Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
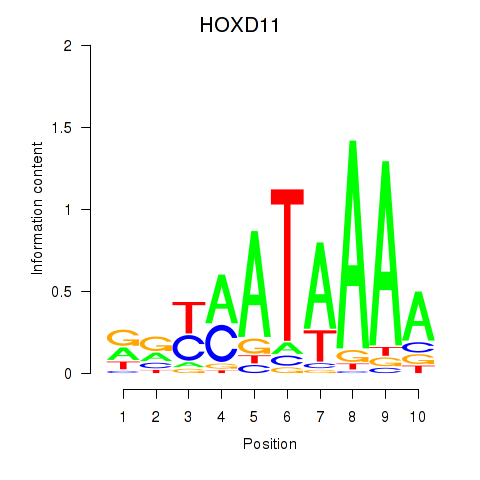
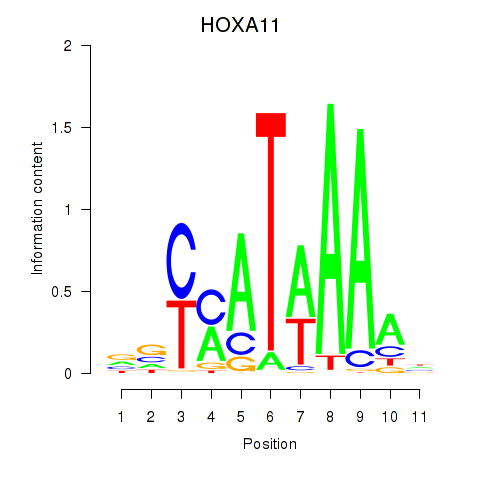
Results for HOXD11_HOXA11
Z-value: 0.85
Transcription factors associated with HOXD11_HOXA11
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXD11
|
ENSG00000128713.11 | homeobox D11 |
|
HOXA11
|
ENSG00000005073.5 | homeobox A11 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA11 | hg19_v2_chr7_-_27224842_27224872, hg19_v2_chr7_-_27224795_27224840 | -0.15 | 2.7e-02 | Click! |
| HOXD11 | hg19_v2_chr2_+_176972000_176972025 | -0.14 | 3.4e-02 | Click! |
Activity profile of HOXD11_HOXA11 motif
Sorted Z-values of HOXD11_HOXA11 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_74728998 | 15.24 |
ENST00000359645.3
ENST00000397875.3 ENST00000397869.3 ENST00000578193.1 ENST00000578873.1 ENST00000397866.4 ENST00000528160.1 ENST00000527041.1 ENST00000526111.1 ENST00000397865.5 ENST00000382582.3 |
MBP
|
myelin basic protein |
| chr4_+_41258786 | 11.81 |
ENST00000503431.1
ENST00000284440.4 ENST00000508768.1 ENST00000512788.1 |
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
| chr16_+_15031300 | 10.74 |
ENST00000328085.6
|
NPIPA1
|
nuclear pore complex interacting protein family, member A1 |
| chr14_-_60097297 | 9.95 |
ENST00000395090.1
|
RTN1
|
reticulon 1 |
| chr11_-_796197 | 9.32 |
ENST00000530360.1
ENST00000528606.1 ENST00000320230.5 |
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr1_-_28527152 | 9.09 |
ENST00000321830.5
|
AL353354.1
|
Uncharacterized protein |
| chr5_+_36608422 | 9.09 |
ENST00000381918.3
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr14_-_60097524 | 8.77 |
ENST00000342503.4
|
RTN1
|
reticulon 1 |
| chr2_-_51259292 | 8.48 |
ENST00000401669.2
|
NRXN1
|
neurexin 1 |
| chr2_-_51259229 | 8.32 |
ENST00000405472.3
|
NRXN1
|
neurexin 1 |
| chr9_+_137979506 | 8.26 |
ENST00000539529.1
ENST00000392991.4 ENST00000371793.3 |
OLFM1
|
olfactomedin 1 |
| chr8_+_107738240 | 7.82 |
ENST00000449762.2
ENST00000297447.6 |
OXR1
|
oxidation resistance 1 |
| chr12_+_6833237 | 7.81 |
ENST00000229251.3
ENST00000539735.1 ENST00000538410.1 |
COPS7A
|
COP9 signalosome subunit 7A |
| chr12_+_6833437 | 7.64 |
ENST00000534947.1
ENST00000541866.1 ENST00000534877.1 ENST00000538753.1 |
COPS7A
|
COP9 signalosome subunit 7A |
| chr2_-_207023918 | 7.26 |
ENST00000455934.2
ENST00000449699.1 ENST00000454195.1 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr5_-_73937244 | 7.10 |
ENST00000302351.4
ENST00000510316.1 ENST00000508331.1 |
ENC1
|
ectodermal-neural cortex 1 (with BTB domain) |
| chr9_-_93405352 | 7.09 |
ENST00000375765.3
|
DIRAS2
|
DIRAS family, GTP-binding RAS-like 2 |
| chrX_-_13835147 | 6.72 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr12_-_6580094 | 6.68 |
ENST00000361716.3
|
VAMP1
|
vesicle-associated membrane protein 1 (synaptobrevin 1) |
| chr12_-_90049828 | 6.62 |
ENST00000261173.2
ENST00000348959.3 |
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr12_-_90049878 | 6.54 |
ENST00000359142.3
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr1_+_84630645 | 6.28 |
ENST00000394839.2
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr5_+_67586465 | 6.25 |
ENST00000336483.5
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr11_-_47447970 | 6.16 |
ENST00000298852.3
ENST00000530912.1 |
PSMC3
|
proteasome (prosome, macropain) 26S subunit, ATPase, 3 |
| chr12_+_79258444 | 6.03 |
ENST00000261205.4
|
SYT1
|
synaptotagmin I |
| chr19_-_54618650 | 5.92 |
ENST00000391757.1
|
TFPT
|
TCF3 (E2A) fusion partner (in childhood Leukemia) |
| chr14_+_73563735 | 5.70 |
ENST00000532192.1
|
RBM25
|
RNA binding motif protein 25 |
| chr2_-_207024134 | 5.50 |
ENST00000457011.1
ENST00000440274.1 ENST00000432169.1 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr7_-_37026108 | 5.42 |
ENST00000396045.3
|
ELMO1
|
engulfment and cell motility 1 |
| chr19_+_19626531 | 5.41 |
ENST00000507754.4
|
NDUFA13
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 13 |
| chr2_-_207024233 | 5.39 |
ENST00000423725.1
ENST00000233190.6 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr5_-_111093167 | 5.38 |
ENST00000446294.2
ENST00000419114.2 |
NREP
|
neuronal regeneration related protein |
| chr19_-_54619006 | 5.30 |
ENST00000391759.1
|
TFPT
|
TCF3 (E2A) fusion partner (in childhood Leukemia) |
| chr3_-_98241358 | 5.02 |
ENST00000503004.1
ENST00000506575.1 ENST00000513452.1 ENST00000515620.1 |
CLDND1
|
claudin domain containing 1 |
| chr11_-_62473706 | 4.99 |
ENST00000403550.1
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr8_-_81083890 | 4.89 |
ENST00000518937.1
|
TPD52
|
tumor protein D52 |
| chrX_+_65382433 | 4.86 |
ENST00000374727.3
|
HEPH
|
hephaestin |
| chr6_+_158733692 | 4.84 |
ENST00000367094.2
ENST00000367097.3 |
TULP4
|
tubby like protein 4 |
| chr4_-_174320687 | 4.78 |
ENST00000296506.3
|
SCRG1
|
stimulator of chondrogenesis 1 |
| chr8_-_22089845 | 4.70 |
ENST00000454243.2
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein |
| chr7_+_73106926 | 4.63 |
ENST00000453316.1
|
WBSCR22
|
Williams Beuren syndrome chromosome region 22 |
| chr17_-_29624343 | 4.62 |
ENST00000247271.4
|
OMG
|
oligodendrocyte myelin glycoprotein |
| chr11_-_47447767 | 4.58 |
ENST00000530651.1
ENST00000524447.2 ENST00000531051.2 ENST00000526993.1 ENST00000602866.1 |
PSMC3
|
proteasome (prosome, macropain) 26S subunit, ATPase, 3 |
| chr5_+_140227048 | 4.55 |
ENST00000532602.1
|
PCDHA9
|
protocadherin alpha 9 |
| chr4_-_102268628 | 4.47 |
ENST00000323055.6
ENST00000512215.1 ENST00000394854.3 |
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr12_+_49621658 | 4.43 |
ENST00000541364.1
|
TUBA1C
|
tubulin, alpha 1c |
| chr19_+_1440838 | 4.37 |
ENST00000594262.1
|
AC027307.3
|
Uncharacterized protein |
| chr16_-_4852915 | 4.37 |
ENST00000322048.7
|
ROGDI
|
rogdi homolog (Drosophila) |
| chr4_-_102268484 | 4.34 |
ENST00000394853.4
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr16_+_22501658 | 4.29 |
ENST00000415833.2
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr16_-_18466642 | 4.25 |
ENST00000541810.1
ENST00000531453.2 |
RP11-1212A22.4
|
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chr2_-_136678123 | 4.21 |
ENST00000422708.1
|
DARS
|
aspartyl-tRNA synthetase |
| chr8_-_18666360 | 4.21 |
ENST00000286485.8
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr1_+_120254510 | 4.18 |
ENST00000369409.4
|
PHGDH
|
phosphoglycerate dehydrogenase |
| chr2_+_31456874 | 4.17 |
ENST00000541626.1
|
EHD3
|
EH-domain containing 3 |
| chr11_-_62473776 | 4.15 |
ENST00000278893.7
ENST00000407022.3 ENST00000421906.1 |
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr12_-_6579808 | 4.13 |
ENST00000535180.1
ENST00000400911.3 |
VAMP1
|
vesicle-associated membrane protein 1 (synaptobrevin 1) |
| chr1_+_229440129 | 4.11 |
ENST00000366688.3
|
SPHAR
|
S-phase response (cyclin related) |
| chr11_-_102576537 | 3.89 |
ENST00000260229.4
|
MMP27
|
matrix metallopeptidase 27 |
| chr18_-_5396271 | 3.84 |
ENST00000579951.1
|
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr9_-_21305312 | 3.79 |
ENST00000259555.4
|
IFNA5
|
interferon, alpha 5 |
| chr2_-_176046391 | 3.76 |
ENST00000392541.3
ENST00000409194.1 |
ATP5G3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr3_-_18480260 | 3.71 |
ENST00000454909.2
|
SATB1
|
SATB homeobox 1 |
| chr14_-_21493123 | 3.70 |
ENST00000556147.1
ENST00000554489.1 ENST00000555657.1 ENST00000557274.1 ENST00000555158.1 ENST00000554833.1 ENST00000555384.1 ENST00000556420.1 ENST00000554893.1 ENST00000553503.1 ENST00000555733.1 ENST00000553867.1 ENST00000397856.3 ENST00000397855.3 ENST00000556008.1 ENST00000557182.1 ENST00000554483.1 ENST00000556688.1 ENST00000397853.3 ENST00000556329.2 ENST00000554143.1 ENST00000397851.2 ENST00000555142.1 ENST00000557676.1 ENST00000556924.1 |
NDRG2
|
NDRG family member 2 |
| chr6_-_38670897 | 3.56 |
ENST00000373365.4
|
GLO1
|
glyoxalase I |
| chr10_+_94352956 | 3.56 |
ENST00000260731.3
|
KIF11
|
kinesin family member 11 |
| chr5_-_176057365 | 3.53 |
ENST00000310112.3
|
SNCB
|
synuclein, beta |
| chr6_-_8102714 | 3.51 |
ENST00000502429.1
ENST00000429723.2 ENST00000507463.1 ENST00000379715.5 |
EEF1E1
|
eukaryotic translation elongation factor 1 epsilon 1 |
| chr3_-_33700933 | 3.49 |
ENST00000480013.1
|
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr15_+_66797627 | 3.48 |
ENST00000565627.1
ENST00000564179.1 |
ZWILCH
|
zwilch kinetochore protein |
| chr4_-_48782259 | 3.40 |
ENST00000507711.1
ENST00000358350.4 ENST00000537810.1 ENST00000264319.7 |
FRYL
|
FRY-like |
| chr17_+_17082842 | 3.39 |
ENST00000579361.1
|
MPRIP
|
myosin phosphatase Rho interacting protein |
| chr3_+_157827841 | 3.35 |
ENST00000295930.3
ENST00000471994.1 ENST00000464171.1 ENST00000312179.6 ENST00000475278.2 |
RSRC1
|
arginine/serine-rich coiled-coil 1 |
| chr10_-_116444371 | 3.33 |
ENST00000533213.2
ENST00000369252.4 |
ABLIM1
|
actin binding LIM protein 1 |
| chr13_-_47471155 | 3.21 |
ENST00000543956.1
ENST00000542664.1 |
HTR2A
|
5-hydroxytryptamine (serotonin) receptor 2A, G protein-coupled |
| chr4_+_170581213 | 3.16 |
ENST00000507875.1
|
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chr1_-_101360331 | 3.15 |
ENST00000416479.1
ENST00000370113.3 |
EXTL2
|
exostosin-like glycosyltransferase 2 |
| chr11_+_19798964 | 3.06 |
ENST00000527559.2
|
NAV2
|
neuron navigator 2 |
| chrX_-_37706815 | 2.98 |
ENST00000378578.4
|
DYNLT3
|
dynein, light chain, Tctex-type 3 |
| chr8_+_26150628 | 2.97 |
ENST00000523925.1
ENST00000315985.7 |
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha |
| chr16_+_16429787 | 2.93 |
ENST00000331436.4
ENST00000541593.1 |
AC138969.4
|
Protein PKD1P1 |
| chr16_-_66764119 | 2.92 |
ENST00000569320.1
|
DYNC1LI2
|
dynein, cytoplasmic 1, light intermediate chain 2 |
| chr15_+_66797455 | 2.92 |
ENST00000446801.2
|
ZWILCH
|
zwilch kinetochore protein |
| chr17_-_4843316 | 2.91 |
ENST00000544061.2
|
SLC25A11
|
solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11 |
| chr5_-_20575959 | 2.84 |
ENST00000507958.1
|
CDH18
|
cadherin 18, type 2 |
| chr13_+_53030107 | 2.84 |
ENST00000490903.1
ENST00000480747.1 |
CKAP2
|
cytoskeleton associated protein 2 |
| chr4_+_83956237 | 2.79 |
ENST00000264389.2
|
COPS4
|
COP9 signalosome subunit 4 |
| chr2_+_101591314 | 2.68 |
ENST00000450763.1
|
NPAS2
|
neuronal PAS domain protein 2 |
| chr8_-_91095099 | 2.66 |
ENST00000265431.3
|
CALB1
|
calbindin 1, 28kDa |
| chr11_-_117748138 | 2.66 |
ENST00000527717.1
|
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr12_-_30887948 | 2.65 |
ENST00000433722.2
|
CAPRIN2
|
caprin family member 2 |
| chr9_+_108463234 | 2.64 |
ENST00000374688.1
|
TMEM38B
|
transmembrane protein 38B |
| chr16_+_16472912 | 2.63 |
ENST00000530217.2
|
NPIPA7
|
nuclear pore complex interacting protein family, member A7 |
| chr4_+_83956312 | 2.62 |
ENST00000509317.1
ENST00000503682.1 ENST00000511653.1 |
COPS4
|
COP9 signalosome subunit 4 |
| chr8_-_95220775 | 2.60 |
ENST00000441892.2
ENST00000521491.1 ENST00000027335.3 |
CDH17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr12_-_10573149 | 2.57 |
ENST00000381904.2
ENST00000381903.2 ENST00000396439.2 |
KLRC3
|
killer cell lectin-like receptor subfamily C, member 3 |
| chr12_-_71551868 | 2.57 |
ENST00000247829.3
|
TSPAN8
|
tetraspanin 8 |
| chr19_+_54619125 | 2.55 |
ENST00000445811.1
ENST00000419967.1 ENST00000445124.1 ENST00000447810.1 |
PRPF31
|
pre-mRNA processing factor 31 |
| chr8_+_104892639 | 2.53 |
ENST00000436393.2
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr11_+_27062502 | 2.51 |
ENST00000263182.3
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chrX_+_65384052 | 2.47 |
ENST00000336279.5
ENST00000458621.1 |
HEPH
|
hephaestin |
| chrX_+_56590002 | 2.46 |
ENST00000338222.5
|
UBQLN2
|
ubiquilin 2 |
| chr1_-_246357029 | 2.45 |
ENST00000391836.2
|
SMYD3
|
SET and MYND domain containing 3 |
| chr2_+_64069459 | 2.45 |
ENST00000445915.2
ENST00000475462.1 |
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr16_+_14805546 | 2.43 |
ENST00000552140.1
|
NPIPA3
|
nuclear pore complex interacting protein family, member A3 |
| chr2_+_74056147 | 2.41 |
ENST00000394070.2
ENST00000536064.1 |
STAMBP
|
STAM binding protein |
| chr14_-_21492113 | 2.40 |
ENST00000554094.1
|
NDRG2
|
NDRG family member 2 |
| chr2_+_64068844 | 2.38 |
ENST00000337130.5
|
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr1_-_154909329 | 2.38 |
ENST00000368467.3
|
PMVK
|
phosphomevalonate kinase |
| chr14_-_65439132 | 2.37 |
ENST00000533601.2
|
RAB15
|
RAB15, member RAS oncogene family |
| chr14_+_53173890 | 2.36 |
ENST00000445930.2
|
PSMC6
|
proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
| chr11_+_121447469 | 2.35 |
ENST00000532694.1
ENST00000534286.1 |
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing |
| chr9_+_70856397 | 2.34 |
ENST00000360171.6
|
CBWD3
|
COBW domain containing 3 |
| chr14_+_24540046 | 2.32 |
ENST00000397016.2
ENST00000537691.1 ENST00000560356.1 ENST00000558450.1 |
CPNE6
|
copine VI (neuronal) |
| chr22_-_30642782 | 2.32 |
ENST00000249075.3
|
LIF
|
leukemia inhibitory factor |
| chr1_+_65730385 | 2.29 |
ENST00000263441.7
ENST00000395325.3 |
DNAJC6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr18_+_32556892 | 2.29 |
ENST00000591734.1
ENST00000413393.1 ENST00000589180.1 ENST00000587359.1 |
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr12_+_4758264 | 2.28 |
ENST00000266544.5
|
NDUFA9
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9, 39kDa |
| chr2_+_163175394 | 2.26 |
ENST00000446271.1
ENST00000429691.2 |
GCA
|
grancalcin, EF-hand calcium binding protein |
| chr6_-_99842041 | 2.25 |
ENST00000254759.3
ENST00000369242.1 |
COQ3
|
coenzyme Q3 methyltransferase |
| chrX_+_77154935 | 2.22 |
ENST00000481445.1
|
COX7B
|
cytochrome c oxidase subunit VIIb |
| chr10_-_62493223 | 2.22 |
ENST00000373827.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr17_-_4843206 | 2.20 |
ENST00000576951.1
|
SLC25A11
|
solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11 |
| chr14_-_21492251 | 2.20 |
ENST00000554398.1
|
NDRG2
|
NDRG family member 2 |
| chrX_+_65382381 | 2.17 |
ENST00000519389.1
|
HEPH
|
hephaestin |
| chr13_-_31736027 | 2.15 |
ENST00000380406.5
ENST00000320027.5 ENST00000380405.4 |
HSPH1
|
heat shock 105kDa/110kDa protein 1 |
| chr6_+_30687978 | 2.15 |
ENST00000327892.8
ENST00000435534.1 |
TUBB
|
tubulin, beta class I |
| chr16_-_15472151 | 2.13 |
ENST00000360151.4
ENST00000543801.1 |
NPIPA5
|
nuclear pore complex interacting protein family, member A5 |
| chr11_+_27062272 | 2.13 |
ENST00000529202.1
ENST00000533566.1 |
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr1_+_73771844 | 2.12 |
ENST00000440762.1
ENST00000444827.1 ENST00000415686.1 ENST00000411903.1 |
RP4-598G3.1
|
RP4-598G3.1 |
| chr18_-_24443151 | 2.11 |
ENST00000440832.3
|
AQP4
|
aquaporin 4 |
| chr17_+_53344945 | 2.10 |
ENST00000575345.1
|
HLF
|
hepatic leukemia factor |
| chr3_+_130569429 | 2.09 |
ENST00000505330.1
ENST00000504381.1 ENST00000507488.2 ENST00000393221.4 |
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr14_+_56127989 | 2.09 |
ENST00000555573.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr9_+_106856831 | 2.05 |
ENST00000303219.8
ENST00000374787.3 |
SMC2
|
structural maintenance of chromosomes 2 |
| chr9_-_130829588 | 2.01 |
ENST00000373078.4
|
NAIF1
|
nuclear apoptosis inducing factor 1 |
| chr12_-_71148357 | 2.00 |
ENST00000378778.1
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr17_+_48823975 | 1.97 |
ENST00000513969.1
ENST00000503728.1 |
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr3_+_35722487 | 1.96 |
ENST00000441454.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr8_-_66546439 | 1.96 |
ENST00000276569.3
|
ARMC1
|
armadillo repeat containing 1 |
| chr15_-_55489097 | 1.94 |
ENST00000260443.4
|
RSL24D1
|
ribosomal L24 domain containing 1 |
| chr16_-_30441293 | 1.94 |
ENST00000565758.1
ENST00000567983.1 ENST00000319285.4 |
DCTPP1
|
dCTP pyrophosphatase 1 |
| chr11_+_27062860 | 1.93 |
ENST00000528583.1
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chrX_-_102943022 | 1.91 |
ENST00000433176.2
|
MORF4L2
|
mortality factor 4 like 2 |
| chr13_+_53029564 | 1.90 |
ENST00000468284.1
ENST00000378034.3 ENST00000258607.5 ENST00000378037.5 |
CKAP2
|
cytoskeleton associated protein 2 |
| chr7_+_141811539 | 1.90 |
ENST00000550469.2
ENST00000477922.3 |
RP11-1220K2.2
|
Putative inactive maltase-glucoamylase-like protein LOC93432 |
| chr11_-_60674037 | 1.89 |
ENST00000541371.1
ENST00000227524.4 |
PRPF19
|
pre-mRNA processing factor 19 |
| chrX_-_148571884 | 1.88 |
ENST00000537071.1
|
IDS
|
iduronate 2-sulfatase |
| chr20_+_36405665 | 1.88 |
ENST00000373469.1
|
CTNNBL1
|
catenin, beta like 1 |
| chr16_+_14844670 | 1.88 |
ENST00000553201.1
|
NPIPA2
|
nuclear pore complex interacting protein family, member A2 |
| chrX_+_65384182 | 1.87 |
ENST00000441993.2
ENST00000419594.1 |
HEPH
|
hephaestin |
| chr17_-_38545799 | 1.87 |
ENST00000577541.1
|
TOP2A
|
topoisomerase (DNA) II alpha 170kDa |
| chr5_+_135496675 | 1.87 |
ENST00000507637.1
|
SMAD5
|
SMAD family member 5 |
| chr12_-_118796910 | 1.86 |
ENST00000541186.1
ENST00000539872.1 |
TAOK3
|
TAO kinase 3 |
| chr19_+_56152262 | 1.83 |
ENST00000325333.5
ENST00000590190.1 |
ZNF580
|
zinc finger protein 580 |
| chr4_-_87281224 | 1.83 |
ENST00000395169.3
ENST00000395161.2 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr16_+_57653989 | 1.82 |
ENST00000567835.1
ENST00000569372.1 ENST00000563548.1 ENST00000562003.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr1_-_226926864 | 1.80 |
ENST00000429204.1
ENST00000366784.1 |
ITPKB
|
inositol-trisphosphate 3-kinase B |
| chr4_+_86396265 | 1.79 |
ENST00000395184.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr11_-_35441524 | 1.78 |
ENST00000395750.1
ENST00000449068.1 |
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr13_-_31736478 | 1.77 |
ENST00000445273.2
|
HSPH1
|
heat shock 105kDa/110kDa protein 1 |
| chr3_-_39321512 | 1.75 |
ENST00000399220.2
|
CX3CR1
|
chemokine (C-X3-C motif) receptor 1 |
| chr1_+_156163880 | 1.74 |
ENST00000359511.4
ENST00000423538.2 |
SLC25A44
|
solute carrier family 25, member 44 |
| chr10_+_47894023 | 1.74 |
ENST00000358474.5
|
FAM21B
|
family with sequence similarity 21, member B |
| chrX_-_135962876 | 1.73 |
ENST00000431446.3
ENST00000570135.1 ENST00000320676.7 ENST00000562646.1 |
RBMX
|
RNA binding motif protein, X-linked |
| chr18_+_19192228 | 1.73 |
ENST00000300413.5
ENST00000579618.1 ENST00000582475.1 |
SNRPD1
|
small nuclear ribonucleoprotein D1 polypeptide 16kDa |
| chr1_-_68962805 | 1.72 |
ENST00000370966.5
|
DEPDC1
|
DEP domain containing 1 |
| chr11_+_27076764 | 1.72 |
ENST00000525090.1
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr14_+_56078695 | 1.72 |
ENST00000416613.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr1_+_66797687 | 1.71 |
ENST00000371045.5
ENST00000531025.1 ENST00000526197.1 |
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr4_-_164534657 | 1.70 |
ENST00000339875.5
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr5_+_167913450 | 1.69 |
ENST00000231572.3
ENST00000538719.1 |
RARS
|
arginyl-tRNA synthetase |
| chr11_-_35440796 | 1.67 |
ENST00000278379.3
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr2_+_170440844 | 1.66 |
ENST00000260970.3
ENST00000433207.1 ENST00000409714.3 ENST00000462903.1 |
PPIG
|
peptidylprolyl isomerase G (cyclophilin G) |
| chr7_-_36634181 | 1.66 |
ENST00000538464.1
|
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chr16_+_57653854 | 1.64 |
ENST00000568908.1
ENST00000568909.1 ENST00000566778.1 ENST00000561988.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr2_-_166930131 | 1.63 |
ENST00000303395.4
ENST00000409050.1 ENST00000423058.2 ENST00000375405.3 |
SCN1A
|
sodium channel, voltage-gated, type I, alpha subunit |
| chr11_+_76745385 | 1.61 |
ENST00000533140.1
ENST00000354301.5 ENST00000528622.1 |
B3GNT6
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 6 (core 3 synthase) |
| chr12_-_100486668 | 1.59 |
ENST00000550544.1
ENST00000551980.1 ENST00000548045.1 ENST00000545232.2 ENST00000551973.1 |
UHRF1BP1L
|
UHRF1 binding protein 1-like |
| chr12_+_1800179 | 1.59 |
ENST00000357103.4
|
ADIPOR2
|
adiponectin receptor 2 |
| chr2_+_173600514 | 1.58 |
ENST00000264111.6
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr1_+_155278539 | 1.57 |
ENST00000447866.1
|
FDPS
|
farnesyl diphosphate synthase |
| chr17_+_58018269 | 1.52 |
ENST00000591035.1
|
RP11-178C3.1
|
Uncharacterized protein |
| chrX_-_100604184 | 1.52 |
ENST00000372902.3
|
TIMM8A
|
translocase of inner mitochondrial membrane 8 homolog A (yeast) |
| chr10_-_50970322 | 1.52 |
ENST00000374103.4
|
OGDHL
|
oxoglutarate dehydrogenase-like |
| chr5_+_140749803 | 1.51 |
ENST00000576222.1
|
PCDHGB3
|
protocadherin gamma subfamily B, 3 |
| chr16_-_21431078 | 1.48 |
ENST00000458643.2
|
NPIPB3
|
nuclear pore complex interacting protein family, member B3 |
| chr12_-_48099773 | 1.48 |
ENST00000432584.3
ENST00000005386.3 |
RPAP3
|
RNA polymerase II associated protein 3 |
| chr12_+_19358228 | 1.48 |
ENST00000424268.1
ENST00000543806.1 |
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr19_-_10420459 | 1.48 |
ENST00000403352.1
ENST00000403903.3 |
ZGLP1
|
zinc finger, GATA-like protein 1 |
| chr9_-_21202204 | 1.48 |
ENST00000239347.3
|
IFNA7
|
interferon, alpha 7 |
| chr2_-_209010874 | 1.47 |
ENST00000260988.4
|
CRYGB
|
crystallin, gamma B |
| chr22_+_41956767 | 1.46 |
ENST00000306149.7
|
CSDC2
|
cold shock domain containing C2, RNA binding |
| chr1_-_149900122 | 1.45 |
ENST00000271628.8
|
SF3B4
|
splicing factor 3b, subunit 4, 49kDa |
| chr13_-_30424821 | 1.45 |
ENST00000380680.4
|
UBL3
|
ubiquitin-like 3 |
| chr6_-_131321863 | 1.44 |
ENST00000528282.1
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr6_+_12958137 | 1.43 |
ENST00000457702.2
ENST00000379345.2 |
PHACTR1
|
phosphatase and actin regulator 1 |
| chr2_-_228497888 | 1.41 |
ENST00000264387.4
ENST00000409066.1 |
C2orf83
|
chromosome 2 open reading frame 83 |
| chr2_+_220143989 | 1.40 |
ENST00000336576.5
|
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr8_-_117886955 | 1.39 |
ENST00000297338.2
|
RAD21
|
RAD21 homolog (S. pombe) |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXD11_HOXA11
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 11.8 | GO:0007412 | axon target recognition(GO:0007412) |
| 3.0 | 15.2 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 2.9 | 8.8 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 2.6 | 13.2 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 2.1 | 16.8 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 2.0 | 6.0 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 2.0 | 2.0 | GO:0030001 | metal ion transport(GO:0030001) |
| 1.6 | 12.5 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 1.4 | 4.2 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 1.4 | 4.2 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 1.3 | 20.9 | GO:0000338 | protein deneddylation(GO:0000338) |
| 1.3 | 6.3 | GO:0097338 | response to clozapine(GO:0097338) |
| 1.0 | 4.0 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.9 | 8.3 | GO:0003190 | atrioventricular valve formation(GO:0003190) |
| 0.9 | 4.4 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.9 | 5.1 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.8 | 2.5 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.8 | 2.4 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.8 | 7.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.8 | 3.2 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.8 | 2.4 | GO:1902997 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.8 | 9.3 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.8 | 7.7 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.8 | 3.8 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.8 | 3.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.7 | 3.6 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.7 | 6.7 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.7 | 2.6 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.6 | 1.9 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.6 | 10.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.6 | 3.2 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.6 | 1.3 | GO:0099543 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) |
| 0.6 | 2.5 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.6 | 5.4 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.6 | 2.9 | GO:0033383 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.6 | 4.7 | GO:1903750 | negative regulation of establishment of protein localization to mitochondrion(GO:1903748) regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.6 | 1.7 | GO:0002881 | negative regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002881) |
| 0.6 | 2.3 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.6 | 2.3 | GO:0060708 | histone H3-K27 acetylation(GO:0043974) leukemia inhibitory factor signaling pathway(GO:0048861) spongiotrophoblast differentiation(GO:0060708) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.6 | 10.7 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.6 | 1.7 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.5 | 3.8 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.5 | 8.3 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.5 | 2.6 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.5 | 1.9 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.5 | 12.0 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.5 | 1.4 | GO:0015888 | thiamine transport(GO:0015888) thiamine transmembrane transport(GO:0071934) |
| 0.5 | 1.8 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.5 | 2.7 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.4 | 6.2 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.4 | 11.4 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.4 | 3.5 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.4 | 2.6 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.4 | 2.5 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.4 | 1.2 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.4 | 1.9 | GO:0009149 | pyrimidine nucleoside triphosphate catabolic process(GO:0009149) pyrimidine deoxyribonucleoside triphosphate catabolic process(GO:0009213) |
| 0.4 | 2.3 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.4 | 2.3 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.4 | 1.9 | GO:0016445 | somatic diversification of immunoglobulins(GO:0016445) |
| 0.4 | 1.5 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.4 | 21.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.4 | 1.8 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.4 | 8.5 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.3 | 2.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.3 | 1.0 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.3 | 4.2 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.3 | 1.4 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.3 | 1.0 | GO:0050923 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 0.3 | 1.3 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.3 | 1.9 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.3 | 1.9 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.3 | 1.2 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 0.3 | 5.8 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.3 | 1.2 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.3 | 3.6 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.3 | 2.5 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.3 | 1.1 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.3 | 1.9 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.3 | 1.3 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.3 | 2.0 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.2 | 3.7 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.2 | 2.0 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.2 | 0.7 | GO:0036079 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.2 | 1.4 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.2 | 1.8 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.2 | 4.0 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.2 | 1.3 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 0.2 | 4.2 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 0.8 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.2 | 0.6 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.2 | 2.0 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.2 | 2.4 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.2 | 1.0 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.2 | 1.6 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.2 | 0.8 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.2 | 0.6 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.2 | 0.8 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.2 | 0.6 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.2 | 0.6 | GO:0018283 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.2 | 3.4 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.2 | 3.6 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.2 | 1.9 | GO:0030263 | resolution of meiotic recombination intermediates(GO:0000712) apoptotic chromosome condensation(GO:0030263) regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.2 | 0.9 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.2 | 0.9 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.2 | 4.7 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.2 | 2.7 | GO:0051775 | response to redox state(GO:0051775) |
| 0.2 | 12.5 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.2 | 1.0 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.2 | 0.8 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.2 | 1.7 | GO:0097267 | omega-hydroxylase P450 pathway(GO:0097267) |
| 0.2 | 3.0 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.2 | 2.5 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.2 | 1.5 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.1 | 0.6 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.1 | 1.2 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 5.4 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.1 | 0.3 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.1 | 2.8 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 0.6 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.1 | 1.0 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 1.5 | GO:0061458 | reproductive system development(GO:0061458) |
| 0.1 | 1.7 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.7 | GO:1900133 | regulation of renin secretion into blood stream(GO:1900133) |
| 0.1 | 3.8 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 2.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.3 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.1 | 1.5 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 0.9 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 | 1.4 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 0.7 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.1 | 0.6 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 4.5 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.1 | 1.9 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 0.6 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.1 | 0.9 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.1 | 2.5 | GO:0006833 | water transport(GO:0006833) |
| 0.1 | 2.4 | GO:0014904 | myotube cell development(GO:0014904) |
| 0.1 | 1.7 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) |
| 0.1 | 2.7 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.1 | 1.0 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.1 | 0.7 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 0.2 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.1 | 0.5 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 2.5 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.1 | 0.6 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.1 | 2.8 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 1.1 | GO:1903206 | negative regulation of hydrogen peroxide-induced cell death(GO:1903206) |
| 0.1 | 0.7 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.1 | 0.7 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 | 3.6 | GO:0042417 | dopamine metabolic process(GO:0042417) |
| 0.1 | 1.9 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 1.0 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.1 | 0.6 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.1 | 0.4 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.1 | 2.0 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 0.6 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.1 | 0.8 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.1 | 1.4 | GO:0017004 | cytochrome complex assembly(GO:0017004) |
| 0.1 | 4.6 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.1 | 0.4 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.1 | 0.3 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 0.5 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 | 0.5 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.1 | 2.7 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.1 | 0.6 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.1 | 1.6 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 0.2 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.0 | 6.6 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.0 | 1.7 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.1 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.0 | 0.3 | GO:1900378 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.4 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 8.2 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 1.3 | GO:0042994 | cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.0 | 0.4 | GO:0009713 | catechol-containing compound biosynthetic process(GO:0009713) catecholamine biosynthetic process(GO:0042423) |
| 0.0 | 6.1 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.8 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.0 | 0.3 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) |
| 0.0 | 0.1 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 0.0 | 2.0 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 1.1 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.1 | GO:0032900 | negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) regulation of neurotrophin production(GO:0032899) negative regulation of neurotrophin production(GO:0032900) negative regulation of transforming growth factor beta1 production(GO:0032911) negative regulation of receptor catabolic process(GO:2000645) |
| 0.0 | 0.7 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 0.2 | GO:0032328 | alanine transport(GO:0032328) |
| 0.0 | 3.8 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.2 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.0 | 0.6 | GO:0007265 | Ras protein signal transduction(GO:0007265) |
| 0.0 | 9.1 | GO:0006310 | DNA recombination(GO:0006310) |
| 0.0 | 6.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.6 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.0 | 1.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 3.2 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 | 0.8 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.7 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 2.3 | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902108) |
| 0.0 | 1.3 | GO:0032480 | negative regulation of type I interferon production(GO:0032480) |
| 0.0 | 0.2 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.4 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 2.9 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.4 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 0.6 | GO:0045143 | homologous chromosome segregation(GO:0045143) |
| 0.0 | 1.8 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 0.8 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.4 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.3 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.2 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 1.8 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.6 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 2.2 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.1 | GO:0043304 | regulation of myeloid leukocyte mediated immunity(GO:0002886) regulation of mast cell activation(GO:0033003) regulation of mast cell activation involved in immune response(GO:0033006) regulation of leukocyte degranulation(GO:0043300) regulation of mast cell degranulation(GO:0043304) |
| 0.0 | 0.9 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 1.7 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 1.2 | GO:0035036 | sperm-egg recognition(GO:0035036) |
| 0.0 | 0.5 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.3 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.2 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.2 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.5 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.0 | 0.6 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.2 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.4 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.4 | GO:0034776 | response to histamine(GO:0034776) cellular response to histamine(GO:0071420) |
| 0.0 | 0.2 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.3 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 0.3 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.3 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.3 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.0 | 0.5 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.1 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.0 | 0.3 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.0 | 0.4 | GO:0010458 | exit from mitosis(GO:0010458) |
| 0.0 | 1.7 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.0 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.0 | 0.1 | GO:1905065 | protein-pyridoxal-5-phosphate linkage(GO:0018352) positive regulation of vascular smooth muscle cell differentiation(GO:1905065) |
| 0.0 | 0.1 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.5 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 1.2 | GO:0006096 | glycolytic process(GO:0006096) ATP generation from ADP(GO:0006757) |
| 0.0 | 0.2 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.2 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 1.1 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.3 | GO:0007183 | SMAD protein complex assembly(GO:0007183) |
| 0.0 | 0.2 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 0.3 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 0.1 | GO:0002003 | angiotensin maturation(GO:0002003) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 18.2 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 2.2 | 15.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 1.6 | 6.4 | GO:1990423 | RZZ complex(GO:1990423) |
| 1.6 | 6.2 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 1.2 | 6.0 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 1.1 | 13.1 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 1.0 | 8.8 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.8 | 4.2 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.6 | 3.9 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.6 | 1.9 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.6 | 13.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.6 | 11.6 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.5 | 2.5 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.5 | 3.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.5 | 9.6 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.5 | 5.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.4 | 23.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.4 | 9.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.4 | 1.9 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.3 | 3.5 | GO:0045180 | basal cortex(GO:0045180) |
| 0.3 | 1.4 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.3 | 1.7 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.3 | 6.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.3 | 5.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.3 | 4.8 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.3 | 1.5 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.3 | 4.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.3 | 3.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.3 | 4.3 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.3 | 11.8 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.2 | 1.5 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.2 | 1.9 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.2 | 3.8 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.2 | 5.6 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.2 | 2.0 | GO:0000796 | condensin complex(GO:0000796) |
| 0.2 | 19.5 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.2 | 8.7 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.2 | 1.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.2 | 4.5 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.2 | 0.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 0.7 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.1 | 3.3 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 8.6 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 2.2 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.1 | 1.2 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.1 | 1.9 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.1 | 0.6 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.1 | 2.0 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.1 | 1.1 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 0.9 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 7.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 16.4 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 4.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 3.6 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 0.8 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 1.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 2.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 1.7 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 1.7 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 0.9 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 2.2 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 9.6 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 0.9 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 1.7 | GO:0032982 | myosin filament(GO:0032982) |
| 0.1 | 11.7 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.1 | 0.3 | GO:0031417 | NatC complex(GO:0031417) |
| 0.1 | 1.1 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 2.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 2.9 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.1 | 1.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 1.7 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 0.3 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 7.7 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.1 | 1.4 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 0.2 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 1.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 17.2 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 7.4 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 2.6 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.6 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.6 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 2.3 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.9 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 1.2 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.5 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 2.4 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.2 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.0 | 0.4 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.4 | GO:0032589 | neuron projection membrane(GO:0032589) |
| 0.0 | 0.4 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 3.4 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 1.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 12.3 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.8 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 4.1 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 10.8 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.8 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 2.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.3 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 1.2 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 2.1 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.7 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 2.7 | GO:0101002 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.0 | 0.2 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 24.9 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 3.0 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.6 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 5.5 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.6 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 2.8 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.2 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.7 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 1.6 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.1 | GO:0070069 | cytochrome complex(GO:0070069) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 11.8 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 2.1 | 8.3 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 2.0 | 21.9 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 1.6 | 4.8 | GO:0051748 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 1.3 | 15.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 1.3 | 11.4 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 1.2 | 6.0 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 1.2 | 13.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 1.1 | 16.8 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 1.0 | 5.1 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.9 | 8.8 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.9 | 17.1 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.8 | 4.2 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.8 | 4.2 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.8 | 2.5 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.8 | 2.4 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.8 | 6.2 | GO:0043559 | insulin binding(GO:0043559) |
| 0.7 | 1.4 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.7 | 2.6 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.6 | 1.9 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.6 | 3.2 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.6 | 5.0 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.6 | 1.9 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.6 | 2.9 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.6 | 1.7 | GO:0016495 | C-X3-C chemokine receptor activity(GO:0016495) |
| 0.6 | 1.7 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.5 | 5.4 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.5 | 1.6 | GO:0047223 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 0.5 | 3.2 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.5 | 15.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.5 | 1.4 | GO:0015403 | thiamine transmembrane transporter activity(GO:0015234) thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.4 | 5.8 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.4 | 6.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.4 | 4.7 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.4 | 1.2 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.4 | 3.0 | GO:0019103 | pyrimidine nucleotide binding(GO:0019103) |
| 0.4 | 2.6 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.3 | 1.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.3 | 1.4 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.3 | 1.3 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.3 | 1.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.3 | 1.8 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.3 | 1.5 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.3 | 1.8 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.3 | 1.2 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.3 | 1.1 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.3 | 1.4 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.3 | 2.4 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.3 | 0.8 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.3 | 3.5 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.3 | 1.0 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.2 | 3.0 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.2 | 1.0 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.2 | 0.7 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.2 | 0.7 | GO:0036080 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.2 | 0.7 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.2 | 5.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.2 | 3.0 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.2 | 2.3 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.2 | 1.2 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.2 | 0.8 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.2 | 9.6 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.2 | 2.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.2 | 8.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.2 | 0.9 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.2 | 0.6 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.2 | 3.1 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.2 | 1.7 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.2 | 4.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 2.1 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.2 | 2.5 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.2 | 0.9 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.2 | 2.6 | GO:0015250 | water channel activity(GO:0015250) |
| 0.2 | 1.7 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.4 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.1 | 2.4 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 1.4 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 2.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 3.8 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 3.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.8 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 1.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 2.3 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.1 | 0.5 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 1.9 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 2.5 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 0.9 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 0.9 | GO:0015116 | secondary active sulfate transmembrane transporter activity(GO:0008271) sulfate transmembrane transporter activity(GO:0015116) |
| 0.1 | 0.9 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 0.7 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 1.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 0.9 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 2.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 2.2 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 1.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 2.2 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.1 | 1.0 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.1 | 0.7 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 2.7 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 4.0 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 2.1 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 1.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 0.5 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 1.9 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 1.6 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 0.5 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 3.7 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 2.0 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 2.2 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.6 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 1.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 3.7 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 1.1 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 0.3 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 1.2 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 2.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.5 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 1.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 3.1 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 0.9 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.1 | 1.9 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 2.7 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 2.1 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 2.3 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 2.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 2.3 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 0.2 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) valine-tRNA ligase activity(GO:0004832) |
| 0.1 | 0.2 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 2.0 | GO:0001637 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.1 | 0.7 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 1.1 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.6 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 1.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 8.3 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.5 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 1.7 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 2.4 | GO:0016776 | phosphotransferase activity, phosphate group as acceptor(GO:0016776) |
| 0.0 | 0.8 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.4 | GO:0008252 | nucleotidase activity(GO:0008252) |
| 0.0 | 0.9 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 2.3 | GO:0005524 | ATP binding(GO:0005524) adenyl nucleotide binding(GO:0030554) adenyl ribonucleotide binding(GO:0032559) |
| 0.0 | 0.5 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 1.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.3 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.2 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 1.9 | GO:0051536 | iron-sulfur cluster binding(GO:0051536) metal cluster binding(GO:0051540) |
| 0.0 | 1.5 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 3.5 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 2.4 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.4 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 9.1 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.6 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.2 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.5 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 1.0 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 3.8 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 4.9 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 2.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.1 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.7 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.7 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 9.5 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.8 | GO:0008289 | lipid binding(GO:0008289) |
| 0.0 | 0.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 1.5 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.8 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.2 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.3 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.6 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.2 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.8 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.2 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.4 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.8 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 1.2 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.5 | GO:0032947 | protein complex scaffold(GO:0032947) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.2 | 6.0 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.2 | 8.8 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.2 | 8.2 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.2 | 1.8 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.2 | 10.1 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 5.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 1.9 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 2.7 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 12.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 4.5 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 6.2 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 1.8 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 3.7 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 0.6 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 6.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 1.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 3.4 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 2.1 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 3.2 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 2.5 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.5 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.5 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.2 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 2.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.1 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.9 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.5 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.5 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.2 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.4 | PID RHOA PATHWAY | RhoA signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 15.5 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.5 | 11.4 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.3 | 27.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.3 | 6.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.3 | 16.0 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.3 | 9.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.2 | 6.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.2 | 6.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 13.0 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.2 | 2.6 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 4.7 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 4.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.2 | 5.1 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 4.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 10.6 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 1.7 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 2.8 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 5.4 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.1 | 3.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.2 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.1 | 5.1 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 1.0 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 2.1 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.1 | 1.7 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 2.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 1.0 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 6.1 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 7.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 2.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.3 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 0.1 | 1.2 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 2.8 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 3.8 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 4.2 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 0.6 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 1.8 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 1.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 1.0 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 1.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 0.9 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 0.5 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 0.7 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 4.6 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.1 | 2.0 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 2.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 0.5 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 3.7 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 0.5 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.1 | 3.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.9 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 1.0 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.7 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.6 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.8 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 1.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.4 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.0 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 1.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 2.5 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.6 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.9 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.9 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.2 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.6 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.3 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.8 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.3 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |