Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
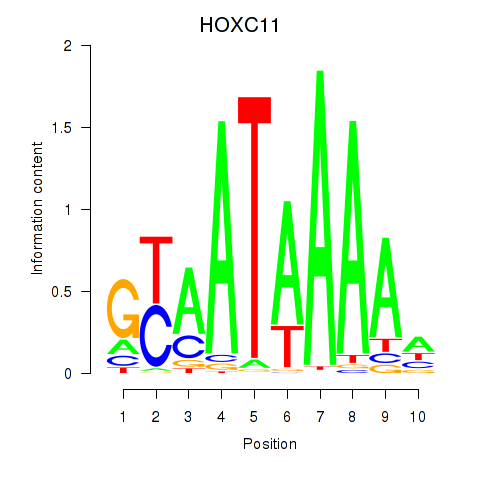
Results for HOXC11
Z-value: 0.84
Transcription factors associated with HOXC11
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXC11
|
ENSG00000123388.4 | homeobox C11 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXC11 | hg19_v2_chr12_+_54366894_54366922 | -0.42 | 7.1e-11 | Click! |
Activity profile of HOXC11 motif
Sorted Z-values of HOXC11 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_190044480 | 32.16 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr18_+_3252265 | 21.69 |
ENST00000580887.1
ENST00000536605.1 |
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr12_-_54071181 | 19.48 |
ENST00000338662.5
|
ATP5G2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C2 (subunit 9) |
| chr2_-_136678123 | 18.79 |
ENST00000422708.1
|
DARS
|
aspartyl-tRNA synthetase |
| chr8_+_26150628 | 14.36 |
ENST00000523925.1
ENST00000315985.7 |
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha |
| chr7_-_93519471 | 11.37 |
ENST00000451238.1
|
TFPI2
|
tissue factor pathway inhibitor 2 |
| chr5_-_111091948 | 11.33 |
ENST00000447165.2
|
NREP
|
neuronal regeneration related protein |
| chr6_+_12290586 | 11.03 |
ENST00000379375.5
|
EDN1
|
endothelin 1 |
| chr2_-_176046391 | 10.87 |
ENST00000392541.3
ENST00000409194.1 |
ATP5G3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr2_+_109204909 | 10.66 |
ENST00000393310.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr7_+_134464376 | 10.43 |
ENST00000454108.1
ENST00000361675.2 |
CALD1
|
caldesmon 1 |
| chr14_+_56127989 | 10.34 |
ENST00000555573.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr2_-_207023918 | 10.07 |
ENST00000455934.2
ENST00000449699.1 ENST00000454195.1 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr7_-_47579188 | 9.89 |
ENST00000398879.1
ENST00000355730.3 ENST00000442536.2 ENST00000458317.2 |
TNS3
|
tensin 3 |
| chr8_+_104831554 | 9.76 |
ENST00000408894.2
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr19_+_36036477 | 9.66 |
ENST00000222284.5
ENST00000392204.2 |
TMEM147
|
transmembrane protein 147 |
| chr7_+_134464414 | 9.58 |
ENST00000361901.2
|
CALD1
|
caldesmon 1 |
| chr6_+_32812568 | 9.37 |
ENST00000414474.1
|
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr19_+_36036583 | 9.30 |
ENST00000392205.1
|
TMEM147
|
transmembrane protein 147 |
| chr7_-_151217166 | 8.65 |
ENST00000496004.1
|
RHEB
|
Ras homolog enriched in brain |
| chr19_-_39924349 | 8.39 |
ENST00000602153.1
|
RPS16
|
ribosomal protein S16 |
| chr2_+_27440229 | 8.30 |
ENST00000264705.4
ENST00000403525.1 |
CAD
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr18_+_3252206 | 8.14 |
ENST00000578562.2
|
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr16_-_2314222 | 7.61 |
ENST00000566397.1
|
RNPS1
|
RNA binding protein S1, serine-rich domain |
| chr1_+_167298281 | 7.42 |
ENST00000367862.5
|
POU2F1
|
POU class 2 homeobox 1 |
| chr17_-_39684550 | 7.40 |
ENST00000455635.1
ENST00000361566.3 |
KRT19
|
keratin 19 |
| chr4_-_74088800 | 6.95 |
ENST00000509867.2
|
ANKRD17
|
ankyrin repeat domain 17 |
| chr10_+_114709999 | 6.80 |
ENST00000355995.4
ENST00000545257.1 ENST00000543371.1 ENST00000536810.1 ENST00000355717.4 ENST00000538897.1 ENST00000534894.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr19_+_19626531 | 6.69 |
ENST00000507754.4
|
NDUFA13
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 13 |
| chr17_-_77924627 | 6.29 |
ENST00000572862.1
ENST00000573782.1 ENST00000574427.1 ENST00000570373.1 ENST00000340848.7 ENST00000576768.1 |
TBC1D16
|
TBC1 domain family, member 16 |
| chr1_-_9129895 | 6.20 |
ENST00000473209.1
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chrX_-_10851762 | 6.12 |
ENST00000380785.1
ENST00000380787.1 |
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr3_-_37216055 | 6.02 |
ENST00000336686.4
|
LRRFIP2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr1_+_23345930 | 5.92 |
ENST00000356634.3
|
KDM1A
|
lysine (K)-specific demethylase 1A |
| chr12_+_1800179 | 5.91 |
ENST00000357103.4
|
ADIPOR2
|
adiponectin receptor 2 |
| chr19_+_49468558 | 5.77 |
ENST00000331825.6
|
FTL
|
ferritin, light polypeptide |
| chr14_-_25479811 | 5.55 |
ENST00000550887.1
|
STXBP6
|
syntaxin binding protein 6 (amisyn) |
| chr11_+_64001962 | 5.38 |
ENST00000309422.2
|
VEGFB
|
vascular endothelial growth factor B |
| chr1_+_23345943 | 5.21 |
ENST00000400181.4
ENST00000542151.1 |
KDM1A
|
lysine (K)-specific demethylase 1A |
| chr19_-_45927622 | 5.14 |
ENST00000300853.3
ENST00000589165.1 |
ERCC1
|
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence) |
| chr16_-_67978016 | 5.04 |
ENST00000264005.5
|
LCAT
|
lecithin-cholesterol acyltransferase |
| chr22_-_30642782 | 4.93 |
ENST00000249075.3
|
LIF
|
leukemia inhibitory factor |
| chr10_+_47894023 | 4.90 |
ENST00000358474.5
|
FAM21B
|
family with sequence similarity 21, member B |
| chr7_-_41742697 | 4.67 |
ENST00000242208.4
|
INHBA
|
inhibin, beta A |
| chr11_-_27723158 | 4.67 |
ENST00000395980.2
|
BDNF
|
brain-derived neurotrophic factor |
| chr4_+_169013666 | 4.63 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chr2_-_145278475 | 4.36 |
ENST00000558170.2
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr5_+_43121698 | 4.08 |
ENST00000505606.2
ENST00000509634.1 ENST00000509341.1 |
ZNF131
|
zinc finger protein 131 |
| chr4_-_25865159 | 4.03 |
ENST00000502949.1
ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr3_-_79816965 | 3.96 |
ENST00000464233.1
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr5_-_82969405 | 3.59 |
ENST00000510978.1
|
HAPLN1
|
hyaluronan and proteoglycan link protein 1 |
| chr16_+_67906919 | 3.53 |
ENST00000358933.5
|
EDC4
|
enhancer of mRNA decapping 4 |
| chr9_+_12693336 | 3.47 |
ENST00000381137.2
ENST00000388918.5 |
TYRP1
|
tyrosinase-related protein 1 |
| chr1_-_94147385 | 3.40 |
ENST00000260502.6
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr6_+_31683117 | 3.31 |
ENST00000375825.3
ENST00000375824.1 |
LY6G6D
|
lymphocyte antigen 6 complex, locus G6D |
| chr19_-_36233332 | 3.28 |
ENST00000592537.1
ENST00000246532.1 ENST00000344990.3 ENST00000588992.1 |
IGFLR1
|
IGF-like family receptor 1 |
| chr15_+_67418047 | 3.16 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr6_+_46761118 | 3.14 |
ENST00000230588.4
|
MEP1A
|
meprin A, alpha (PABA peptide hydrolase) |
| chr1_+_12524965 | 3.03 |
ENST00000471923.1
|
VPS13D
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr4_+_144354644 | 2.91 |
ENST00000512843.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr17_-_79876010 | 2.83 |
ENST00000328666.6
|
SIRT7
|
sirtuin 7 |
| chr7_-_27205136 | 2.78 |
ENST00000396345.1
ENST00000343483.6 |
HOXA9
|
homeobox A9 |
| chrX_-_133792480 | 2.76 |
ENST00000359237.4
|
PLAC1
|
placenta-specific 1 |
| chr6_+_15246501 | 2.72 |
ENST00000341776.2
|
JARID2
|
jumonji, AT rich interactive domain 2 |
| chr10_+_17271266 | 2.71 |
ENST00000224237.5
|
VIM
|
vimentin |
| chr10_+_115614370 | 2.61 |
ENST00000369301.3
|
NHLRC2
|
NHL repeat containing 2 |
| chr1_-_9129631 | 2.51 |
ENST00000377414.3
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr7_+_22766766 | 2.47 |
ENST00000426291.1
ENST00000401651.1 ENST00000258743.5 ENST00000420258.2 ENST00000407492.1 ENST00000401630.3 ENST00000406575.1 |
IL6
|
interleukin 6 (interferon, beta 2) |
| chr2_+_211421262 | 2.43 |
ENST00000233072.5
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial |
| chr8_+_134203273 | 2.43 |
ENST00000250160.6
|
WISP1
|
WNT1 inducible signaling pathway protein 1 |
| chrX_-_107682702 | 2.42 |
ENST00000372216.4
|
COL4A6
|
collagen, type IV, alpha 6 |
| chr17_+_58018269 | 2.32 |
ENST00000591035.1
|
RP11-178C3.1
|
Uncharacterized protein |
| chr10_-_96829246 | 2.28 |
ENST00000371270.3
ENST00000535898.1 ENST00000539050.1 |
CYP2C8
|
cytochrome P450, family 2, subfamily C, polypeptide 8 |
| chr12_+_64173583 | 2.23 |
ENST00000261234.6
|
TMEM5
|
transmembrane protein 5 |
| chr2_+_208423891 | 2.19 |
ENST00000448277.1
ENST00000457101.1 |
CREB1
|
cAMP responsive element binding protein 1 |
| chr5_+_148521381 | 2.07 |
ENST00000504238.1
|
ABLIM3
|
actin binding LIM protein family, member 3 |
| chr4_+_160188889 | 1.96 |
ENST00000264431.4
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr2_+_228678550 | 1.95 |
ENST00000409189.3
ENST00000358813.4 |
CCL20
|
chemokine (C-C motif) ligand 20 |
| chr5_-_88119580 | 1.67 |
ENST00000539796.1
|
MEF2C
|
myocyte enhancer factor 2C |
| chrX_+_65382433 | 1.65 |
ENST00000374727.3
|
HEPH
|
hephaestin |
| chr3_+_173116225 | 1.62 |
ENST00000457714.1
|
NLGN1
|
neuroligin 1 |
| chr16_+_15031300 | 1.59 |
ENST00000328085.6
|
NPIPA1
|
nuclear pore complex interacting protein family, member A1 |
| chr13_-_46679185 | 1.47 |
ENST00000439329.3
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr15_+_49715293 | 1.46 |
ENST00000267843.4
ENST00000560270.1 |
FGF7
|
fibroblast growth factor 7 |
| chr6_+_6588902 | 1.44 |
ENST00000230568.4
|
LY86
|
lymphocyte antigen 86 |
| chr1_-_204135450 | 1.42 |
ENST00000272190.8
ENST00000367195.2 |
REN
|
renin |
| chr8_+_76452097 | 1.35 |
ENST00000396423.2
|
HNF4G
|
hepatocyte nuclear factor 4, gamma |
| chr1_+_87794150 | 1.30 |
ENST00000370544.5
|
LMO4
|
LIM domain only 4 |
| chr3_+_190105909 | 1.26 |
ENST00000456423.1
|
CLDN16
|
claudin 16 |
| chrX_-_105282712 | 1.26 |
ENST00000372563.1
|
SERPINA7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr12_+_25205568 | 1.23 |
ENST00000548766.1
ENST00000556887.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr7_-_122840015 | 1.22 |
ENST00000194130.2
|
SLC13A1
|
solute carrier family 13 (sodium/sulfate symporter), member 1 |
| chr13_-_46679144 | 1.22 |
ENST00000181383.4
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr12_-_28124903 | 1.20 |
ENST00000395872.1
ENST00000354417.3 ENST00000201015.4 |
PTHLH
|
parathyroid hormone-like hormone |
| chr6_-_41703952 | 1.16 |
ENST00000358871.2
ENST00000403298.4 |
TFEB
|
transcription factor EB |
| chr10_-_115614127 | 1.06 |
ENST00000369305.1
|
DCLRE1A
|
DNA cross-link repair 1A |
| chr19_+_1440838 | 1.01 |
ENST00000594262.1
|
AC027307.3
|
Uncharacterized protein |
| chr5_+_145718587 | 0.95 |
ENST00000230732.4
|
POU4F3
|
POU class 4 homeobox 3 |
| chr7_-_81399355 | 0.95 |
ENST00000457544.2
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr6_-_64029879 | 0.92 |
ENST00000370658.5
ENST00000485906.2 ENST00000370657.4 |
LGSN
|
lengsin, lens protein with glutamine synthetase domain |
| chr12_+_25205666 | 0.88 |
ENST00000547044.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr15_-_68497657 | 0.86 |
ENST00000448060.2
ENST00000467889.1 |
CALML4
|
calmodulin-like 4 |
| chr17_-_46806540 | 0.83 |
ENST00000290295.7
|
HOXB13
|
homeobox B13 |
| chr2_+_176987088 | 0.71 |
ENST00000249499.6
|
HOXD9
|
homeobox D9 |
| chr16_-_21663950 | 0.69 |
ENST00000268389.4
|
IGSF6
|
immunoglobulin superfamily, member 6 |
| chr3_+_15045419 | 0.65 |
ENST00000406272.2
|
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr17_+_7461613 | 0.61 |
ENST00000438470.1
ENST00000436057.1 |
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chrX_+_99899180 | 0.58 |
ENST00000373004.3
|
SRPX2
|
sushi-repeat containing protein, X-linked 2 |
| chr1_-_9129598 | 0.56 |
ENST00000535586.1
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr2_-_40739501 | 0.54 |
ENST00000403092.1
ENST00000402441.1 ENST00000448531.1 |
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr13_-_103719196 | 0.45 |
ENST00000245312.3
|
SLC10A2
|
solute carrier family 10 (sodium/bile acid cotransporter), member 2 |
| chr16_-_21663919 | 0.44 |
ENST00000569602.1
|
IGSF6
|
immunoglobulin superfamily, member 6 |
| chr1_+_62439037 | 0.40 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr22_+_40573921 | 0.38 |
ENST00000454349.2
ENST00000335727.9 |
TNRC6B
|
trinucleotide repeat containing 6B |
| chr6_-_31745085 | 0.36 |
ENST00000375686.3
ENST00000447450.1 |
VWA7
|
von Willebrand factor A domain containing 7 |
| chr7_-_151217001 | 0.35 |
ENST00000262187.5
|
RHEB
|
Ras homolog enriched in brain |
| chr12_-_24102576 | 0.35 |
ENST00000537393.1
ENST00000309359.1 ENST00000381381.2 ENST00000451604.2 |
SOX5
|
SRY (sex determining region Y)-box 5 |
| chr11_-_102576537 | 0.35 |
ENST00000260229.4
|
MMP27
|
matrix metallopeptidase 27 |
| chr8_+_104831472 | 0.34 |
ENST00000262231.10
ENST00000507740.1 |
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr2_-_166930131 | 0.20 |
ENST00000303395.4
ENST00000409050.1 ENST00000423058.2 ENST00000375405.3 |
SCN1A
|
sodium channel, voltage-gated, type I, alpha subunit |
| chr8_-_57123815 | 0.18 |
ENST00000316981.3
ENST00000423799.2 ENST00000429357.2 |
PLAG1
|
pleiomorphic adenoma gene 1 |
| chrX_+_107683096 | 0.16 |
ENST00000328300.6
ENST00000361603.2 |
COL4A5
|
collagen, type IV, alpha 5 |
| chr3_+_89156674 | 0.12 |
ENST00000336596.2
|
EPHA3
|
EPH receptor A3 |
| chr9_-_73736511 | 0.12 |
ENST00000377110.3
ENST00000377111.2 |
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr12_-_47219733 | 0.12 |
ENST00000547477.1
ENST00000447411.1 ENST00000266579.4 |
SLC38A4
|
solute carrier family 38, member 4 |
| chr12_-_52779433 | 0.05 |
ENST00000257951.3
|
KRT84
|
keratin 84 |
| chr8_-_59412717 | 0.03 |
ENST00000301645.3
|
CYP7A1
|
cytochrome P450, family 7, subfamily A, polypeptide 1 |
| chr7_-_81399438 | 0.02 |
ENST00000222390.5
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXC11
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.0 | 32.2 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 6.3 | 18.8 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 3.7 | 11.1 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 3.7 | 11.0 | GO:0060584 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 2.8 | 8.3 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 1.7 | 7.0 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 1.6 | 4.7 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 1.6 | 4.7 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 1.3 | 4.0 | GO:0021823 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 1.3 | 5.0 | GO:0090107 | regulation of high-density lipoprotein particle assembly(GO:0090107) |
| 1.2 | 4.9 | GO:0060708 | histone H3-K27 acetylation(GO:0043974) leukemia inhibitory factor signaling pathway(GO:0048861) spongiotrophoblast differentiation(GO:0060708) regulation of histone H3-K27 acetylation(GO:1901674) |
| 1.2 | 8.4 | GO:0097319 | fructose transport(GO:0015755) fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 1.1 | 10.1 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 1.1 | 4.4 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 1.0 | 9.0 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.9 | 30.3 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.9 | 2.8 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.9 | 5.1 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) positive regulation of t-circle formation(GO:1904431) |
| 0.8 | 5.8 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.8 | 2.5 | GO:0002384 | hepatic immune response(GO:0002384) response to prolactin(GO:1990637) regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 0.8 | 14.4 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.7 | 6.7 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.7 | 5.9 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.7 | 3.5 | GO:1900378 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.7 | 2.7 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.7 | 2.0 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.7 | 2.0 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.6 | 3.2 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.6 | 3.7 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.6 | 5.5 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.6 | 2.4 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) response to oleic acid(GO:0034201) |
| 0.6 | 5.4 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.5 | 1.6 | GO:0099543 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.5 | 1.5 | GO:0060437 | lung growth(GO:0060437) |
| 0.5 | 6.1 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.4 | 3.5 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.4 | 1.7 | GO:0060025 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) regulation of synaptic activity(GO:0060025) |
| 0.4 | 2.2 | GO:1904550 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.4 | 19.0 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.4 | 11.4 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.3 | 2.3 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.3 | 29.8 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.3 | 10.7 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.3 | 1.4 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.3 | 0.8 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.3 | 2.8 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.2 | 9.9 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.2 | 2.7 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.2 | 2.8 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.2 | 0.9 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.2 | 7.6 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.2 | 7.4 | GO:0060706 | cell differentiation involved in embryonic placenta development(GO:0060706) |
| 0.2 | 8.4 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.2 | 2.7 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.2 | 2.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.2 | 1.4 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.2 | 6.3 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.1 | 1.3 | GO:0042659 | regulation of cell fate specification(GO:0042659) |
| 0.1 | 3.3 | GO:0095500 | acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.1 | 1.2 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.1 | 10.1 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.1 | 1.0 | GO:0021562 | vestibulocochlear nerve development(GO:0021562) |
| 0.1 | 0.6 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 1.3 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 1.1 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 | 1.3 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.1 | 9.4 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 7.4 | GO:0009301 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.1 | 1.8 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.1 | 0.3 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.1 | 0.5 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.1 | 1.2 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.7 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.0 | 10.3 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 2.9 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 2.6 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 2.1 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.4 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 | 0.7 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 1.2 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 11.2 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.2 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.0 | 0.2 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 2.1 | GO:0006903 | vesicle targeting(GO:0006903) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.7 | 32.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 2.8 | 11.0 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 2.0 | 10.1 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 1.7 | 30.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 1.6 | 4.7 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 1.4 | 5.8 | GO:0008043 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 1.3 | 20.0 | GO:0030478 | actin cap(GO:0030478) |
| 1.2 | 7.4 | GO:1990357 | terminal web(GO:1990357) |
| 1.2 | 9.4 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.9 | 18.8 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.8 | 7.6 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.7 | 5.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) ERCC4-ERCC1 complex(GO:0070522) |
| 0.6 | 3.7 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.6 | 2.5 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.4 | 14.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.4 | 3.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.3 | 2.6 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.3 | 28.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.3 | 5.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.2 | 11.1 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.2 | 10.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.2 | 3.5 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 1.6 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 5.0 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 8.4 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 2.7 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 8.3 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 6.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 8.4 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.1 | 0.3 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 2.2 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 5.8 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 2.1 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 2.4 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 3.3 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 2.1 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 2.2 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 23.3 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 9.0 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 15.4 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 3.2 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.5 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 4.3 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 3.9 | GO:0044452 | nucleolar part(GO:0044452) |
| 0.0 | 0.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 1.6 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.6 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 12.9 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 3.4 | GO:0005769 | early endosome(GO:0005769) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 18.8 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 3.7 | 11.1 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 3.6 | 10.7 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 2.2 | 11.0 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 1.6 | 4.7 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 1.6 | 4.7 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 1.3 | 5.0 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 1.2 | 8.4 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 1.1 | 30.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.9 | 2.8 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.9 | 5.4 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.8 | 14.4 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.7 | 5.1 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.7 | 20.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.7 | 4.0 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.7 | 2.0 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.6 | 3.2 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.6 | 3.3 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.5 | 2.0 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.5 | 49.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.5 | 5.8 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.5 | 10.1 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.5 | 2.3 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.5 | 2.7 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.4 | 4.9 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.3 | 9.4 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.3 | 5.9 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.3 | 2.5 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.3 | 5.5 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.3 | 2.2 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.2 | 9.1 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.2 | 0.9 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.2 | 10.3 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.2 | 3.7 | GO:0070016 | gamma-catenin binding(GO:0045295) armadillo repeat domain binding(GO:0070016) |
| 0.2 | 0.5 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.2 | 1.2 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.1 | 1.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 1.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 9.0 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 0.5 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 4.4 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 4.6 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 1.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 14.9 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 2.7 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.1 | 1.4 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.1 | 6.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 1.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 1.4 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 6.3 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 1.5 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 5.1 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 2.2 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.2 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 1.2 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 8.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 3.5 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.0 | 1.1 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 3.2 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 9.4 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 1.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 34.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.3 | 5.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.2 | 4.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.2 | 11.1 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.2 | 2.5 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.2 | 7.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.2 | 9.0 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.2 | 12.8 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.2 | 11.0 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 14.3 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 9.0 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 5.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 8.3 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 1.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 3.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 3.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 3.7 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 2.9 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 2.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 2.8 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 6.2 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 4.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 3.6 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 34.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.5 | 18.8 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.4 | 9.0 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.4 | 8.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.4 | 10.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.3 | 4.7 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.3 | 20.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.3 | 5.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.2 | 8.3 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.2 | 3.5 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.2 | 5.0 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.2 | 5.1 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.2 | 14.4 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.2 | 7.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.2 | 16.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.2 | 2.9 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.2 | 7.6 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.2 | 4.0 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 2.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 9.4 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.1 | 2.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 2.2 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.1 | 5.4 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 8.4 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 3.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 2.7 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 2.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 1.5 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.1 | 11.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 10.8 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 1.7 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 1.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.4 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.5 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |