Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for IRX6_IRX4
Z-value: 0.34
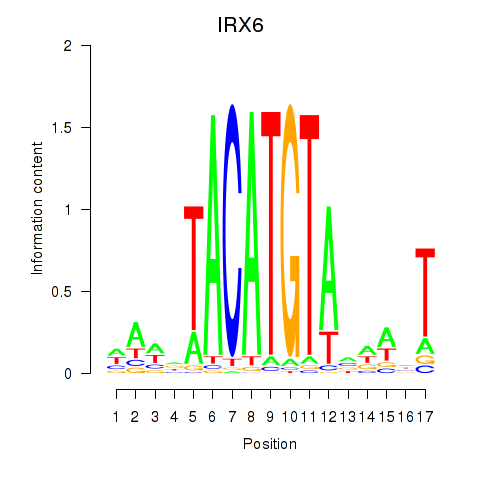
Transcription factors associated with IRX6_IRX4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IRX6
|
ENSG00000159387.7 | iroquois homeobox 6 |
|
IRX4
|
ENSG00000113430.5 | iroquois homeobox 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IRX4 | hg19_v2_chr5_-_1882858_1883003 | 0.08 | 2.3e-01 | Click! |
Activity profile of IRX6_IRX4 motif
Sorted Z-values of IRX6_IRX4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_46716969 | 6.82 |
ENST00000435666.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr21_-_48024986 | 6.42 |
ENST00000291700.4
ENST00000367071.4 |
S100B
|
S100 calcium binding protein B |
| chrX_+_49294472 | 3.26 |
ENST00000361446.5
|
GAGE12B
|
G antigen 12B |
| chr15_+_66585555 | 2.26 |
ENST00000319194.5
ENST00000525134.2 ENST00000441424.2 |
DIS3L
|
DIS3 mitotic control homolog (S. cerevisiae)-like |
| chr15_+_66585879 | 2.25 |
ENST00000319212.4
|
DIS3L
|
DIS3 mitotic control homolog (S. cerevisiae)-like |
| chr2_+_58655461 | 1.91 |
ENST00000429095.1
ENST00000429664.1 ENST00000452840.1 |
AC007092.1
|
long intergenic non-protein coding RNA 1122 |
| chr1_+_26644441 | 1.71 |
ENST00000374213.2
|
CD52
|
CD52 molecule |
| chr4_+_158142750 | 1.66 |
ENST00000505888.1
ENST00000449365.1 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr20_+_10199468 | 1.49 |
ENST00000254976.2
ENST00000304886.2 |
SNAP25
|
synaptosomal-associated protein, 25kDa |
| chr3_-_9994021 | 1.47 |
ENST00000411976.2
ENST00000412055.1 |
PRRT3
|
proline-rich transmembrane protein 3 |
| chr4_+_158141899 | 1.41 |
ENST00000264426.9
ENST00000506284.1 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr17_-_29641084 | 1.31 |
ENST00000544462.1
|
EVI2B
|
ecotropic viral integration site 2B |
| chr14_+_56127989 | 1.30 |
ENST00000555573.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr7_-_16844611 | 1.27 |
ENST00000401412.1
ENST00000419304.2 |
AGR2
|
anterior gradient 2 |
| chr17_-_29641104 | 1.27 |
ENST00000577894.1
ENST00000330927.4 |
EVI2B
|
ecotropic viral integration site 2B |
| chr12_+_32655048 | 1.22 |
ENST00000427716.2
ENST00000266482.3 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr1_+_67673297 | 1.20 |
ENST00000425614.1
ENST00000395227.1 |
IL23R
|
interleukin 23 receptor |
| chrX_-_15333736 | 1.12 |
ENST00000380470.3
|
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr5_+_54320078 | 1.11 |
ENST00000231009.2
|
GZMK
|
granzyme K (granzyme 3; tryptase II) |
| chr14_-_21737551 | 1.09 |
ENST00000554891.1
ENST00000555883.1 ENST00000553753.1 ENST00000555914.1 ENST00000557336.1 ENST00000555215.1 ENST00000556628.1 ENST00000555137.1 ENST00000556226.1 ENST00000555309.1 ENST00000556142.1 ENST00000554969.1 ENST00000554455.1 ENST00000556513.1 ENST00000557201.1 ENST00000420743.2 ENST00000557768.1 ENST00000553300.1 ENST00000554383.1 ENST00000554539.1 |
HNRNPC
|
heterogeneous nuclear ribonucleoprotein C (C1/C2) |
| chr4_-_143481822 | 1.00 |
ENST00000510812.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr22_-_24096562 | 0.98 |
ENST00000398465.3
|
VPREB3
|
pre-B lymphocyte 3 |
| chr4_-_153303658 | 0.97 |
ENST00000296555.5
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chrX_-_19988382 | 0.97 |
ENST00000356980.3
ENST00000379687.3 ENST00000379682.4 |
CXorf23
|
chromosome X open reading frame 23 |
| chr12_-_76462713 | 0.95 |
ENST00000552056.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr8_+_84824920 | 0.92 |
ENST00000523678.1
|
RP11-120I21.2
|
RP11-120I21.2 |
| chr8_-_67976509 | 0.92 |
ENST00000518747.1
|
COPS5
|
COP9 signalosome subunit 5 |
| chr12_-_10282836 | 0.78 |
ENST00000304084.8
ENST00000353231.5 ENST00000525605.1 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr1_+_220267429 | 0.76 |
ENST00000366922.1
ENST00000302637.5 |
IARS2
|
isoleucyl-tRNA synthetase 2, mitochondrial |
| chr1_+_53308398 | 0.68 |
ENST00000371528.1
|
ZYG11A
|
zyg-11 family member A, cell cycle regulator |
| chr8_-_17555164 | 0.61 |
ENST00000297488.6
|
MTUS1
|
microtubule associated tumor suppressor 1 |
| chr14_-_23623577 | 0.57 |
ENST00000422941.2
ENST00000453702.1 |
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8 |
| chr4_+_40194609 | 0.57 |
ENST00000508513.1
|
RHOH
|
ras homolog family member H |
| chr8_+_27631903 | 0.54 |
ENST00000305188.8
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr3_+_171561127 | 0.52 |
ENST00000334567.5
ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr12_-_10282742 | 0.48 |
ENST00000298523.5
ENST00000396484.2 ENST00000310002.4 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr10_+_118350468 | 0.48 |
ENST00000358834.4
ENST00000528052.1 ENST00000442761.1 |
PNLIPRP1
|
pancreatic lipase-related protein 1 |
| chr1_-_201438282 | 0.45 |
ENST00000367311.3
ENST00000367309.1 |
PHLDA3
|
pleckstrin homology-like domain, family A, member 3 |
| chr10_+_118350522 | 0.45 |
ENST00000530319.1
ENST00000527980.1 ENST00000471549.1 ENST00000534537.1 |
PNLIPRP1
|
pancreatic lipase-related protein 1 |
| chr5_-_177207634 | 0.38 |
ENST00000513554.1
ENST00000440605.3 |
FAM153A
|
family with sequence similarity 153, member A |
| chr4_+_113558612 | 0.34 |
ENST00000505034.1
ENST00000324052.6 |
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr9_-_37465396 | 0.33 |
ENST00000307750.4
|
ZBTB5
|
zinc finger and BTB domain containing 5 |
| chr6_-_52705641 | 0.29 |
ENST00000370989.2
|
GSTA5
|
glutathione S-transferase alpha 5 |
| chr2_-_231989808 | 0.24 |
ENST00000258400.3
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr5_+_175490540 | 0.24 |
ENST00000515817.1
|
FAM153B
|
family with sequence similarity 153, member B |
| chrX_+_15767971 | 0.23 |
ENST00000479740.1
ENST00000454127.2 |
CA5B
|
carbonic anhydrase VB, mitochondrial |
| chr1_-_3566590 | 0.21 |
ENST00000424367.1
ENST00000378322.3 |
WRAP73
|
WD repeat containing, antisense to TP73 |
| chr8_+_67976593 | 0.20 |
ENST00000262210.5
ENST00000412460.1 |
CSPP1
|
centrosome and spindle pole associated protein 1 |
| chr1_-_152086556 | 0.19 |
ENST00000368804.1
|
TCHH
|
trichohyalin |
| chr2_+_113670548 | 0.18 |
ENST00000263326.3
ENST00000352179.3 ENST00000349806.3 ENST00000353225.3 |
IL37
|
interleukin 37 |
| chr2_-_183106641 | 0.14 |
ENST00000346717.4
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr14_-_106845789 | 0.13 |
ENST00000390617.2
|
IGHV3-35
|
immunoglobulin heavy variable 3-35 (non-functional) |
| chrX_+_99899180 | 0.12 |
ENST00000373004.3
|
SRPX2
|
sushi-repeat containing protein, X-linked 2 |
| chr7_-_142232071 | 0.12 |
ENST00000390364.3
|
TRBV10-1
|
T cell receptor beta variable 10-1(gene/pseudogene) |
| chr21_-_34185944 | 0.12 |
ENST00000479548.1
|
C21orf62
|
chromosome 21 open reading frame 62 |
| chr3_-_27498235 | 0.11 |
ENST00000295736.5
ENST00000428386.1 ENST00000428179.1 |
SLC4A7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr14_+_22993296 | 0.09 |
ENST00000390517.1
|
TRAJ20
|
T cell receptor alpha joining 20 |
| chr1_-_3566627 | 0.09 |
ENST00000419924.2
ENST00000270708.7 |
WRAP73
|
WD repeat containing, antisense to TP73 |
| chr6_-_49834240 | 0.08 |
ENST00000335847.4
|
CRISP1
|
cysteine-rich secretory protein 1 |
| chr6_-_49834209 | 0.08 |
ENST00000507853.1
|
CRISP1
|
cysteine-rich secretory protein 1 |
| chr21_-_34186006 | 0.07 |
ENST00000490358.1
|
C21orf62
|
chromosome 21 open reading frame 62 |
| chr4_-_104119528 | 0.00 |
ENST00000380026.3
ENST00000503705.1 ENST00000265148.3 |
CENPE
|
centromere protein E, 312kDa |
Network of associatons between targets according to the STRING database.
First level regulatory network of IRX6_IRX4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 6.4 | GO:2001015 | negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 0.4 | 6.8 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.3 | 4.5 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.3 | 1.2 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.3 | 1.5 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.3 | 0.8 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.2 | 1.3 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.2 | 1.0 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.1 | 3.1 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 1.3 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.1 | 0.9 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.2 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 1.1 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.0 | 1.7 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.5 | GO:0071168 | protein localization to chromatin(GO:0071168) |
| 0.0 | 1.0 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 1.2 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.3 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.4 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.1 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.6 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.1 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.4 | 1.5 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.3 | 6.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.3 | 1.2 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.3 | 4.5 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 1.0 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 0.7 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.5 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.9 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 1.1 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 7.6 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 1.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.6 | GO:0001772 | immunological synapse(GO:0001772) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.4 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.3 | 1.2 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.3 | 3.1 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.3 | 0.8 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.2 | 1.0 | GO:0052828 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.1 | 4.5 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.1 | 1.0 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.1 | 0.5 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.9 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 1.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 1.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 1.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 1.3 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.5 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.9 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.4 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.3 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.6 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 1.3 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 6.6 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.2 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 1.2 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.1 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.2 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.0 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.9 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 6.7 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.1 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.4 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 3.1 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.1 | 1.5 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 1.0 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.8 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.0 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.2 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |