Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
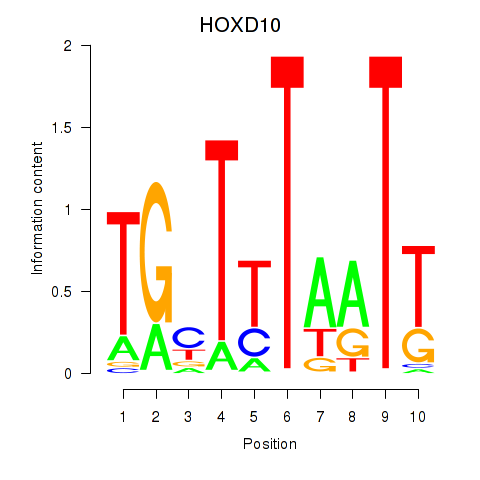
Results for HOXD10
Z-value: 1.06
Transcription factors associated with HOXD10
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXD10
|
ENSG00000128710.5 | homeobox D10 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXD10 | hg19_v2_chr2_+_176981307_176981307 | -0.42 | 8.5e-11 | Click! |
Activity profile of HOXD10 motif
Sorted Z-values of HOXD10 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_84035905 | 24.09 |
ENST00000311507.4
|
PLAC8
|
placenta-specific 8 |
| chr4_-_84035868 | 22.77 |
ENST00000426923.2
ENST00000509973.1 |
PLAC8
|
placenta-specific 8 |
| chr1_-_149900122 | 21.39 |
ENST00000271628.8
|
SF3B4
|
splicing factor 3b, subunit 4, 49kDa |
| chr3_-_185641681 | 16.88 |
ENST00000259043.7
|
TRA2B
|
transformer 2 beta homolog (Drosophila) |
| chr5_+_162930114 | 16.79 |
ENST00000280969.5
|
MAT2B
|
methionine adenosyltransferase II, beta |
| chr1_+_158978768 | 16.74 |
ENST00000447473.2
|
IFI16
|
interferon, gamma-inducible protein 16 |
| chr1_+_198608146 | 16.37 |
ENST00000367376.2
ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chr4_+_40198527 | 15.30 |
ENST00000381799.5
|
RHOH
|
ras homolog family member H |
| chr8_-_101719159 | 15.00 |
ENST00000520868.1
ENST00000522658.1 |
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr5_+_167913450 | 14.69 |
ENST00000231572.3
ENST00000538719.1 |
RARS
|
arginyl-tRNA synthetase |
| chr1_+_41204506 | 14.49 |
ENST00000525290.1
ENST00000530965.1 ENST00000416859.2 ENST00000308733.5 |
NFYC
|
nuclear transcription factor Y, gamma |
| chr11_-_104827425 | 14.27 |
ENST00000393150.3
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chrX_-_109590174 | 12.93 |
ENST00000372054.1
|
GNG5P2
|
guanine nucleotide binding protein (G protein), gamma 5 pseudogene 2 |
| chr9_+_42671887 | 12.62 |
ENST00000456520.1
ENST00000377391.3 |
CBWD7
|
COBW domain containing 7 |
| chr11_+_60223225 | 12.43 |
ENST00000524807.1
ENST00000345732.4 |
MS4A1
|
membrane-spanning 4-domains, subfamily A, member 1 |
| chr2_+_102456277 | 12.39 |
ENST00000421882.1
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr7_-_99698338 | 11.94 |
ENST00000354230.3
ENST00000425308.1 |
MCM7
|
minichromosome maintenance complex component 7 |
| chr6_-_32557610 | 11.93 |
ENST00000360004.5
|
HLA-DRB1
|
major histocompatibility complex, class II, DR beta 1 |
| chr6_-_150067632 | 11.83 |
ENST00000460354.2
ENST00000367404.4 ENST00000543637.1 |
NUP43
|
nucleoporin 43kDa |
| chr15_-_55562479 | 11.60 |
ENST00000564609.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr8_-_101718991 | 11.28 |
ENST00000517990.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr1_-_111743285 | 11.07 |
ENST00000357640.4
|
DENND2D
|
DENN/MADD domain containing 2D |
| chr12_+_20963647 | 10.97 |
ENST00000381545.3
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chrX_+_119737806 | 10.61 |
ENST00000371317.5
|
MCTS1
|
malignant T cell amplified sequence 1 |
| chr17_+_48823896 | 10.57 |
ENST00000511974.1
|
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr12_+_20963632 | 10.40 |
ENST00000540853.1
ENST00000261196.2 |
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr18_+_3252265 | 10.17 |
ENST00000580887.1
ENST00000536605.1 |
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr6_-_150067696 | 10.06 |
ENST00000340413.2
ENST00000367403.3 |
NUP43
|
nucleoporin 43kDa |
| chr14_+_73563735 | 9.97 |
ENST00000532192.1
|
RBM25
|
RNA binding motif protein 25 |
| chr6_-_36515177 | 9.90 |
ENST00000229812.7
|
STK38
|
serine/threonine kinase 38 |
| chr16_-_24942273 | 9.65 |
ENST00000571406.1
|
ARHGAP17
|
Rho GTPase activating protein 17 |
| chr1_+_174844645 | 9.23 |
ENST00000486220.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr12_-_9102549 | 9.21 |
ENST00000000412.3
|
M6PR
|
mannose-6-phosphate receptor (cation dependent) |
| chr6_-_32498046 | 9.02 |
ENST00000374975.3
|
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 5 |
| chr6_-_24667232 | 8.84 |
ENST00000378198.4
|
TDP2
|
tyrosyl-DNA phosphodiesterase 2 |
| chr13_-_47012325 | 8.75 |
ENST00000409879.2
|
KIAA0226L
|
KIAA0226-like |
| chr2_+_118572226 | 8.70 |
ENST00000263239.2
|
DDX18
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 18 |
| chr8_+_26150628 | 8.55 |
ENST00000523925.1
ENST00000315985.7 |
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha |
| chr12_-_118797475 | 8.55 |
ENST00000541786.1
ENST00000419821.2 ENST00000541878.1 |
TAOK3
|
TAO kinase 3 |
| chr2_+_68961905 | 8.46 |
ENST00000295381.3
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chrX_-_100872911 | 8.43 |
ENST00000361910.4
ENST00000539247.1 ENST00000538627.1 |
ARMCX6
|
armadillo repeat containing, X-linked 6 |
| chr6_+_26365443 | 8.39 |
ENST00000527422.1
ENST00000356386.2 ENST00000396934.3 ENST00000377708.2 ENST00000396948.1 ENST00000508906.2 |
BTN3A2
|
butyrophilin, subfamily 3, member A2 |
| chr2_+_68961934 | 8.37 |
ENST00000409202.3
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr11_-_104905840 | 8.36 |
ENST00000526568.1
ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1
|
caspase 1, apoptosis-related cysteine peptidase |
| chr12_+_16109519 | 8.31 |
ENST00000526530.1
|
DERA
|
deoxyribose-phosphate aldolase (putative) |
| chr11_+_60223312 | 8.09 |
ENST00000532491.1
ENST00000532073.1 ENST00000534668.1 ENST00000528313.1 ENST00000533306.1 |
MS4A1
|
membrane-spanning 4-domains, subfamily A, member 1 |
| chr12_-_49333446 | 8.01 |
ENST00000537495.1
|
AC073610.5
|
Uncharacterized protein |
| chr17_+_48823975 | 7.80 |
ENST00000513969.1
ENST00000503728.1 |
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr15_+_64680003 | 7.76 |
ENST00000261884.3
|
TRIP4
|
thyroid hormone receptor interactor 4 |
| chr6_-_24936170 | 7.74 |
ENST00000538035.1
|
FAM65B
|
family with sequence similarity 65, member B |
| chr2_-_89521942 | 7.72 |
ENST00000482769.1
|
IGKV2-28
|
immunoglobulin kappa variable 2-28 |
| chr2_+_90458201 | 7.70 |
ENST00000603238.1
|
CH17-132F21.1
|
Uncharacterized protein |
| chr9_+_70856397 | 7.69 |
ENST00000360171.6
|
CBWD3
|
COBW domain containing 3 |
| chr11_-_104972158 | 7.57 |
ENST00000598974.1
ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1
CARD16
CARD17
|
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr3_+_136649311 | 7.55 |
ENST00000469404.1
ENST00000467911.1 |
NCK1
|
NCK adaptor protein 1 |
| chr11_-_64647144 | 7.55 |
ENST00000359393.2
ENST00000433803.1 ENST00000411683.1 |
EHD1
|
EH-domain containing 1 |
| chr6_-_5261141 | 7.44 |
ENST00000330636.4
ENST00000500576.2 |
LYRM4
|
LYR motif containing 4 |
| chr7_+_77469439 | 7.27 |
ENST00000450574.1
ENST00000416283.2 ENST00000248550.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr6_-_5260963 | 7.26 |
ENST00000464010.1
ENST00000468929.1 ENST00000480566.1 |
LYRM4
|
LYR motif containing 4 |
| chr8_+_74903580 | 7.21 |
ENST00000284818.2
ENST00000518893.1 |
LY96
|
lymphocyte antigen 96 |
| chr6_-_8102714 | 7.18 |
ENST00000502429.1
ENST00000429723.2 ENST00000507463.1 ENST00000379715.5 |
EEF1E1
|
eukaryotic translation elongation factor 1 epsilon 1 |
| chr12_+_21525818 | 7.08 |
ENST00000240652.3
ENST00000542023.1 ENST00000537593.1 |
IAPP
|
islet amyloid polypeptide |
| chr2_+_122513109 | 7.06 |
ENST00000389682.3
ENST00000536142.1 |
TSN
|
translin |
| chr2_+_187371440 | 7.05 |
ENST00000445547.1
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr1_-_54405773 | 7.02 |
ENST00000371376.1
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr10_+_51576285 | 6.95 |
ENST00000443446.1
|
NCOA4
|
nuclear receptor coactivator 4 |
| chr10_+_11047259 | 6.87 |
ENST00000379261.4
ENST00000416382.2 |
CELF2
|
CUGBP, Elav-like family member 2 |
| chr2_+_68592305 | 6.83 |
ENST00000234313.7
|
PLEK
|
pleckstrin |
| chr17_-_38978847 | 6.77 |
ENST00000269576.5
|
KRT10
|
keratin 10 |
| chr12_-_10022735 | 6.67 |
ENST00000228438.2
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr13_-_50367057 | 6.58 |
ENST00000261667.3
|
KPNA3
|
karyopherin alpha 3 (importin alpha 4) |
| chr1_+_236558694 | 6.49 |
ENST00000359362.5
|
EDARADD
|
EDAR-associated death domain |
| chr2_-_90538397 | 6.22 |
ENST00000443397.3
|
RP11-685N3.1
|
Uncharacterized protein |
| chr2_+_87565634 | 6.04 |
ENST00000421835.2
|
IGKV3OR2-268
|
immunoglobulin kappa variable 3/OR2-268 (non-functional) |
| chr18_+_32556892 | 5.84 |
ENST00000591734.1
ENST00000413393.1 ENST00000589180.1 ENST00000587359.1 |
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr1_-_40042073 | 5.67 |
ENST00000372858.3
|
PABPC4
|
poly(A) binding protein, cytoplasmic 4 (inducible form) |
| chr14_+_72399114 | 5.64 |
ENST00000553525.1
ENST00000555571.1 |
RGS6
|
regulator of G-protein signaling 6 |
| chr3_+_196466710 | 5.57 |
ENST00000327134.3
|
PAK2
|
p21 protein (Cdc42/Rac)-activated kinase 2 |
| chr5_+_96211643 | 5.47 |
ENST00000437043.3
ENST00000510373.1 |
ERAP2
|
endoplasmic reticulum aminopeptidase 2 |
| chr5_+_156607829 | 5.46 |
ENST00000422843.3
|
ITK
|
IL2-inducible T-cell kinase |
| chr22_+_41258250 | 5.45 |
ENST00000544094.1
|
XPNPEP3
|
X-prolyl aminopeptidase (aminopeptidase P) 3, putative |
| chr3_+_30647994 | 5.45 |
ENST00000295754.5
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa) |
| chr10_-_43892279 | 5.42 |
ENST00000443950.2
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F |
| chr6_-_24667180 | 5.40 |
ENST00000545995.1
|
TDP2
|
tyrosyl-DNA phosphodiesterase 2 |
| chr17_-_55038375 | 5.28 |
ENST00000240316.4
|
COIL
|
coilin |
| chr3_+_130569429 | 5.24 |
ENST00000505330.1
ENST00000504381.1 ENST00000507488.2 ENST00000393221.4 |
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr2_+_86333301 | 5.22 |
ENST00000254630.7
|
PTCD3
|
pentatricopeptide repeat domain 3 |
| chr6_+_24667257 | 5.18 |
ENST00000537591.1
ENST00000230048.4 |
ACOT13
|
acyl-CoA thioesterase 13 |
| chrX_+_37639264 | 5.10 |
ENST00000378588.4
|
CYBB
|
cytochrome b-245, beta polypeptide |
| chr7_-_38305279 | 5.09 |
ENST00000443402.2
|
TRGC1
|
T cell receptor gamma constant 1 |
| chr5_+_1801503 | 5.01 |
ENST00000274137.5
ENST00000469176.1 |
NDUFS6
|
NADH dehydrogenase (ubiquinone) Fe-S protein 6, 13kDa (NADH-coenzyme Q reductase) |
| chr16_+_30751953 | 4.90 |
ENST00000483578.1
|
RP11-2C24.4
|
RP11-2C24.4 |
| chr2_+_47630255 | 4.88 |
ENST00000406134.1
|
MSH2
|
mutS homolog 2 |
| chr2_+_90273679 | 4.79 |
ENST00000423080.2
|
IGKV3D-7
|
immunoglobulin kappa variable 3D-7 |
| chr14_-_107049312 | 4.78 |
ENST00000390627.2
|
IGHV3-53
|
immunoglobulin heavy variable 3-53 |
| chr8_-_117886955 | 4.78 |
ENST00000297338.2
|
RAD21
|
RAD21 homolog (S. pombe) |
| chr1_+_79115503 | 4.71 |
ENST00000370747.4
ENST00000438486.1 ENST00000545124.1 |
IFI44
|
interferon-induced protein 44 |
| chr17_+_73663402 | 4.56 |
ENST00000355423.3
|
SAP30BP
|
SAP30 binding protein |
| chr3_+_30648066 | 4.44 |
ENST00000359013.4
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa) |
| chr2_+_89998789 | 4.42 |
ENST00000453166.2
|
IGKV2D-28
|
immunoglobulin kappa variable 2D-28 |
| chr4_-_72649763 | 4.40 |
ENST00000513476.1
|
GC
|
group-specific component (vitamin D binding protein) |
| chrX_+_37639302 | 4.32 |
ENST00000545017.1
ENST00000536160.1 |
CYBB
|
cytochrome b-245, beta polypeptide |
| chr2_+_90198535 | 4.23 |
ENST00000390276.2
|
IGKV1D-12
|
immunoglobulin kappa variable 1D-12 |
| chr1_-_197036364 | 4.11 |
ENST00000367412.1
|
F13B
|
coagulation factor XIII, B polypeptide |
| chr15_+_57511609 | 4.08 |
ENST00000543579.1
ENST00000537840.1 ENST00000343827.3 |
TCF12
|
transcription factor 12 |
| chr7_-_115608304 | 4.00 |
ENST00000457268.1
|
TFEC
|
transcription factor EC |
| chr3_-_185538849 | 3.98 |
ENST00000421047.2
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr4_-_102268628 | 3.93 |
ENST00000323055.6
ENST00000512215.1 ENST00000394854.3 |
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr8_+_132952112 | 3.89 |
ENST00000520362.1
ENST00000519656.1 |
EFR3A
|
EFR3 homolog A (S. cerevisiae) |
| chr8_+_21823726 | 3.81 |
ENST00000433566.4
|
XPO7
|
exportin 7 |
| chr7_-_115670804 | 3.80 |
ENST00000320239.7
|
TFEC
|
transcription factor EC |
| chr1_+_145883868 | 3.76 |
ENST00000447947.2
|
GPR89C
|
G protein-coupled receptor 89C |
| chr1_+_154229547 | 3.73 |
ENST00000428595.1
|
UBAP2L
|
ubiquitin associated protein 2-like |
| chr5_+_54603566 | 3.73 |
ENST00000230640.5
|
SKIV2L2
|
superkiller viralicidic activity 2-like 2 (S. cerevisiae) |
| chr5_-_96518907 | 3.62 |
ENST00000508447.1
ENST00000283109.3 |
RIOK2
|
RIO kinase 2 |
| chr18_+_3252206 | 3.60 |
ENST00000578562.2
|
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr1_+_158801095 | 3.53 |
ENST00000368141.4
|
MNDA
|
myeloid cell nuclear differentiation antigen |
| chr15_-_64673665 | 3.52 |
ENST00000300035.4
|
KIAA0101
|
KIAA0101 |
| chr5_-_43557129 | 3.52 |
ENST00000514514.1
ENST00000504075.1 ENST00000306846.3 ENST00000436644.2 |
PAIP1
|
poly(A) binding protein interacting protein 1 |
| chr7_-_115670792 | 3.50 |
ENST00000265440.7
ENST00000393485.1 |
TFEC
|
transcription factor EC |
| chr14_+_89060739 | 3.44 |
ENST00000318308.6
|
ZC3H14
|
zinc finger CCCH-type containing 14 |
| chr3_+_138340049 | 3.42 |
ENST00000464668.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr6_+_25963020 | 3.37 |
ENST00000357085.3
|
TRIM38
|
tripartite motif containing 38 |
| chr1_+_74701062 | 3.34 |
ENST00000326637.3
|
TNNI3K
|
TNNI3 interacting kinase |
| chr16_-_15736881 | 3.33 |
ENST00000540441.2
|
KIAA0430
|
KIAA0430 |
| chr3_+_120461484 | 3.26 |
ENST00000484715.1
ENST00000469772.1 ENST00000283875.5 ENST00000492959.1 |
GTF2E1
|
general transcription factor IIE, polypeptide 1, alpha 56kDa |
| chr12_-_7656357 | 3.24 |
ENST00000396620.3
ENST00000432237.2 ENST00000359156.4 |
CD163
|
CD163 molecule |
| chr3_+_63953415 | 3.23 |
ENST00000484332.1
|
ATXN7
|
ataxin 7 |
| chr7_+_101460882 | 3.16 |
ENST00000292535.7
ENST00000549414.2 ENST00000550008.2 ENST00000546411.2 ENST00000556210.1 |
CUX1
|
cut-like homeobox 1 |
| chr3_+_158288999 | 3.16 |
ENST00000482628.1
ENST00000478894.2 ENST00000392822.3 ENST00000466246.1 |
MLF1
|
myeloid leukemia factor 1 |
| chr3_+_43328004 | 3.15 |
ENST00000454177.1
ENST00000429705.2 ENST00000296088.7 ENST00000437827.1 |
SNRK
|
SNF related kinase |
| chr16_-_15180257 | 3.14 |
ENST00000540462.1
|
RRN3
|
RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae) |
| chr12_+_75784850 | 3.14 |
ENST00000550916.1
ENST00000435775.1 ENST00000378689.2 ENST00000378692.3 ENST00000320460.4 ENST00000547164.1 |
GLIPR1L2
|
GLI pathogenesis-related 1 like 2 |
| chr19_+_54619125 | 3.10 |
ENST00000445811.1
ENST00000419967.1 ENST00000445124.1 ENST00000447810.1 |
PRPF31
|
pre-mRNA processing factor 31 |
| chr5_-_150460914 | 3.05 |
ENST00000389378.2
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr1_-_231560790 | 3.02 |
ENST00000366641.3
|
EGLN1
|
egl-9 family hypoxia-inducible factor 1 |
| chr2_-_165424973 | 2.89 |
ENST00000543549.1
|
GRB14
|
growth factor receptor-bound protein 14 |
| chr19_-_39881669 | 2.88 |
ENST00000221266.7
|
PAF1
|
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae) |
| chr14_-_106453155 | 2.86 |
ENST00000390594.2
|
IGHV1-2
|
immunoglobulin heavy variable 1-2 |
| chr19_+_49588677 | 2.79 |
ENST00000598984.1
ENST00000598441.1 |
SNRNP70
|
small nuclear ribonucleoprotein 70kDa (U1) |
| chr2_+_183580954 | 2.78 |
ENST00000264065.7
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10 |
| chr1_+_244214577 | 2.78 |
ENST00000358704.4
|
ZBTB18
|
zinc finger and BTB domain containing 18 |
| chr1_-_155942086 | 2.74 |
ENST00000368315.4
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
| chr8_-_79717750 | 2.57 |
ENST00000263851.4
ENST00000379113.2 |
IL7
|
interleukin 7 |
| chr5_-_64064508 | 2.52 |
ENST00000513458.4
|
SREK1IP1
|
SREK1-interacting protein 1 |
| chr3_+_169629354 | 2.52 |
ENST00000428432.2
ENST00000335556.3 |
SAMD7
|
sterile alpha motif domain containing 7 |
| chr2_-_190044480 | 2.52 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr10_-_75226166 | 2.50 |
ENST00000544628.1
|
PPP3CB
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr3_+_158288960 | 2.49 |
ENST00000484955.1
ENST00000359117.5 ENST00000498592.1 ENST00000477042.1 ENST00000471745.1 ENST00000469452.1 |
MLF1
|
myeloid leukemia factor 1 |
| chr5_+_141346385 | 2.45 |
ENST00000513019.1
ENST00000356143.1 |
RNF14
|
ring finger protein 14 |
| chr6_+_158733692 | 2.42 |
ENST00000367094.2
ENST00000367097.3 |
TULP4
|
tubby like protein 4 |
| chr22_-_29107919 | 2.41 |
ENST00000434810.1
ENST00000456369.1 |
CHEK2
|
checkpoint kinase 2 |
| chr16_+_4896659 | 2.41 |
ENST00000592120.1
|
UBN1
|
ubinuclein 1 |
| chr2_+_90211643 | 2.41 |
ENST00000390277.2
|
IGKV3D-11
|
immunoglobulin kappa variable 3D-11 |
| chr22_+_32871224 | 2.41 |
ENST00000452138.1
ENST00000382058.3 ENST00000397426.1 |
FBXO7
|
F-box protein 7 |
| chr11_+_120973375 | 2.30 |
ENST00000264037.2
|
TECTA
|
tectorin alpha |
| chr6_-_133055896 | 2.27 |
ENST00000367927.5
ENST00000425515.2 ENST00000207771.3 ENST00000392393.3 ENST00000450865.2 ENST00000392394.2 |
VNN3
|
vanin 3 |
| chr5_-_180665195 | 2.22 |
ENST00000509148.1
|
GNB2L1
|
guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 |
| chr3_-_27498235 | 2.21 |
ENST00000295736.5
ENST00000428386.1 ENST00000428179.1 |
SLC4A7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr9_-_36401155 | 2.20 |
ENST00000377885.2
|
RNF38
|
ring finger protein 38 |
| chr14_+_75988851 | 2.19 |
ENST00000555504.1
|
BATF
|
basic leucine zipper transcription factor, ATF-like |
| chr15_+_41549105 | 2.18 |
ENST00000560965.1
|
CHP1
|
calcineurin-like EF-hand protein 1 |
| chr1_+_207262881 | 2.14 |
ENST00000451804.2
|
C4BPB
|
complement component 4 binding protein, beta |
| chr12_-_13248732 | 2.13 |
ENST00000396302.3
|
GSG1
|
germ cell associated 1 |
| chrX_+_12924732 | 2.12 |
ENST00000218032.6
ENST00000311912.5 |
TLR8
|
toll-like receptor 8 |
| chr5_-_39203093 | 2.12 |
ENST00000515010.1
|
FYB
|
FYN binding protein |
| chr2_+_185463093 | 2.07 |
ENST00000302277.6
|
ZNF804A
|
zinc finger protein 804A |
| chr9_+_82187487 | 2.07 |
ENST00000435650.1
ENST00000414465.1 ENST00000376537.4 ENST00000376534.4 |
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr3_+_158288942 | 2.02 |
ENST00000491767.1
ENST00000355893.5 |
MLF1
|
myeloid leukemia factor 1 |
| chr8_+_124084899 | 2.01 |
ENST00000287380.1
ENST00000309336.3 ENST00000519418.1 ENST00000327098.5 ENST00000522420.1 ENST00000521676.1 ENST00000378080.2 |
TBC1D31
|
TBC1 domain family, member 31 |
| chr3_+_108541608 | 1.96 |
ENST00000426646.1
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr12_-_25055177 | 1.91 |
ENST00000538118.1
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr10_+_35456444 | 1.74 |
ENST00000361599.4
|
CREM
|
cAMP responsive element modulator |
| chr6_-_25830785 | 1.73 |
ENST00000468082.1
|
SLC17A1
|
solute carrier family 17 (organic anion transporter), member 1 |
| chr12_-_10588539 | 1.66 |
ENST00000381902.2
ENST00000381901.1 ENST00000539033.1 |
KLRC2
NKG2-E
|
killer cell lectin-like receptor subfamily C, member 2 Uncharacterized protein |
| chrX_+_149887090 | 1.65 |
ENST00000538506.1
|
MTMR1
|
myotubularin related protein 1 |
| chr12_-_13248705 | 1.63 |
ENST00000396310.2
|
GSG1
|
germ cell associated 1 |
| chr4_-_76928641 | 1.53 |
ENST00000264888.5
|
CXCL9
|
chemokine (C-X-C motif) ligand 9 |
| chr6_-_134639180 | 1.53 |
ENST00000367858.5
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr3_-_37216055 | 1.52 |
ENST00000336686.4
|
LRRFIP2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr6_-_133055815 | 1.52 |
ENST00000509351.1
ENST00000417437.2 ENST00000414302.2 ENST00000423615.2 ENST00000427187.2 ENST00000275223.3 ENST00000519686.2 |
VNN3
|
vanin 3 |
| chr6_+_5261225 | 1.48 |
ENST00000324331.6
|
FARS2
|
phenylalanyl-tRNA synthetase 2, mitochondrial |
| chr5_-_58882219 | 1.48 |
ENST00000505453.1
ENST00000360047.5 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr3_+_108541545 | 1.47 |
ENST00000295756.6
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr4_-_102268484 | 1.43 |
ENST00000394853.4
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr12_-_13248562 | 1.42 |
ENST00000457134.2
ENST00000537302.1 |
GSG1
|
germ cell associated 1 |
| chr5_-_111312622 | 1.37 |
ENST00000395634.3
|
NREP
|
neuronal regeneration related protein |
| chr4_-_70080449 | 1.30 |
ENST00000446444.1
|
UGT2B11
|
UDP glucuronosyltransferase 2 family, polypeptide B11 |
| chr9_+_82187630 | 1.30 |
ENST00000265284.6
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr2_+_166326157 | 1.28 |
ENST00000421875.1
ENST00000314499.7 ENST00000409664.1 |
CSRNP3
|
cysteine-serine-rich nuclear protein 3 |
| chr6_-_100016527 | 1.27 |
ENST00000523985.1
ENST00000518714.1 ENST00000520371.1 |
CCNC
|
cyclin C |
| chr16_+_1728257 | 1.25 |
ENST00000248098.3
ENST00000562684.1 ENST00000561516.1 ENST00000382711.5 ENST00000566742.1 |
HN1L
|
hematological and neurological expressed 1-like |
| chr18_+_55816546 | 1.22 |
ENST00000435432.2
ENST00000357895.5 ENST00000586263.1 |
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr12_+_60083118 | 1.20 |
ENST00000261187.4
ENST00000543448.1 |
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr11_-_30608413 | 1.19 |
ENST00000528686.1
|
MPPED2
|
metallophosphoesterase domain containing 2 |
| chr13_+_78109884 | 1.17 |
ENST00000377246.3
ENST00000349847.3 |
SCEL
|
sciellin |
| chr7_-_7782204 | 1.13 |
ENST00000418534.2
|
AC007161.5
|
AC007161.5 |
| chr14_+_75988768 | 1.11 |
ENST00000286639.6
|
BATF
|
basic leucine zipper transcription factor, ATF-like |
| chr12_-_131323754 | 1.09 |
ENST00000261653.6
|
STX2
|
syntaxin 2 |
| chr7_+_107224364 | 1.06 |
ENST00000491150.1
|
BCAP29
|
B-cell receptor-associated protein 29 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXD10
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 14.7 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 4.2 | 16.9 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 4.2 | 16.8 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 4.0 | 15.9 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 3.5 | 10.6 | GO:0002188 | translation reinitiation(GO:0002188) |
| 3.3 | 9.9 | GO:0002661 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 3.3 | 16.4 | GO:0048539 | immunoglobulin biosynthetic process(GO:0002378) bone marrow development(GO:0048539) |
| 2.9 | 26.3 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 2.6 | 46.9 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 2.5 | 7.5 | GO:1903676 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 2.4 | 11.9 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 1.9 | 11.6 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 1.9 | 9.4 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 1.8 | 5.4 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 1.6 | 14.3 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 1.4 | 5.6 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 1.4 | 8.3 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 1.2 | 24.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 1.2 | 3.6 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 1.2 | 11.9 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 1.1 | 6.8 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 1.1 | 7.5 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 1.0 | 7.2 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 1.0 | 3.1 | GO:0044501 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 1.0 | 4.9 | GO:0010520 | meiotic gene conversion(GO:0006311) regulation of reciprocal meiotic recombination(GO:0010520) |
| 1.0 | 5.8 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.9 | 9.2 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.8 | 2.4 | GO:1903925 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.8 | 4.8 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.8 | 7.1 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.7 | 5.7 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.7 | 2.7 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.7 | 25.2 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.6 | 2.5 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 0.6 | 3.7 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.5 | 7.1 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.5 | 7.0 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.5 | 12.4 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.5 | 2.1 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.5 | 3.1 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.5 | 3.0 | GO:0051344 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.5 | 21.4 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.5 | 21.9 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.5 | 8.6 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.4 | 3.1 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.4 | 7.2 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.4 | 5.5 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.4 | 2.5 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.4 | 8.4 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.4 | 2.3 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.4 | 3.8 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.4 | 3.4 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.4 | 1.5 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.4 | 1.1 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.3 | 16.7 | GO:0045824 | negative regulation of innate immune response(GO:0045824) |
| 0.3 | 1.0 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.3 | 1.0 | GO:2000360 | positive regulation of female gonad development(GO:2000196) negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.3 | 2.9 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.3 | 1.9 | GO:0009099 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.3 | 7.0 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.3 | 15.3 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.3 | 3.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.3 | 8.7 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.3 | 2.2 | GO:0070885 | positive regulation of sodium:proton antiporter activity(GO:0032417) negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.3 | 2.3 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.2 | 1.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.2 | 3.4 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.2 | 3.4 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.2 | 14.5 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.2 | 1.5 | GO:0003096 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.2 | 1.1 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.2 | 20.9 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.2 | 10.0 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.2 | 9.2 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.2 | 4.4 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.2 | 3.3 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.2 | 2.8 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.2 | 1.7 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.2 | 5.0 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.2 | 0.5 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.2 | 7.8 | GO:1901998 | toxin transport(GO:1901998) |
| 0.2 | 2.4 | GO:1903599 | positive regulation of mitophagy(GO:1903599) |
| 0.2 | 1.2 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 21.0 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 2.2 | GO:0060117 | auditory receptor cell development(GO:0060117) |
| 0.1 | 1.3 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 3.2 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.1 | 0.5 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.1 | 0.5 | GO:0019249 | lactate biosynthetic process(GO:0019249) |
| 0.1 | 0.9 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.8 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 9.1 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.1 | 1.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 3.7 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.1 | 0.4 | GO:1903961 | positive regulation of anion channel activity(GO:1901529) positive regulation of anion transmembrane transport(GO:1903961) |
| 0.1 | 0.3 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.1 | 4.7 | GO:0009615 | response to virus(GO:0009615) |
| 0.1 | 1.5 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.1 | 9.0 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.1 | 3.5 | GO:0048255 | mRNA stabilization(GO:0048255) |
| 0.1 | 2.9 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.1 | 7.1 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.1 | 8.6 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.1 | 0.2 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 5.4 | GO:0071349 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.1 | 16.4 | GO:0038095 | Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.1 | 5.2 | GO:0043407 | negative regulation of MAP kinase activity(GO:0043407) |
| 0.1 | 3.2 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.1 | 0.7 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 | 1.6 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 3.2 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.1 | 5.2 | GO:0070126 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 0.1 | 2.5 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.1 | 12.5 | GO:0006302 | double-strand break repair(GO:0006302) |
| 0.1 | 6.8 | GO:0098773 | skin epidermis development(GO:0098773) |
| 0.1 | 6.5 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.1 | 4.0 | GO:0042035 | regulation of cytokine biosynthetic process(GO:0042035) |
| 0.1 | 2.4 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-independent nucleosome organization(GO:0034724) |
| 0.1 | 2.4 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.1 | 1.0 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 | 1.0 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 0.8 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 2.3 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 4.8 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 1.0 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.0 | 0.4 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.6 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 3.3 | GO:0042795 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 3.9 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 1.7 | GO:0002228 | natural killer cell mediated immunity(GO:0002228) |
| 0.0 | 0.9 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 1.7 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 1.3 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.0 | 0.7 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.3 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.1 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.0 | 2.4 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 6.9 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 0.9 | GO:0002223 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) |
| 0.0 | 0.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 1.3 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 2.2 | GO:0046546 | male gonad development(GO:0008584) development of primary male sexual characteristics(GO:0046546) |
| 0.0 | 0.2 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.0 | 0.8 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.1 | GO:1904251 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.6 | 16.8 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 5.0 | 30.2 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 1.6 | 4.9 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 1.5 | 7.5 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 1.4 | 7.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 1.4 | 26.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 1.4 | 21.9 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 1.3 | 14.5 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 1.2 | 4.8 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 1.1 | 9.9 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 1.0 | 21.9 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 1.0 | 20.9 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 1.0 | 11.6 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.9 | 16.7 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.9 | 11.9 | GO:0042555 | MCM complex(GO:0042555) |
| 0.9 | 7.9 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.8 | 2.5 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.8 | 3.1 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.8 | 20.1 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.6 | 9.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.5 | 7.0 | GO:0044754 | autolysosome(GO:0044754) |
| 0.5 | 3.6 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.4 | 7.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.4 | 2.4 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.3 | 1.0 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.3 | 8.0 | GO:0030904 | retromer complex(GO:0030904) |
| 0.3 | 7.0 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.3 | 32.5 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.3 | 3.3 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.3 | 2.9 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.3 | 14.2 | GO:0016235 | aggresome(GO:0016235) |
| 0.3 | 15.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.3 | 1.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.3 | 8.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 1.0 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.2 | 5.8 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.2 | 3.7 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.2 | 14.8 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.2 | 3.4 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 2.8 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 10.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 3.7 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.1 | 12.1 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 0.2 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 2.1 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.1 | 5.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 10.4 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 5.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 5.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 15.4 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.1 | 0.5 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 2.2 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 0.7 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.1 | 0.9 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 20.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 46.9 | GO:0016604 | nuclear body(GO:0016604) |
| 0.1 | 2.8 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 2.7 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 2.3 | GO:0044216 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 3.8 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 6.2 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 1.3 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 5.8 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 4.9 | GO:1904813 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.0 | 0.4 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 2.3 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.2 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 2.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.5 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.8 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 14.7 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 4.7 | 14.2 | GO:0070260 | tyrosyl-RNA phosphodiesterase activity(GO:0036317) 5'-tyrosyl-DNA phosphodiesterase activity(GO:0070260) |
| 2.8 | 8.3 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 2.5 | 9.9 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 2.2 | 30.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 1.9 | 5.7 | GO:0017130 | poly(C) RNA binding(GO:0017130) |
| 1.8 | 29.8 | GO:0008494 | translation activator activity(GO:0008494) |
| 1.6 | 4.9 | GO:0032181 | heteroduplex DNA loop binding(GO:0000404) double-strand/single-strand DNA junction binding(GO:0000406) dinucleotide repeat insertion binding(GO:0032181) |
| 1.5 | 12.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 1.5 | 7.5 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 1.0 | 3.1 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 1.0 | 32.5 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 1.0 | 21.4 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.8 | 7.2 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.8 | 7.9 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.8 | 3.8 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.7 | 4.4 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.7 | 20.3 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.7 | 15.3 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.5 | 9.9 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.5 | 5.9 | GO:0070990 | snRNP binding(GO:0070990) |
| 0.5 | 6.8 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.5 | 9.4 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.5 | 9.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.4 | 11.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.4 | 5.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.4 | 11.7 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.4 | 1.5 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.3 | 3.8 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.3 | 1.7 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.3 | 1.9 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.3 | 3.3 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.3 | 0.9 | GO:0000773 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.3 | 1.2 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.3 | 4.6 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.3 | 5.4 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.3 | 6.8 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.3 | 9.0 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.3 | 1.1 | GO:0035276 | aldehyde oxidase activity(GO:0004031) ethanol binding(GO:0035276) |
| 0.3 | 11.1 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.3 | 3.0 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.3 | 7.8 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.2 | 5.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.2 | 2.8 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.2 | 0.7 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.2 | 1.7 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.2 | 6.6 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.2 | 3.4 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.2 | 5.8 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.2 | 12.9 | GO:0008026 | ATP-dependent RNA helicase activity(GO:0004004) ATP-dependent helicase activity(GO:0008026) RNA-dependent ATPase activity(GO:0008186) purine NTP-dependent helicase activity(GO:0070035) |
| 0.2 | 4.1 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.2 | 5.6 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.2 | 5.6 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.2 | 9.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.2 | 28.5 | GO:0003823 | antigen binding(GO:0003823) |
| 0.2 | 10.6 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.2 | 39.4 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.2 | 1.2 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 9.4 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 7.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 12.7 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.1 | 6.9 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.1 | 7.5 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.1 | 5.5 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.1 | 1.6 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 1.7 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 2.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 2.7 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 1.0 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) acrosin binding(GO:0032190) |
| 0.1 | 52.3 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.1 | 34.7 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 15.4 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 0.4 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.1 | 1.5 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.1 | 3.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.6 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 3.1 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 1.1 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 0.5 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 2.9 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 3.2 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 1.5 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.1 | 0.4 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.1 | 0.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 4.1 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 4.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 11.3 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 4.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 1.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 2.6 | GO:0070851 | growth factor receptor binding(GO:0070851) |
| 0.0 | 0.7 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.9 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.5 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 1.0 | GO:0008289 | lipid binding(GO:0008289) |
| 0.0 | 0.2 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 15.9 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.4 | 34.5 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.4 | 20.0 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.3 | 14.4 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.2 | 7.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 9.9 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.2 | 14.6 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.2 | 11.9 | PID ATR PATHWAY | ATR signaling pathway |
| 0.2 | 11.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.2 | 9.4 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.2 | 9.8 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 16.1 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 3.0 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 4.4 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 10.9 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 10.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 4.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 7.0 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 5.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 2.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 3.1 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 2.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 3.4 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.8 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.8 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.7 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 4.9 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.1 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 3.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 37.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 1.3 | 20.8 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.9 | 29.8 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.7 | 11.9 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.7 | 29.4 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.6 | 20.7 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.6 | 21.9 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.6 | 7.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.6 | 21.9 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.4 | 16.8 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.4 | 9.9 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.4 | 21.7 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.3 | 2.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.2 | 7.1 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.2 | 38.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.2 | 9.4 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.2 | 9.2 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.2 | 3.4 | REACTOME TCR SIGNALING | Genes involved in TCR signaling |
| 0.2 | 2.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 7.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 6.6 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 8.6 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 1.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 2.4 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 3.2 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 4.4 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 5.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 3.3 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 1.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 3.1 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 8.2 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 1.5 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.1 | 1.9 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 4.1 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 5.5 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 2.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 2.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 5.4 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.0 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 3.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 1.7 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.4 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |