Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
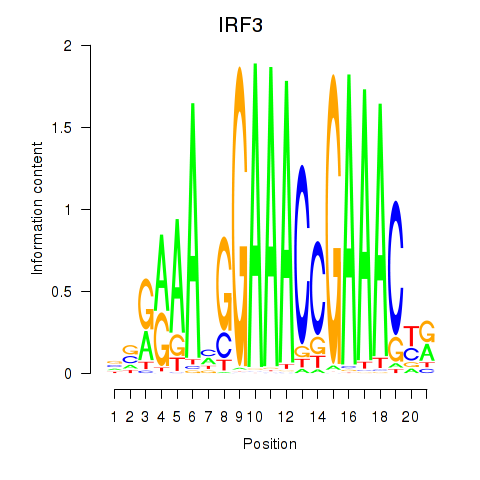
Results for IRF3
Z-value: 0.87
Transcription factors associated with IRF3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IRF3
|
ENSG00000126456.11 | interferon regulatory factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IRF3 | hg19_v2_chr19_-_50168861_50168873 | 0.39 | 3.8e-09 | Click! |
Activity profile of IRF3 motif
Sorted Z-values of IRF3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_32821924 | 20.03 |
ENST00000374859.2
ENST00000453265.2 |
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr2_+_127413677 | 13.73 |
ENST00000356887.7
|
GYPC
|
glycophorin C (Gerbich blood group) |
| chr2_+_127413481 | 13.61 |
ENST00000259254.4
|
GYPC
|
glycophorin C (Gerbich blood group) |
| chr2_+_127413704 | 13.28 |
ENST00000409836.3
|
GYPC
|
glycophorin C (Gerbich blood group) |
| chr14_+_24605361 | 11.80 |
ENST00000206451.6
ENST00000559123.1 |
PSME1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr12_-_10022735 | 11.78 |
ENST00000228438.2
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr19_-_17516449 | 11.61 |
ENST00000252593.6
|
BST2
|
bone marrow stromal cell antigen 2 |
| chr2_-_231084820 | 11.21 |
ENST00000258382.5
ENST00000338556.3 |
SP110
|
SP110 nuclear body protein |
| chr14_+_24605389 | 11.11 |
ENST00000382708.3
ENST00000561435.1 |
PSME1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr2_-_231084617 | 10.21 |
ENST00000409815.2
|
SP110
|
SP110 nuclear body protein |
| chr6_-_32821599 | 10.01 |
ENST00000354258.4
|
TAP1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr11_-_615570 | 9.97 |
ENST00000525445.1
ENST00000348655.6 ENST00000397566.1 |
IRF7
|
interferon regulatory factor 7 |
| chr6_+_26440700 | 9.88 |
ENST00000494393.1
ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3
|
butyrophilin, subfamily 3, member A3 |
| chr21_+_42792442 | 9.79 |
ENST00000398600.2
|
MX1
|
myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) |
| chr11_-_615942 | 9.69 |
ENST00000397562.3
ENST00000330243.5 ENST00000397570.1 ENST00000397574.2 |
IRF7
|
interferon regulatory factor 7 |
| chr19_+_10196781 | 9.62 |
ENST00000253110.11
|
C19orf66
|
chromosome 19 open reading frame 66 |
| chr11_-_57335280 | 9.17 |
ENST00000287156.4
|
UBE2L6
|
ubiquitin-conjugating enzyme E2L 6 |
| chr19_+_18284477 | 9.12 |
ENST00000407280.3
|
IFI30
|
interferon, gamma-inducible protein 30 |
| chr1_+_948803 | 8.83 |
ENST00000379389.4
|
ISG15
|
ISG15 ubiquitin-like modifier |
| chr2_-_231084659 | 8.80 |
ENST00000258381.6
ENST00000358662.4 ENST00000455674.1 ENST00000392048.3 |
SP110
|
SP110 nuclear body protein |
| chr16_-_67970990 | 8.53 |
ENST00000358514.4
|
PSMB10
|
proteasome (prosome, macropain) subunit, beta type, 10 |
| chr15_-_62352570 | 8.52 |
ENST00000261517.5
ENST00000395896.4 ENST00000395898.3 |
VPS13C
|
vacuolar protein sorting 13 homolog C (S. cerevisiae) |
| chr8_-_67525473 | 8.31 |
ENST00000522677.3
|
MYBL1
|
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
| chr1_+_79115503 | 7.67 |
ENST00000370747.4
ENST00000438486.1 ENST00000545124.1 |
IFI44
|
interferon-induced protein 44 |
| chr4_-_74847800 | 7.59 |
ENST00000296029.3
|
PF4
|
platelet factor 4 |
| chr7_-_138794081 | 6.99 |
ENST00000464606.1
|
ZC3HAV1
|
zinc finger CCCH-type, antiviral 1 |
| chr3_-_172241250 | 6.43 |
ENST00000420541.2
ENST00000241261.2 |
TNFSF10
|
tumor necrosis factor (ligand) superfamily, member 10 |
| chr6_+_26365443 | 6.35 |
ENST00000527422.1
ENST00000356386.2 ENST00000396934.3 ENST00000377708.2 ENST00000396948.1 ENST00000508906.2 |
BTN3A2
|
butyrophilin, subfamily 3, member A2 |
| chr7_-_92747269 | 6.08 |
ENST00000446617.1
ENST00000379958.2 |
SAMD9
|
sterile alpha motif domain containing 9 |
| chr12_+_113416191 | 5.90 |
ENST00000342315.4
ENST00000392583.2 |
OAS2
|
2'-5'-oligoadenylate synthetase 2, 69/71kDa |
| chr22_+_31892261 | 5.78 |
ENST00000432498.1
ENST00000540643.1 ENST00000443326.1 ENST00000414585.1 |
SFI1
|
Sfi1 homolog, spindle assembly associated (yeast) |
| chr5_-_95158644 | 5.72 |
ENST00000237858.6
|
GLRX
|
glutaredoxin (thioltransferase) |
| chr19_+_49977466 | 5.67 |
ENST00000596435.1
ENST00000344019.3 ENST00000597551.1 ENST00000204637.2 ENST00000600429.1 |
FLT3LG
|
fms-related tyrosine kinase 3 ligand |
| chr6_+_26402465 | 5.60 |
ENST00000476549.2
ENST00000289361.6 ENST00000450085.2 ENST00000425234.2 ENST00000427334.1 ENST00000506698.1 |
BTN3A1
|
butyrophilin, subfamily 3, member A1 |
| chr12_+_6561190 | 5.54 |
ENST00000544021.1
ENST00000266556.7 |
TAPBPL
|
TAP binding protein-like |
| chr3_+_63898275 | 5.19 |
ENST00000538065.1
|
ATXN7
|
ataxin 7 |
| chr4_-_169239921 | 4.91 |
ENST00000514995.1
ENST00000393743.3 |
DDX60
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 |
| chr7_-_138794394 | 4.64 |
ENST00000242351.5
ENST00000471652.1 |
ZC3HAV1
|
zinc finger CCCH-type, antiviral 1 |
| chr10_+_91087651 | 4.54 |
ENST00000371818.4
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr2_+_201981527 | 4.48 |
ENST00000441224.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr9_-_95087838 | 4.33 |
ENST00000442668.2
ENST00000421075.2 ENST00000536624.1 |
NOL8
|
nucleolar protein 8 |
| chr22_-_36556821 | 4.30 |
ENST00000531095.1
ENST00000397293.2 ENST00000349314.2 |
APOL3
|
apolipoprotein L, 3 |
| chr15_+_45003675 | 4.21 |
ENST00000558401.1
ENST00000559916.1 ENST00000544417.1 |
B2M
|
beta-2-microglobulin |
| chr9_-_95087604 | 4.16 |
ENST00000542613.1
ENST00000542053.1 ENST00000358855.4 ENST00000545558.1 ENST00000432670.2 ENST00000433029.2 ENST00000411621.2 |
NOL8
|
nucleolar protein 8 |
| chr19_+_49977818 | 4.13 |
ENST00000594009.1
ENST00000595510.1 |
FLT3LG
|
fms-related tyrosine kinase 3 ligand |
| chr15_-_55611306 | 4.00 |
ENST00000563262.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr17_+_36861735 | 3.94 |
ENST00000378137.5
ENST00000325718.7 |
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6 |
| chr22_+_31892373 | 3.94 |
ENST00000443011.1
ENST00000400289.1 ENST00000444859.1 ENST00000400288.2 |
SFI1
|
Sfi1 homolog, spindle assembly associated (yeast) |
| chr14_+_73525144 | 3.93 |
ENST00000261973.7
ENST00000540173.1 |
RBM25
|
RNA binding motif protein 25 |
| chr3_+_63897605 | 3.90 |
ENST00000487717.1
|
ATXN7
|
ataxin 7 |
| chr4_-_87855851 | 3.84 |
ENST00000473559.1
|
C4orf36
|
chromosome 4 open reading frame 36 |
| chr5_-_96143602 | 3.84 |
ENST00000443439.2
ENST00000503921.1 ENST00000508227.1 ENST00000507154.1 |
ERAP1
|
endoplasmic reticulum aminopeptidase 1 |
| chr17_+_25958174 | 3.78 |
ENST00000313648.6
ENST00000577392.1 ENST00000584661.1 ENST00000413914.2 |
LGALS9
|
lectin, galactoside-binding, soluble, 9 |
| chr14_+_73525229 | 3.70 |
ENST00000527432.1
ENST00000531500.1 ENST00000525321.1 ENST00000526754.1 |
RBM25
|
RNA binding motif protein 25 |
| chr19_+_10196981 | 3.60 |
ENST00000591813.1
|
C19orf66
|
chromosome 19 open reading frame 66 |
| chr2_+_201980827 | 3.50 |
ENST00000309955.3
ENST00000443227.1 ENST00000341222.6 ENST00000355558.4 ENST00000340870.5 ENST00000341582.6 |
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr5_-_77656175 | 3.49 |
ENST00000513755.1
ENST00000421004.3 |
CTD-2037K23.2
|
CTD-2037K23.2 |
| chr22_+_36649056 | 3.37 |
ENST00000397278.3
ENST00000422706.1 ENST00000426053.1 ENST00000319136.4 |
APOL1
|
apolipoprotein L, 1 |
| chr11_-_4414880 | 3.27 |
ENST00000254436.7
ENST00000543625.1 |
TRIM21
|
tripartite motif containing 21 |
| chr9_-_32526184 | 3.23 |
ENST00000545044.1
ENST00000379868.1 |
DDX58
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| chr14_-_24615805 | 3.15 |
ENST00000560410.1
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr5_-_95158375 | 3.07 |
ENST00000512469.2
ENST00000379979.4 ENST00000505427.1 ENST00000508780.1 |
GLRX
|
glutaredoxin (thioltransferase) |
| chr6_+_26402517 | 3.01 |
ENST00000414912.2
|
BTN3A1
|
butyrophilin, subfamily 3, member A1 |
| chr4_+_87928140 | 2.83 |
ENST00000307808.6
|
AFF1
|
AF4/FMR2 family, member 1 |
| chr4_+_118955500 | 2.79 |
ENST00000296499.5
|
NDST3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr2_-_157198860 | 2.77 |
ENST00000409572.1
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr4_+_87856129 | 2.63 |
ENST00000395146.4
ENST00000507468.1 |
AFF1
|
AF4/FMR2 family, member 1 |
| chr17_+_6659153 | 2.61 |
ENST00000441631.1
ENST00000438512.1 ENST00000346752.4 ENST00000361842.3 |
XAF1
|
XIAP associated factor 1 |
| chr5_-_135701164 | 2.60 |
ENST00000355180.3
ENST00000426057.2 ENST00000513104.1 |
TRPC7
|
transient receptor potential cation channel, subfamily C, member 7 |
| chr6_-_41039567 | 2.55 |
ENST00000468811.1
|
OARD1
|
O-acyl-ADP-ribose deacylase 1 |
| chr2_-_163175133 | 2.50 |
ENST00000421365.2
ENST00000263642.2 |
IFIH1
|
interferon induced with helicase C domain 1 |
| chr4_-_156875003 | 2.47 |
ENST00000433477.3
|
CTSO
|
cathepsin O |
| chr12_-_99288536 | 2.41 |
ENST00000549797.1
ENST00000333732.7 ENST00000341752.7 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr12_+_133614119 | 2.39 |
ENST00000327668.7
|
ZNF84
|
zinc finger protein 84 |
| chr1_+_145292806 | 2.33 |
ENST00000448873.2
|
NBPF10
|
neuroblastoma breakpoint family, member 10 |
| chr17_-_63052929 | 2.32 |
ENST00000439174.2
|
GNA13
|
guanine nucleotide binding protein (G protein), alpha 13 |
| chrX_-_109561294 | 2.31 |
ENST00000372059.2
ENST00000262844.5 |
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
| chr11_+_5646213 | 2.30 |
ENST00000429814.2
|
TRIM34
|
tripartite motif containing 34 |
| chr6_-_32806506 | 2.29 |
ENST00000374897.2
ENST00000452392.2 |
TAP2
TAP2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) Uncharacterized protein |
| chr10_-_14996017 | 2.28 |
ENST00000378241.1
ENST00000456122.1 ENST00000418843.1 ENST00000378249.1 ENST00000396817.2 ENST00000378255.1 ENST00000378254.1 ENST00000378278.2 ENST00000357717.2 |
DCLRE1C
|
DNA cross-link repair 1C |
| chr9_-_140082983 | 2.23 |
ENST00000323927.2
|
ANAPC2
|
anaphase promoting complex subunit 2 |
| chr1_-_25747283 | 2.22 |
ENST00000346452.4
ENST00000340849.4 ENST00000349438.4 ENST00000294413.7 ENST00000413854.1 ENST00000455194.1 ENST00000243186.6 ENST00000425135.1 |
RHCE
|
Rh blood group, CcEe antigens |
| chr22_+_39436862 | 2.19 |
ENST00000381565.2
ENST00000452957.2 |
APOBEC3F
APOBEC3G
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3F apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3G |
| chr17_-_40264692 | 2.14 |
ENST00000591220.1
ENST00000251642.3 |
DHX58
|
DEXH (Asp-Glu-X-His) box polypeptide 58 |
| chr7_-_32931387 | 2.03 |
ENST00000304056.4
|
KBTBD2
|
kelch repeat and BTB (POZ) domain containing 2 |
| chr9_-_32526299 | 1.87 |
ENST00000379882.1
ENST00000379883.2 |
DDX58
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| chr16_-_28223166 | 1.83 |
ENST00000304658.5
|
XPO6
|
exportin 6 |
| chr12_+_133613878 | 1.76 |
ENST00000392319.2
ENST00000543758.1 |
ZNF84
|
zinc finger protein 84 |
| chr12_+_133613937 | 1.71 |
ENST00000539354.1
ENST00000542874.1 ENST00000438628.2 |
ZNF84
|
zinc finger protein 84 |
| chr3_+_42190714 | 1.70 |
ENST00000449246.1
|
TRAK1
|
trafficking protein, kinesin binding 1 |
| chr2_+_201981119 | 1.63 |
ENST00000395148.2
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr10_-_14996070 | 1.61 |
ENST00000378258.1
ENST00000453695.2 ENST00000378246.2 |
DCLRE1C
|
DNA cross-link repair 1C |
| chr3_-_53079281 | 1.57 |
ENST00000394750.1
|
SFMBT1
|
Scm-like with four mbt domains 1 |
| chr20_+_388935 | 1.53 |
ENST00000382181.2
ENST00000400247.3 |
RBCK1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr19_+_50887585 | 1.48 |
ENST00000440232.2
ENST00000601098.1 ENST00000599857.1 ENST00000593887.1 |
POLD1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr14_-_91976794 | 1.46 |
ENST00000555462.1
|
SMEK1
|
SMEK homolog 1, suppressor of mek1 (Dictyostelium) |
| chr15_+_55611128 | 1.37 |
ENST00000164305.5
ENST00000539642.1 |
PIGB
|
phosphatidylinositol glycan anchor biosynthesis, class B |
| chr14_+_73525265 | 1.36 |
ENST00000525161.1
|
RBM25
|
RNA binding motif protein 25 |
| chr2_-_73869508 | 1.31 |
ENST00000272425.3
|
NAT8
|
N-acetyltransferase 8 (GCN5-related, putative) |
| chrX_-_48937531 | 1.29 |
ENST00000473974.1
ENST00000475880.1 ENST00000396681.4 ENST00000553851.1 ENST00000471338.1 ENST00000476728.1 ENST00000376368.2 ENST00000485908.1 ENST00000376372.3 ENST00000376358.3 |
WDR45
AF196779.12
|
WD repeat domain 45 WD repeat domain phosphoinositide-interacting protein 4 |
| chr2_+_99771418 | 1.23 |
ENST00000393473.2
ENST00000393477.3 ENST00000393474.3 ENST00000340066.1 ENST00000393471.2 ENST00000449211.1 ENST00000434566.1 ENST00000410042.1 |
LIPT1
MRPL30
|
lipoyltransferase 1 39S ribosomal protein L30, mitochondrial |
| chr12_-_8218997 | 1.23 |
ENST00000307637.4
|
C3AR1
|
complement component 3a receptor 1 |
| chr4_+_89378261 | 1.12 |
ENST00000264350.3
|
HERC5
|
HECT and RLD domain containing E3 ubiquitin protein ligase 5 |
| chr6_+_3068606 | 1.08 |
ENST00000259808.4
|
RIPK1
|
receptor (TNFRSF)-interacting serine-threonine kinase 1 |
| chr20_+_388679 | 1.02 |
ENST00000356286.5
ENST00000475269.1 |
RBCK1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr1_-_159046617 | 1.02 |
ENST00000368130.4
|
AIM2
|
absent in melanoma 2 |
| chr16_-_11350036 | 0.99 |
ENST00000332029.2
|
SOCS1
|
suppressor of cytokine signaling 1 |
| chr3_-_160283348 | 0.99 |
ENST00000334256.4
|
KPNA4
|
karyopherin alpha 4 (importin alpha 3) |
| chr8_+_145438870 | 0.93 |
ENST00000527931.1
|
FAM203B
|
family with sequence similarity 203, member B |
| chr17_+_79373540 | 0.92 |
ENST00000307745.7
|
RP11-1055B8.7
|
BAH and coiled-coil domain-containing protein 1 |
| chr1_+_221054584 | 0.92 |
ENST00000549319.1
|
HLX
|
H2.0-like homeobox |
| chr16_+_58283814 | 0.91 |
ENST00000443128.2
ENST00000219299.4 |
CCDC113
|
coiled-coil domain containing 113 |
| chr14_-_24615523 | 0.91 |
ENST00000559056.1
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr4_+_100495864 | 0.86 |
ENST00000265517.5
ENST00000422897.2 |
MTTP
|
microsomal triglyceride transfer protein |
| chr6_-_90529418 | 0.85 |
ENST00000439638.1
ENST00000369393.3 ENST00000428876.1 |
MDN1
|
MDN1, midasin homolog (yeast) |
| chr6_+_31540056 | 0.81 |
ENST00000418386.2
|
LTA
|
lymphotoxin alpha |
| chr20_+_61436146 | 0.76 |
ENST00000290291.6
|
OGFR
|
opioid growth factor receptor |
| chr5_-_96143796 | 0.74 |
ENST00000296754.3
|
ERAP1
|
endoplasmic reticulum aminopeptidase 1 |
| chr16_-_29875057 | 0.73 |
ENST00000219789.6
|
CDIPT
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase |
| chr19_-_47735918 | 0.72 |
ENST00000449228.1
ENST00000300880.7 ENST00000341983.4 |
BBC3
|
BCL2 binding component 3 |
| chr17_-_61517572 | 0.70 |
ENST00000582997.1
|
CYB561
|
cytochrome b561 |
| chr1_-_89664595 | 0.69 |
ENST00000355754.6
|
GBP4
|
guanylate binding protein 4 |
| chr22_+_36044411 | 0.69 |
ENST00000409652.4
|
APOL6
|
apolipoprotein L, 6 |
| chr9_-_100954910 | 0.68 |
ENST00000375077.4
|
CORO2A
|
coronin, actin binding protein, 2A |
| chr2_+_113672770 | 0.66 |
ENST00000311328.2
|
IL37
|
interleukin 37 |
| chr1_+_25598989 | 0.65 |
ENST00000454452.2
|
RHD
|
Rh blood group, D antigen |
| chr1_+_25598872 | 0.59 |
ENST00000328664.4
|
RHD
|
Rh blood group, D antigen |
| chr15_-_88799948 | 0.57 |
ENST00000394480.2
|
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr16_-_29874462 | 0.56 |
ENST00000566113.1
ENST00000569956.1 ENST00000570016.1 |
CDIPT
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase |
| chr19_+_50432400 | 0.46 |
ENST00000423777.2
ENST00000600336.1 ENST00000597227.1 |
ATF5
|
activating transcription factor 5 |
| chr12_-_48963829 | 0.45 |
ENST00000301046.2
ENST00000549817.1 |
LALBA
|
lactalbumin, alpha- |
| chr9_+_140083099 | 0.45 |
ENST00000322310.5
|
SSNA1
|
Sjogren syndrome nuclear autoantigen 1 |
| chr11_+_124735282 | 0.44 |
ENST00000397801.1
|
ROBO3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr6_-_30181133 | 0.41 |
ENST00000454678.2
ENST00000434785.1 |
TRIM26
|
tripartite motif containing 26 |
| chr2_+_69240302 | 0.40 |
ENST00000303714.4
|
ANTXR1
|
anthrax toxin receptor 1 |
| chr2_+_69240415 | 0.40 |
ENST00000409829.3
|
ANTXR1
|
anthrax toxin receptor 1 |
| chr11_+_66008698 | 0.39 |
ENST00000531597.1
|
PACS1
|
phosphofurin acidic cluster sorting protein 1 |
| chr6_-_30181156 | 0.31 |
ENST00000418026.1
ENST00000416596.1 ENST00000453195.1 |
TRIM26
|
tripartite motif containing 26 |
| chr8_+_22446763 | 0.28 |
ENST00000450780.2
ENST00000430850.2 ENST00000447849.1 |
AC037459.4
|
Uncharacterized protein |
| chr1_-_154580616 | 0.23 |
ENST00000368474.4
|
ADAR
|
adenosine deaminase, RNA-specific |
| chr3_+_187086120 | 0.22 |
ENST00000259030.2
|
RTP4
|
receptor (chemosensory) transporter protein 4 |
| chr1_+_76190357 | 0.22 |
ENST00000370834.5
ENST00000541113.1 ENST00000543667.1 ENST00000420607.2 |
ACADM
|
acyl-CoA dehydrogenase, C-4 to C-12 straight chain |
| chr14_-_20881579 | 0.20 |
ENST00000556935.1
ENST00000262715.5 ENST00000556549.1 |
TEP1
|
telomerase-associated protein 1 |
| chr7_+_134832808 | 0.18 |
ENST00000275767.3
|
TMEM140
|
transmembrane protein 140 |
| chr12_-_322821 | 0.16 |
ENST00000359674.4
|
SLC6A12
|
solute carrier family 6 (neurotransmitter transporter), member 12 |
| chrX_+_119384607 | 0.16 |
ENST00000326624.2
ENST00000557385.1 |
ZBTB33
|
zinc finger and BTB domain containing 33 |
| chr1_+_25599018 | 0.12 |
ENST00000417538.2
ENST00000357542.4 ENST00000568195.1 ENST00000342055.5 ENST00000423810.2 |
RHD
|
Rh blood group, D antigen |
| chr6_-_27840099 | 0.10 |
ENST00000328488.2
|
HIST1H3I
|
histone cluster 1, H3i |
| chrX_+_154610428 | 0.02 |
ENST00000354514.4
|
H2AFB2
|
H2A histone family, member B2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of IRF3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 19.7 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) negative regulation of macrophage apoptotic process(GO:2000110) |
| 3.9 | 11.6 | GO:0002731 | negative regulation of dendritic cell cytokine production(GO:0002731) negative regulation of intracellular transport of viral material(GO:1901253) |
| 3.3 | 9.8 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 2.8 | 8.5 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 2.7 | 10.9 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 2.4 | 19.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 2.1 | 4.2 | GO:0002428 | antigen processing and presentation of peptide antigen via MHC class Ib(GO:0002428) |
| 1.9 | 7.6 | GO:0034344 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 1.8 | 7.0 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 1.5 | 7.6 | GO:0045918 | negative regulation of cytolysis(GO:0045918) |
| 1.3 | 3.8 | GO:0032829 | regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 1.1 | 8.5 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.9 | 2.8 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.9 | 9.8 | GO:0003373 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 0.9 | 8.8 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.8 | 5.9 | GO:0018377 | protein myristoylation(GO:0018377) |
| 0.8 | 5.0 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.8 | 9.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.8 | 3.3 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.8 | 2.4 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.8 | 4.6 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.7 | 40.6 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.7 | 4.0 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.6 | 15.0 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.6 | 5.7 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.6 | 55.5 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.5 | 1.6 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) |
| 0.5 | 8.8 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.4 | 2.2 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 0.4 | 1.1 | GO:0032242 | regulation of nucleoside transport(GO:0032242) |
| 0.3 | 6.4 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.3 | 2.8 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.3 | 0.8 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.3 | 2.2 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.3 | 0.8 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 0.3 | 2.5 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.3 | 2.8 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.2 | 1.5 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.2 | 2.3 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.2 | 0.6 | GO:0052026 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.2 | 1.7 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.2 | 0.9 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.1 | 0.4 | GO:0005989 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.1 | 1.0 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.1 | 0.9 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 | 2.2 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.1 | 5.0 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.1 | 1.0 | GO:0010533 | regulation of activation of Janus kinase activity(GO:0010533) |
| 0.1 | 2.6 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.1 | 1.2 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 9.7 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 3.6 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.1 | 7.4 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 0.2 | GO:1900368 | regulation of RNA interference(GO:1900368) negative regulation of RNA interference(GO:1900369) |
| 0.1 | 0.4 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.1 | 7.9 | GO:0002456 | T cell mediated immunity(GO:0002456) |
| 0.1 | 1.3 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 1.2 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.1 | 2.6 | GO:0035456 | response to interferon-beta(GO:0035456) |
| 0.1 | 0.2 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.1 | 1.4 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.1 | 2.7 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 0.7 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.1 | 0.4 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.1 | 1.0 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 0.8 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.7 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 2.3 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.0 | 1.0 | GO:0019054 | modulation by virus of host process(GO:0019054) |
| 0.0 | 1.8 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 1.6 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.0 | 1.3 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 0.7 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.8 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.2 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 3.2 | GO:0042157 | lipoprotein metabolic process(GO:0042157) |
| 0.0 | 0.4 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.1 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 27.0 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 3.6 | 28.6 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 2.5 | 7.6 | GO:0097679 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) other organism cytoplasm(GO:0097679) |
| 1.8 | 10.9 | GO:0042825 | TAP complex(GO:0042825) |
| 0.8 | 4.2 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.7 | 2.2 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.5 | 9.9 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.4 | 2.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.4 | 9.8 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.4 | 1.5 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.3 | 4.0 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.3 | 40.6 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.3 | 3.9 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.2 | 1.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.2 | 11.6 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 1.0 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.1 | 5.5 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 2.7 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.1 | 0.9 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 9.7 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 9.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 2.2 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 3.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 1.3 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 7.0 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 2.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.2 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 2.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 8.9 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 1.7 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 5.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 5.8 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 2.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.8 | GO:0031527 | lamellipodium membrane(GO:0031258) filopodium membrane(GO:0031527) |
| 0.0 | 1.2 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 9.8 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 3.4 | GO:0031968 | mitochondrial outer membrane(GO:0005741) outer membrane(GO:0019867) organelle outer membrane(GO:0031968) |
| 0.0 | 0.4 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 1.2 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 1.0 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.8 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 1.8 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 9.8 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.9 | GO:0043235 | receptor complex(GO:0043235) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 27.0 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 1.7 | 10.3 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 1.7 | 10.0 | GO:0046979 | TAP2 binding(GO:0046979) |
| 1.5 | 7.6 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 1.3 | 3.8 | GO:0048030 | disaccharide binding(GO:0048030) |
| 1.1 | 4.6 | GO:0005151 | interleukin-1, Type II receptor binding(GO:0005151) |
| 1.0 | 28.6 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.9 | 8.8 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.7 | 8.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.7 | 8.8 | GO:0031386 | protein tag(GO:0031386) |
| 0.5 | 3.9 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.5 | 2.8 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.5 | 11.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.4 | 2.2 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.4 | 0.9 | GO:0046977 | TAP binding(GO:0046977) TAP1 binding(GO:0046978) |
| 0.3 | 1.0 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.3 | 1.4 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.3 | 9.9 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.3 | 1.2 | GO:0016979 | lipoate-protein ligase activity(GO:0016979) |
| 0.3 | 1.3 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.2 | 5.0 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.2 | 22.4 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.2 | 2.6 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.2 | 7.2 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.2 | 9.1 | GO:0016667 | oxidoreductase activity, acting on a sulfur group of donors(GO:0016667) |
| 0.2 | 0.7 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.2 | 4.0 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 1.5 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.1 | 2.3 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.1 | 9.8 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 0.4 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.1 | 3.6 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 13.0 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.1 | 1.2 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.1 | 1.7 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 6.9 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.1 | 2.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.8 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 0.6 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 1.3 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 32.7 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.7 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 1.8 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 1.0 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 11.1 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 5.2 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.0 | 9.0 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 9.8 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 1.0 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 1.0 | GO:0003690 | double-stranded DNA binding(GO:0003690) |
| 0.0 | 5.2 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 2.7 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.2 | GO:0015185 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 1.5 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.2 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 25.7 | GO:0004871 | signal transducer activity(GO:0004871) |
| 0.0 | 0.2 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 4.3 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 16.3 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.2 | 7.6 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 3.9 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 2.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 19.7 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 10.1 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 15.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 4.2 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.1 | 0.8 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 5.7 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 8.5 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.9 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.6 | PID SHP2 PATHWAY | SHP2 signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 19.7 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.8 | 7.6 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.8 | 55.5 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.6 | 16.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.5 | 11.0 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.5 | 7.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.4 | 19.1 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.3 | 8.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.2 | 2.2 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.2 | 4.6 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.2 | 2.6 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.2 | 4.0 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 10.6 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 2.2 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.1 | 1.5 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.1 | 2.3 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.1 | 2.8 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 2.1 | REACTOME RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS | Genes involved in RIG-I/MDA5 mediated induction of IFN-alpha/beta pathways |
| 0.1 | 1.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.9 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.4 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.7 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 2.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 2.5 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.7 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |