GO:0004359) |
| 1.6 | 11.4 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 1.5 | 10.5 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 1.5 | 7.4 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 1.5 | 34.9 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 1.4 | 10.1 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 1.4 | 5.5 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 1.3 | 4.0 | GO:0017129 | triglyceride binding(GO:0017129) |
| 1.2 | 155.0 | GO:0005262 | calcium channel activity(GO:0005262) |
| 1.2 | 2.4 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 1.2 | 52.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 1.2 | 5.8 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 1.1 | 9.0 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 1.1 | 6.7 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 1.1 | 4.5 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 1.1 | 53.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 1.1 | 33.9 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 1.1 | 15.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 1.1 | 7.6 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 1.1 | 5.4 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 1.1 | 3.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 1.1 | 3.2 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 1.0 | 18.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 1.0 | 11.0 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 1.0 | 7.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 1.0 | 20.1 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 1.0 | 4.8 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.9 | 3.8 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.9 | 20.4 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.9 | 2.7 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.9 | 7.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.9 | 6.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.9 | 11.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.9 | 136.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.8 | 3.4 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.8 | 7.5 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.8 | 3.3 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.8 | 1.6 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.8 | 14.5 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.8 | 2.3 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.8 | 3.0 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.7 | 5.9 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.7 | 10.0 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.7 | 5.0 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.7 | 19.6 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.7 | 4.2 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.7 | 2.1 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.7 | 27.6 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.7 | 11.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.7 | 17.2 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.7 | 2.6 | GO:0030305 | heparanase activity(GO:0030305) |
| 0.7 | 10.5 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.6 | 4.5 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.6 | 5.2 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.6 | 9.0 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.6 | 4.4 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.6 | 7.0 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.6 | 4.4 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.6 | 6.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.6 | 4.9 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.6 | 3.0 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.6 | 5.5 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.6 | 123.9 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.6 | 17.4 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.6 | 10.4 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.6 | 11.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.6 | 8.5 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.5 | 43.9 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.5 | 21.9 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.5 | 3.6 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.5 | 2.6 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.5 | 3.0 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.5 | 5.4 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.5 | 6.8 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.5 | 3.9 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.5 | 1.4 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.5 | 9.7 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.5 | 3.2 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.5 | 3.6 | GO:0022821 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) potassium ion antiporter activity(GO:0022821) |
| 0.5 | 1.4 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.4 | 7.4 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.4 | 2.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.4 | 5.9 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.4 | 4.9 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.4 | 14.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.4 | 3.2 | GO:0005534 | galactose binding(GO:0005534) |
| 0.4 | 8.6 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.4 | 2.7 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.4 | 1.6 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.4 | 64.6 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.4 | 5.4 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.4 | 5.6 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.4 | 1.4 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.4 | 5.0 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.3 | 48.1 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.3 | 1.0 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.3 | 4.0 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.3 | 7.7 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.3 | 5.4 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.3 | 3.1 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.3 | 2.8 | GO:0031432 | titin binding(GO:0031432) |
| 0.3 | 1.2 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.3 | 50.9 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.3 | 34.0 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.3 | 2.6 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.3 | 3.3 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.3 | 2.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.3 | 2.8 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.3 | 0.8 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.2 | 8.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.2 | 2.0 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.2 | 3.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.2 | 3.7 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.2 | 4.5 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.2 | 1.6 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.2 | 10.1 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.2 | 12.9 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.2 | 18.5 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.2 | 137.3 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.2 | 1.9 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.2 | 26.7 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.2 | 1.3 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.2 | 2.4 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.2 | 2.0 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.2 | 5.4 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.2 | 3.3 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.2 | 0.4 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.2 | 4.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.2 | 8.9 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.2 | 9.4 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.2 | 6.2 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.2 | 1.8 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.2 | 2.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.2 | 1.0 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.2 | 0.8 | GO:0097108 | patched binding(GO:0005113) hedgehog family protein binding(GO:0097108) |
| 0.2 | 4.6 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.2 | 3.6 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.2 | 3.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 1.5 | GO:0047035 | alcohol dehydrogenase (NAD) activity(GO:0004022) testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.1 | 36.2 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 4.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 1.9 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.1 | 0.8 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.1 | 5.9 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 1.5 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 1.8 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 2.2 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.1 | 5.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.9 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.1 | 7.2 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 2.3 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 0.8 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 0.7 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.1 | 4.7 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.1 | 1.5 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 3.3 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.1 | 2.8 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 1.1 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.1 | 8.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.7 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 2.4 | GO:0008144 | drug binding(GO:0008144) |
| 0.1 | 2.0 | GO:0032813 | tumor necrosis factor receptor superfamily binding(GO:0032813) |
| 0.1 | 15.4 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.1 | 0.6 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.1 | 6.9 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 6.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 2.9 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 0.8 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.1 | 0.4 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 1.6 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 6.1 | GO:0002020 | protease binding(GO:0002020) |
| 0.1 | 3.2 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.1 | 1.7 | GO:0030291 | protein serine/threonine kinase inhibitor activity(GO:0030291) |
| 0.0 | 3.2 | GO:0008186 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 4.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.6 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 1.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 1.2 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.7 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 1.6 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 1.7 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.9 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.3 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.8 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 1.5 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.8 | GO:0022843 | voltage-gated cation channel activity(GO:0022843) |
| 0.0 | 0.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 134.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 2.0 | 116.3 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 1.9 | 1.9 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 1.8 | 93.0 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 1.4 | 166.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 1.3 | 88.1 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 1.2 | 63.5 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 1.0 | 71.1 | PID FGF PATHWAY | FGF signaling pathway |
| 0.7 | 27.1 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.7 | 41.2 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.7 | 20.2 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.6 | 13.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.6 | 8.4 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.6 | 11.0 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.6 | 3.9 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.5 | 31.9 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.5 | 40.4 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.5 | 5.5 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.5 | 2.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.5 | 25.6 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.4 | 5.4 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.4 | 7.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.4 | 15.9 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.4 | 21.6 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.3 | 10.0 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.3 | 2.3 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.3 | 13.1 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.3 | 0.6 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.3 | 12.7 | PID MYC PATHWAY | C-MYC pathway |
| 0.3 | 15.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.3 | 10.9 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.2 | 15.6 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.2 | 3.9 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.2 | 5.8 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.2 | 51.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.2 | 16.1 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.2 | 9.3 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.2 | 10.1 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.2 | 3.0 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.2 | 2.8 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 1.3 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 3.2 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 4.7 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 0.5 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.1 | 21.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.4 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 5.4 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 3.8 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 2.7 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 2.4 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 3.2 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 3.2 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 1.8 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 3.0 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 3.7 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 14.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.3 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.7 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.7 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.1 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.2 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.5 | 98.5 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 5.5 | 295.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 5.0 | 136.0 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 4.5 | 13.4 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 3.6 | 110.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 2.6 | 71.4 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 2.5 | 83.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 2.4 | 61.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 1.4 | 41.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 1.3 | 15.1 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 1.2 | 18.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 1.2 | 22.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 1.1 | 4.4 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 1.1 | 9.7 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 1.0 | 11.0 | REACTOME OPSINS | Genes involved in Opsins |
| 1.0 | 24.6 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 1.0 | 24.3 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.9 | 18.0 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.9 | 20.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.8 | 12.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.8 | 11.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.7 | 10.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.7 | 26.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.6 | 19.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.6 | 20.1 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.6 | 12.7 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.6 | 8.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.6 | 9.7 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.6 | 20.4 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.6 | 11.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.5 | 5.3 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.5 | 16.8 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.5 | 6.9 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.4 | 27.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.4 | 6.1 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.4 | 7.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.4 | 3.8 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.4 | 4.9 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.3 | 14.0 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.3 | 2.7 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.3 | 5.6 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.3 | 55.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.3 | 6.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.3 | 6.3 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.3 | 6.9 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.3 | 11.4 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.3 | 5.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.3 | 9.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.3 | 8.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.3 | 15.6 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.3 | 14.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.3 | 12.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.3 | 6.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.3 | 4.0 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.3 | 6.4 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.3 | 6.0 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.2 | 7.0 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 17.5 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.2 | 13.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.2 | 10.7 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.2 | 7.4 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.2 | 3.2 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.2 | 3.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.2 | 1.8 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.2 | 8.8 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.2 | 2.0 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 2.5 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 7.1 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 14.8 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 23.4 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.1 | 3.1 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 3.3 | REACTOME AXON GUIDANCE | Genes involved in Axon guidance |
| 0.1 | 1.6 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.1 | 7.1 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 8.1 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.1 | 5.5 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 2.6 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.1 | 10.6 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 1.8 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 2.0 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 2.7 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 3.4 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 1.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 0.9 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.1 | 0.7 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 0.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.1 | 2.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 5.0 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 1.1 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.4 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 1.9 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 1.5 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.8 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 0.6 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.1 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |