Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
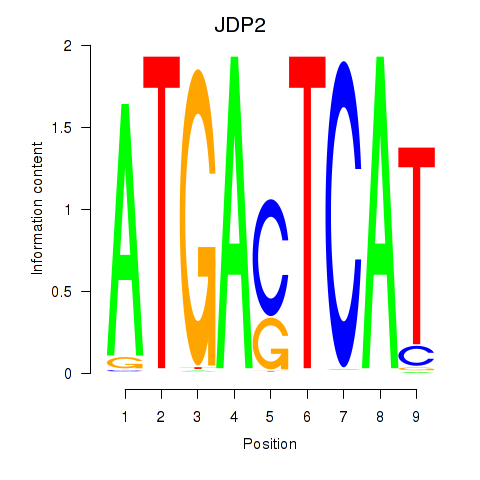
Results for JDP2
Z-value: 0.82
Transcription factors associated with JDP2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
JDP2
|
ENSG00000140044.8 | Jun dimerization protein 2 |
Activity profile of JDP2 motif
Sorted Z-values of JDP2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_175712270 | 22.94 |
ENST00000295497.7
ENST00000444394.1 |
CHN1
|
chimerin 1 |
| chr20_-_62129163 | 17.70 |
ENST00000298049.7
|
EEF1A2
|
eukaryotic translation elongation factor 1 alpha 2 |
| chr12_+_7023735 | 15.35 |
ENST00000538763.1
ENST00000544774.1 ENST00000545045.2 |
ENO2
|
enolase 2 (gamma, neuronal) |
| chr12_+_7023491 | 14.88 |
ENST00000541477.1
ENST00000229277.1 |
ENO2
|
enolase 2 (gamma, neuronal) |
| chr7_+_97361218 | 14.18 |
ENST00000319273.5
|
TAC1
|
tachykinin, precursor 1 |
| chr3_-_98241760 | 13.95 |
ENST00000507874.1
ENST00000502299.1 ENST00000508659.1 ENST00000510545.1 ENST00000511667.1 ENST00000394185.2 ENST00000394181.2 ENST00000508902.1 ENST00000341181.6 ENST00000437922.1 ENST00000394180.2 |
CLDND1
|
claudin domain containing 1 |
| chr7_+_97361388 | 13.82 |
ENST00000350485.4
ENST00000346867.4 |
TAC1
|
tachykinin, precursor 1 |
| chr16_+_15596123 | 13.55 |
ENST00000452191.2
|
C16orf45
|
chromosome 16 open reading frame 45 |
| chr13_-_36429763 | 10.87 |
ENST00000379893.1
|
DCLK1
|
doublecortin-like kinase 1 |
| chr8_-_27472198 | 10.83 |
ENST00000519472.1
ENST00000523589.1 ENST00000522413.1 ENST00000523396.1 ENST00000560366.1 |
CLU
|
clusterin |
| chr20_+_4667094 | 10.67 |
ENST00000424424.1
ENST00000457586.1 |
PRNP
|
prion protein |
| chr12_+_10365404 | 10.51 |
ENST00000266458.5
ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr20_+_4666882 | 10.04 |
ENST00000379440.4
ENST00000430350.2 |
PRNP
|
prion protein |
| chr3_-_98241358 | 9.72 |
ENST00000503004.1
ENST00000506575.1 ENST00000513452.1 ENST00000515620.1 |
CLDND1
|
claudin domain containing 1 |
| chr13_-_67802549 | 7.15 |
ENST00000328454.5
ENST00000377865.2 |
PCDH9
|
protocadherin 9 |
| chr16_+_57662138 | 7.11 |
ENST00000562414.1
ENST00000561969.1 ENST00000562631.1 ENST00000563445.1 ENST00000565338.1 ENST00000567702.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr16_+_57662419 | 6.31 |
ENST00000388812.4
ENST00000538815.1 ENST00000456916.1 ENST00000567154.1 ENST00000388813.5 ENST00000562558.1 ENST00000566271.2 |
GPR56
|
G protein-coupled receptor 56 |
| chr12_+_57914742 | 6.26 |
ENST00000551351.1
|
MBD6
|
methyl-CpG binding domain protein 6 |
| chr14_+_103800513 | 5.28 |
ENST00000560338.1
ENST00000560763.1 ENST00000558316.1 ENST00000558265.1 |
EIF5
|
eukaryotic translation initiation factor 5 |
| chr9_-_35112376 | 5.25 |
ENST00000488109.2
|
FAM214B
|
family with sequence similarity 214, member B |
| chr12_+_124997766 | 5.16 |
ENST00000543970.1
|
RP11-83B20.1
|
RP11-83B20.1 |
| chr2_-_145275228 | 5.01 |
ENST00000427902.1
ENST00000409487.3 ENST00000470879.1 ENST00000435831.1 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr16_+_89988259 | 4.61 |
ENST00000554444.1
ENST00000556565.1 |
TUBB3
|
Tubulin beta-3 chain |
| chr3_+_183894566 | 4.51 |
ENST00000439647.1
|
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit |
| chr6_-_56819385 | 4.49 |
ENST00000370754.5
ENST00000449297.2 |
DST
|
dystonin |
| chr7_+_48128316 | 4.44 |
ENST00000341253.4
|
UPP1
|
uridine phosphorylase 1 |
| chr1_-_67142710 | 4.36 |
ENST00000502413.2
|
AL139147.1
|
Uncharacterized protein |
| chr7_+_48128194 | 4.28 |
ENST00000416681.1
ENST00000331803.4 ENST00000432131.1 |
UPP1
|
uridine phosphorylase 1 |
| chr3_+_155860751 | 4.26 |
ENST00000471742.1
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr2_-_220173685 | 4.24 |
ENST00000423636.2
ENST00000442029.1 ENST00000412847.1 |
PTPRN
|
protein tyrosine phosphatase, receptor type, N |
| chr18_+_21693306 | 4.14 |
ENST00000540918.2
|
TTC39C
|
tetratricopeptide repeat domain 39C |
| chr12_+_75784850 | 4.07 |
ENST00000550916.1
ENST00000435775.1 ENST00000378689.2 ENST00000378692.3 ENST00000320460.4 ENST00000547164.1 |
GLIPR1L2
|
GLI pathogenesis-related 1 like 2 |
| chr12_-_71182695 | 4.05 |
ENST00000342084.4
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr3_-_122512619 | 3.93 |
ENST00000383659.1
ENST00000306103.2 |
HSPBAP1
|
HSPB (heat shock 27kDa) associated protein 1 |
| chr9_-_91793675 | 3.89 |
ENST00000375835.4
ENST00000375830.1 |
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3 |
| chr3_+_69134080 | 3.87 |
ENST00000273258.3
|
ARL6IP5
|
ADP-ribosylation-like factor 6 interacting protein 5 |
| chr7_+_30960915 | 3.67 |
ENST00000441328.2
ENST00000409899.1 ENST00000409611.1 |
AQP1
|
aquaporin 1 (Colton blood group) |
| chr12_+_107168342 | 3.60 |
ENST00000392837.4
|
RIC8B
|
RIC8 guanine nucleotide exchange factor B |
| chr3_-_18466026 | 3.51 |
ENST00000417717.2
|
SATB1
|
SATB homeobox 1 |
| chr1_+_27719148 | 3.45 |
ENST00000374024.3
|
GPR3
|
G protein-coupled receptor 3 |
| chr3_-_114343768 | 3.40 |
ENST00000393785.2
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr8_-_141774467 | 3.27 |
ENST00000520151.1
ENST00000519024.1 ENST00000519465.1 |
PTK2
|
protein tyrosine kinase 2 |
| chr12_+_107168418 | 3.24 |
ENST00000392839.2
ENST00000548914.1 ENST00000355478.2 ENST00000552619.1 ENST00000549643.1 |
RIC8B
|
RIC8 guanine nucleotide exchange factor B |
| chr3_+_69134124 | 3.22 |
ENST00000478935.1
|
ARL6IP5
|
ADP-ribosylation-like factor 6 interacting protein 5 |
| chr17_+_76311791 | 3.17 |
ENST00000586321.1
|
AC061992.2
|
AC061992.2 |
| chr1_+_213224572 | 3.17 |
ENST00000543470.1
ENST00000366960.3 ENST00000366959.3 ENST00000543354.1 |
RPS6KC1
|
ribosomal protein S6 kinase, 52kDa, polypeptide 1 |
| chr17_-_8263538 | 3.09 |
ENST00000535173.1
|
AC135178.1
|
HCG1985372; Uncharacterized protein; cDNA FLJ37541 fis, clone BRCAN2026340 |
| chr1_-_244006528 | 3.07 |
ENST00000336199.5
ENST00000263826.5 |
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 |
| chr7_-_80551671 | 2.99 |
ENST00000419255.2
ENST00000544525.1 |
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr8_-_38386175 | 2.98 |
ENST00000437935.2
ENST00000358138.1 |
C8orf86
|
chromosome 8 open reading frame 86 |
| chr10_+_1102721 | 2.97 |
ENST00000263150.4
|
WDR37
|
WD repeat domain 37 |
| chr9_-_23826298 | 2.96 |
ENST00000380117.1
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr17_+_66539369 | 2.93 |
ENST00000600820.1
|
AC079210.1
|
Uncharacterized protein; cDNA FLJ45097 fis, clone BRAWH3031054 |
| chr9_+_34958254 | 2.93 |
ENST00000242315.3
|
KIAA1045
|
KIAA1045 |
| chr12_-_719573 | 2.92 |
ENST00000397265.3
|
NINJ2
|
ninjurin 2 |
| chr5_-_149492904 | 2.88 |
ENST00000286301.3
ENST00000511344.1 |
CSF1R
|
colony stimulating factor 1 receptor |
| chr7_+_147830776 | 2.87 |
ENST00000538075.1
|
CNTNAP2
|
contactin associated protein-like 2 |
| chr13_+_32838801 | 2.81 |
ENST00000542859.1
|
FRY
|
furry homolog (Drosophila) |
| chr12_-_57914275 | 2.80 |
ENST00000547303.1
ENST00000552740.1 ENST00000547526.1 ENST00000551116.1 ENST00000346473.3 |
DDIT3
|
DNA-damage-inducible transcript 3 |
| chr7_-_135433460 | 2.69 |
ENST00000415751.1
|
FAM180A
|
family with sequence similarity 180, member A |
| chr20_-_48782639 | 2.68 |
ENST00000435301.2
|
RP11-112L6.3
|
RP11-112L6.3 |
| chr4_-_36246060 | 2.66 |
ENST00000303965.4
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr3_-_18466787 | 2.64 |
ENST00000338745.6
ENST00000450898.1 |
SATB1
|
SATB homeobox 1 |
| chr12_+_6644443 | 2.63 |
ENST00000396858.1
|
GAPDH
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr11_-_2924720 | 2.60 |
ENST00000455942.2
|
SLC22A18AS
|
solute carrier family 22 (organic cation transporter), member 18 antisense |
| chr7_-_14942283 | 2.60 |
ENST00000402815.1
|
DGKB
|
diacylglycerol kinase, beta 90kDa |
| chr10_+_88428206 | 2.53 |
ENST00000429277.2
ENST00000458213.2 ENST00000352360.5 |
LDB3
|
LIM domain binding 3 |
| chr10_-_120514720 | 2.48 |
ENST00000369151.3
ENST00000340214.4 |
CACUL1
|
CDK2-associated, cullin domain 1 |
| chr3_-_149293990 | 2.46 |
ENST00000472417.1
|
WWTR1
|
WW domain containing transcription regulator 1 |
| chr12_-_48152611 | 2.42 |
ENST00000389212.3
|
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr12_-_48152853 | 2.41 |
ENST00000171000.4
|
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr17_+_30771279 | 2.31 |
ENST00000261712.3
ENST00000578213.1 ENST00000457654.2 ENST00000579451.1 |
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr3_+_183903811 | 2.29 |
ENST00000429586.2
ENST00000292808.5 |
ABCF3
|
ATP-binding cassette, sub-family F (GCN20), member 3 |
| chr9_-_124991124 | 2.23 |
ENST00000394319.4
ENST00000340587.3 |
LHX6
|
LIM homeobox 6 |
| chr12_-_56101647 | 2.21 |
ENST00000347027.6
ENST00000257879.6 ENST00000257880.7 ENST00000394230.2 ENST00000394229.2 |
ITGA7
|
integrin, alpha 7 |
| chr16_+_28996416 | 2.19 |
ENST00000395456.2
ENST00000454369.2 |
LAT
|
linker for activation of T cells |
| chr11_-_82708519 | 2.18 |
ENST00000534301.1
|
RAB30
|
RAB30, member RAS oncogene family |
| chr19_+_50706866 | 2.15 |
ENST00000440075.2
ENST00000376970.2 ENST00000425460.1 ENST00000599920.1 ENST00000601313.1 |
MYH14
|
myosin, heavy chain 14, non-muscle |
| chr20_-_17539456 | 2.12 |
ENST00000544874.1
ENST00000377868.2 |
BFSP1
|
beaded filament structural protein 1, filensin |
| chr8_-_110988070 | 2.09 |
ENST00000524391.1
|
KCNV1
|
potassium channel, subfamily V, member 1 |
| chr4_+_170581213 | 2.08 |
ENST00000507875.1
|
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chr9_-_21335240 | 2.07 |
ENST00000537938.1
|
KLHL9
|
kelch-like family member 9 |
| chrY_+_15016725 | 2.04 |
ENST00000336079.3
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chr1_+_154377669 | 2.01 |
ENST00000368485.3
ENST00000344086.4 |
IL6R
|
interleukin 6 receptor |
| chr17_+_79650962 | 1.98 |
ENST00000329138.4
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate |
| chr9_-_21335356 | 1.96 |
ENST00000359039.4
|
KLHL9
|
kelch-like family member 9 |
| chr19_-_4455290 | 1.90 |
ENST00000394765.3
ENST00000592515.1 |
UBXN6
|
UBX domain protein 6 |
| chrX_-_48901012 | 1.89 |
ENST00000315869.7
|
TFE3
|
transcription factor binding to IGHM enhancer 3 |
| chr9_+_140135665 | 1.86 |
ENST00000340384.4
|
TUBB4B
|
tubulin, beta 4B class IVb |
| chr17_-_73775839 | 1.84 |
ENST00000592643.1
ENST00000591890.1 ENST00000587171.1 ENST00000254810.4 ENST00000589599.1 |
H3F3B
|
H3 histone, family 3B (H3.3B) |
| chr11_-_75201791 | 1.79 |
ENST00000529721.1
|
GDPD5
|
glycerophosphodiester phosphodiesterase domain containing 5 |
| chr6_+_10521574 | 1.77 |
ENST00000495262.1
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr6_+_151042224 | 1.74 |
ENST00000358517.2
|
PLEKHG1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr6_-_161695042 | 1.69 |
ENST00000366908.5
ENST00000366911.5 ENST00000366905.3 |
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr11_-_2924970 | 1.68 |
ENST00000533594.1
|
SLC22A18AS
|
solute carrier family 22 (organic cation transporter), member 18 antisense |
| chr12_-_8218997 | 1.68 |
ENST00000307637.4
|
C3AR1
|
complement component 3a receptor 1 |
| chr4_+_71588372 | 1.65 |
ENST00000536664.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr7_+_28725585 | 1.56 |
ENST00000396298.2
|
CREB5
|
cAMP responsive element binding protein 5 |
| chr16_+_30077098 | 1.56 |
ENST00000395240.3
ENST00000566846.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr4_+_120056939 | 1.54 |
ENST00000307128.5
|
MYOZ2
|
myozenin 2 |
| chr19_-_5903714 | 1.54 |
ENST00000586349.1
ENST00000585661.1 ENST00000308961.4 ENST00000592634.1 ENST00000418389.2 ENST00000252675.5 |
AC024592.12
NDUFA11
FUT5
|
Uncharacterized protein NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 11, 14.7kDa fucosyltransferase 5 (alpha (1,3) fucosyltransferase) |
| chr15_+_89182156 | 1.52 |
ENST00000379224.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr16_+_30077055 | 1.52 |
ENST00000564595.2
ENST00000569798.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr1_-_153521597 | 1.50 |
ENST00000368712.1
|
S100A3
|
S100 calcium binding protein A3 |
| chr11_-_84028180 | 1.48 |
ENST00000280241.8
|
DLG2
|
discs, large homolog 2 (Drosophila) |
| chrX_-_15288154 | 1.48 |
ENST00000380483.3
ENST00000380485.3 ENST00000380488.4 |
ASB9
|
ankyrin repeat and SOCS box containing 9 |
| chr11_-_66103932 | 1.48 |
ENST00000311320.4
|
RIN1
|
Ras and Rab interactor 1 |
| chr10_+_88428370 | 1.46 |
ENST00000372066.3
ENST00000263066.6 ENST00000372056.4 ENST00000310944.6 ENST00000361373.4 ENST00000542786.1 |
LDB3
|
LIM domain binding 3 |
| chr6_-_161695074 | 1.45 |
ENST00000457520.2
ENST00000366906.5 ENST00000320285.4 |
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr8_-_42623924 | 1.43 |
ENST00000276410.2
|
CHRNA6
|
cholinergic receptor, nicotinic, alpha 6 (neuronal) |
| chr1_-_153521714 | 1.32 |
ENST00000368713.3
|
S100A3
|
S100 calcium binding protein A3 |
| chr17_+_25799008 | 1.30 |
ENST00000583370.1
ENST00000398988.3 ENST00000268763.6 |
KSR1
|
kinase suppressor of ras 1 |
| chr16_+_28996114 | 1.29 |
ENST00000395461.3
|
LAT
|
linker for activation of T cells |
| chr17_+_66245341 | 1.28 |
ENST00000577985.1
|
AMZ2
|
archaelysin family metallopeptidase 2 |
| chr21_-_36421626 | 1.28 |
ENST00000300305.3
|
RUNX1
|
runt-related transcription factor 1 |
| chr21_-_36421535 | 1.28 |
ENST00000416754.1
ENST00000437180.1 ENST00000455571.1 |
RUNX1
|
runt-related transcription factor 1 |
| chr12_-_7818474 | 1.28 |
ENST00000229304.4
|
APOBEC1
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1 |
| chr1_+_17634689 | 1.23 |
ENST00000375453.1
ENST00000375448.4 |
PADI4
|
peptidyl arginine deiminase, type IV |
| chr1_-_51796987 | 1.20 |
ENST00000262676.5
|
TTC39A
|
tetratricopeptide repeat domain 39A |
| chr6_-_49755019 | 1.18 |
ENST00000304801.3
|
PGK2
|
phosphoglycerate kinase 2 |
| chr7_+_134528635 | 1.14 |
ENST00000445569.2
|
CALD1
|
caldesmon 1 |
| chr5_-_75919253 | 1.10 |
ENST00000296641.4
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2 |
| chr17_-_33760269 | 1.07 |
ENST00000452764.3
|
SLFN12
|
schlafen family member 12 |
| chr4_+_3388057 | 1.02 |
ENST00000538395.1
|
RGS12
|
regulator of G-protein signaling 12 |
| chr1_-_160231451 | 1.01 |
ENST00000495887.1
|
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chr6_+_106534192 | 1.00 |
ENST00000369091.2
ENST00000369096.4 |
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr16_+_9449445 | 1.00 |
ENST00000564305.1
|
RP11-243A14.1
|
RP11-243A14.1 |
| chr9_-_21207142 | 0.96 |
ENST00000357374.2
|
IFNA10
|
interferon, alpha 10 |
| chr22_+_39916558 | 0.92 |
ENST00000337304.2
ENST00000396680.1 |
ATF4
|
activating transcription factor 4 |
| chr8_+_15397732 | 0.91 |
ENST00000382020.4
ENST00000506802.1 ENST00000509380.1 ENST00000503731.1 |
TUSC3
|
tumor suppressor candidate 3 |
| chrX_+_1733876 | 0.80 |
ENST00000381241.3
|
ASMT
|
acetylserotonin O-methyltransferase |
| chr4_+_175839551 | 0.79 |
ENST00000404450.4
ENST00000514159.1 |
ADAM29
|
ADAM metallopeptidase domain 29 |
| chr5_+_35856951 | 0.76 |
ENST00000303115.3
ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr1_-_204121013 | 0.76 |
ENST00000367201.3
|
ETNK2
|
ethanolamine kinase 2 |
| chr11_+_5009424 | 0.76 |
ENST00000300762.1
|
MMP26
|
matrix metallopeptidase 26 |
| chr16_+_31366536 | 0.71 |
ENST00000562522.1
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr11_-_102826434 | 0.70 |
ENST00000340273.4
ENST00000260302.3 |
MMP13
|
matrix metallopeptidase 13 (collagenase 3) |
| chr2_+_233562015 | 0.69 |
ENST00000427233.1
ENST00000373566.3 ENST00000373563.4 ENST00000428883.1 ENST00000456491.1 ENST00000409480.1 ENST00000421433.1 ENST00000425040.1 ENST00000430720.1 ENST00000409547.1 ENST00000423659.1 ENST00000409196.3 ENST00000409451.3 ENST00000429187.1 ENST00000440945.1 |
GIGYF2
|
GRB10 interacting GYF protein 2 |
| chr22_+_20861858 | 0.69 |
ENST00000414658.1
ENST00000432052.1 ENST00000425759.2 ENST00000292733.7 ENST00000542773.1 ENST00000263205.7 ENST00000406969.1 ENST00000382974.2 |
MED15
|
mediator complex subunit 15 |
| chr8_-_42623747 | 0.68 |
ENST00000534622.1
|
CHRNA6
|
cholinergic receptor, nicotinic, alpha 6 (neuronal) |
| chr1_-_204121102 | 0.66 |
ENST00000367202.4
|
ETNK2
|
ethanolamine kinase 2 |
| chr11_-_66103867 | 0.65 |
ENST00000424433.2
|
RIN1
|
Ras and Rab interactor 1 |
| chr17_+_55162453 | 0.64 |
ENST00000575322.1
ENST00000337714.3 ENST00000314126.3 |
AKAP1
|
A kinase (PRKA) anchor protein 1 |
| chr4_+_175839506 | 0.63 |
ENST00000505141.1
ENST00000359240.3 ENST00000445694.1 |
ADAM29
|
ADAM metallopeptidase domain 29 |
| chr2_+_152214098 | 0.62 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr18_+_2846972 | 0.61 |
ENST00000254528.3
|
EMILIN2
|
elastin microfibril interfacer 2 |
| chr3_+_48507210 | 0.60 |
ENST00000433541.1
ENST00000422277.2 ENST00000436480.2 ENST00000444177.1 |
TREX1
|
three prime repair exonuclease 1 |
| chr17_+_74381343 | 0.60 |
ENST00000392496.3
|
SPHK1
|
sphingosine kinase 1 |
| chr4_-_122085469 | 0.60 |
ENST00000057513.3
|
TNIP3
|
TNFAIP3 interacting protein 3 |
| chr11_-_66104237 | 0.58 |
ENST00000530056.1
|
RIN1
|
Ras and Rab interactor 1 |
| chr11_-_102668879 | 0.53 |
ENST00000315274.6
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase) |
| chr21_-_36421401 | 0.52 |
ENST00000486278.2
|
RUNX1
|
runt-related transcription factor 1 |
| chr10_+_52152766 | 0.51 |
ENST00000596442.1
|
AC069547.2
|
Uncharacterized protein |
| chr1_-_111148241 | 0.50 |
ENST00000440270.1
|
KCNA2
|
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chr20_+_30946106 | 0.50 |
ENST00000375687.4
ENST00000542461.1 |
ASXL1
|
additional sex combs like 1 (Drosophila) |
| chr7_+_127233689 | 0.49 |
ENST00000265825.5
ENST00000420086.2 |
FSCN3
|
fascin homolog 3, actin-bundling protein, testicular (Strongylocentrotus purpuratus) |
| chrX_+_57618269 | 0.47 |
ENST00000374888.1
|
ZXDB
|
zinc finger, X-linked, duplicated B |
| chr9_-_34662651 | 0.47 |
ENST00000259631.4
|
CCL27
|
chemokine (C-C motif) ligand 27 |
| chr12_+_52695617 | 0.47 |
ENST00000293525.5
|
KRT86
|
keratin 86 |
| chr4_-_100356551 | 0.44 |
ENST00000209665.4
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr16_+_2083265 | 0.42 |
ENST00000565855.1
ENST00000566198.1 |
SLC9A3R2
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
| chr1_-_204121298 | 0.41 |
ENST00000367199.2
|
ETNK2
|
ethanolamine kinase 2 |
| chr5_+_177540444 | 0.39 |
ENST00000274605.5
|
N4BP3
|
NEDD4 binding protein 3 |
| chr16_+_28996364 | 0.39 |
ENST00000564277.1
|
LAT
|
linker for activation of T cells |
| chr2_+_69201705 | 0.38 |
ENST00000377938.2
|
GKN1
|
gastrokine 1 |
| chr2_+_166326157 | 0.36 |
ENST00000421875.1
ENST00000314499.7 ENST00000409664.1 |
CSRNP3
|
cysteine-serine-rich nuclear protein 3 |
| chr1_-_13117736 | 0.33 |
ENST00000376192.5
ENST00000376182.1 |
PRAMEF6
|
PRAME family member 6 |
| chr6_-_31745037 | 0.30 |
ENST00000375688.4
|
VWA7
|
von Willebrand factor A domain containing 7 |
| chr6_+_72596406 | 0.29 |
ENST00000491071.2
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr1_+_13359819 | 0.29 |
ENST00000376168.1
|
PRAMEF5
|
PRAME family member 5 |
| chr10_-_21463116 | 0.28 |
ENST00000417816.2
|
NEBL
|
nebulette |
| chr4_-_100356291 | 0.26 |
ENST00000476959.1
ENST00000482593.1 |
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr1_+_151227179 | 0.26 |
ENST00000368884.3
ENST00000368881.4 |
PSMD4
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 4 |
| chr6_+_122720681 | 0.25 |
ENST00000368455.4
ENST00000452194.1 |
HSF2
|
heat shock transcription factor 2 |
| chr11_+_67250490 | 0.25 |
ENST00000528641.2
ENST00000279146.3 |
AIP
|
aryl hydrocarbon receptor interacting protein |
| chr11_-_102714534 | 0.25 |
ENST00000299855.5
|
MMP3
|
matrix metallopeptidase 3 (stromelysin 1, progelatinase) |
| chr18_+_61442629 | 0.23 |
ENST00000398019.2
ENST00000540675.1 |
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr3_-_194090460 | 0.23 |
ENST00000428839.1
ENST00000347624.3 |
LRRC15
|
leucine rich repeat containing 15 |
| chr4_-_987217 | 0.19 |
ENST00000361661.2
ENST00000398516.2 |
SLC26A1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr2_+_202122703 | 0.19 |
ENST00000447616.1
ENST00000358485.4 |
CASP8
|
caspase 8, apoptosis-related cysteine peptidase |
| chr4_-_100356844 | 0.18 |
ENST00000437033.2
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr3_+_19988885 | 0.17 |
ENST00000422242.1
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chr16_+_31366455 | 0.15 |
ENST00000268296.4
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr7_-_155604967 | 0.15 |
ENST00000297261.2
|
SHH
|
sonic hedgehog |
| chr5_-_75919217 | 0.14 |
ENST00000504899.1
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2 |
| chr1_-_13007420 | 0.14 |
ENST00000376189.1
|
PRAMEF6
|
PRAME family member 6 |
| chr3_+_48507621 | 0.09 |
ENST00000456089.1
|
TREX1
|
three prime repair exonuclease 1 |
| chrX_+_1734051 | 0.05 |
ENST00000381229.4
ENST00000381233.3 |
ASMT
|
acetylserotonin O-methyltransferase |
| chr11_+_82783097 | 0.02 |
ENST00000501011.2
ENST00000527627.1 ENST00000526795.1 ENST00000533528.1 ENST00000533708.1 ENST00000534499.1 |
RAB30-AS1
|
RAB30 antisense RNA 1 (head to head) |
| chr9_+_138453595 | 0.02 |
ENST00000479141.1
ENST00000371766.2 ENST00000277508.5 ENST00000433563.1 |
PAEP
|
progestagen-associated endometrial protein |
| chr20_+_43029911 | 0.01 |
ENST00000443598.2
ENST00000316099.4 ENST00000415691.2 |
HNF4A
|
hepatocyte nuclear factor 4, alpha |
| chr5_+_179247759 | 0.00 |
ENST00000389805.4
ENST00000504627.1 ENST00000402874.3 ENST00000510187.1 |
SQSTM1
|
sequestosome 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of JDP2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.3 | 28.0 | GO:2000854 | positive regulation of corticosterone secretion(GO:2000854) |
| 6.9 | 20.7 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 2.7 | 13.5 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 2.2 | 8.7 | GO:0046108 | uridine catabolic process(GO:0006218) uridine metabolic process(GO:0046108) |
| 1.8 | 10.8 | GO:0061518 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 1.4 | 4.2 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 1.3 | 5.0 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 1.2 | 3.7 | GO:0072019 | carbon dioxide transmembrane transport(GO:0035378) proximal convoluted tubule development(GO:0072019) metanephric proximal convoluted tubule development(GO:0072229) |
| 1.2 | 4.8 | GO:0033123 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 1.0 | 3.0 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 1.0 | 2.9 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.9 | 17.7 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.9 | 7.1 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.9 | 2.6 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.7 | 2.8 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.7 | 33.3 | GO:0061718 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.7 | 4.0 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.7 | 2.0 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.7 | 22.9 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.6 | 10.5 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.5 | 2.7 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.5 | 4.5 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.5 | 10.9 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.4 | 2.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.4 | 3.3 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.4 | 6.2 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.4 | 2.2 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.4 | 2.8 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.3 | 2.9 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) cell-cell junction maintenance(GO:0045217) |
| 0.3 | 0.9 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.3 | 0.9 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.3 | 0.9 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.3 | 4.3 | GO:1902260 | diaphragm development(GO:0060539) negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.3 | 1.8 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.2 | 1.2 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.2 | 1.9 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.2 | 2.5 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.2 | 1.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) DNA cytosine deamination(GO:0070383) |
| 0.2 | 3.1 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.2 | 1.0 | GO:0033078 | extrathymic T cell differentiation(GO:0033078) |
| 0.2 | 0.6 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.2 | 3.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.2 | 3.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.2 | 1.8 | GO:0048505 | regulation of development, heterochronic(GO:0040034) regulation of timing of cell differentiation(GO:0048505) |
| 0.2 | 0.5 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.2 | 2.6 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.2 | 4.7 | GO:1904376 | negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.1 | 2.1 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.1 | 2.0 | GO:1903543 | positive regulation of exosomal secretion(GO:1903543) |
| 0.1 | 1.5 | GO:0042355 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.1 | 0.5 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.1 | 0.9 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 1.7 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.1 | 1.2 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 2.9 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.1 | 3.0 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.1 | 3.4 | GO:0040020 | regulation of meiotic nuclear division(GO:0040020) |
| 0.1 | 0.9 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.1 | 1.8 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 1.1 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.1 | 1.2 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 2.6 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 2.1 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 0.2 | GO:0052053 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.1 | 0.2 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.1 | 1.0 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.1 | 3.9 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.1 | 1.5 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.1 | 0.3 | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 1.6 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 7.6 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 7.2 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 3.4 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) |
| 0.0 | 0.2 | GO:0050428 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.0 | 0.8 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.0 | 0.2 | GO:0051036 | regulation of endosome size(GO:0051036) |
| 0.0 | 8.5 | GO:0007411 | axon guidance(GO:0007411) |
| 0.0 | 0.7 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 1.9 | GO:0042267 | natural killer cell mediated cytotoxicity(GO:0042267) |
| 0.0 | 0.7 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 0.7 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 0.6 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 1.9 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 4.0 | GO:0000910 | cytokinesis(GO:0000910) |
| 0.0 | 0.6 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 1.2 | GO:0030317 | sperm motility(GO:0030317) |
| 0.0 | 0.3 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.8 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.0 | 1.3 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 5.9 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.3 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.7 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.7 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 30.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 1.8 | 17.7 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 1.3 | 20.7 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 1.2 | 3.7 | GO:0020003 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 1.1 | 4.5 | GO:0031673 | H zone(GO:0031673) |
| 0.9 | 2.8 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.7 | 5.0 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.7 | 10.8 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.7 | 2.0 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.5 | 2.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.5 | 3.0 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.5 | 2.9 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.4 | 2.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.4 | 10.5 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.4 | 2.6 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.3 | 6.3 | GO:0010369 | chromocenter(GO:0010369) |
| 0.3 | 3.4 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.2 | 4.5 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.2 | 2.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.2 | 6.2 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.2 | 4.0 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.2 | 0.9 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.1 | 1.8 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 3.9 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 1.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 2.1 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 1.5 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 1.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.9 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 1.6 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 5.3 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 3.1 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 6.6 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.1 | 0.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 4.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 27.2 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 10.9 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 1.0 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 2.8 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 2.1 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 8.3 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 1.5 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.7 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 3.3 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 2.1 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 17.2 | GO:0015630 | microtubule cytoskeleton(GO:0015630) |
| 0.0 | 0.9 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 0.3 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.2 | GO:0097342 | ripoptosome(GO:0097342) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 30.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 3.5 | 20.7 | GO:1903135 | cupric ion binding(GO:1903135) |
| 2.2 | 8.7 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 1.2 | 3.7 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.8 | 10.5 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.7 | 28.0 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.7 | 2.0 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.7 | 2.6 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) aspartic-type endopeptidase inhibitor activity(GO:0019828) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.7 | 17.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.6 | 1.2 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.5 | 10.8 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.5 | 22.9 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.4 | 1.8 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.4 | 1.2 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.4 | 4.8 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.3 | 2.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.3 | 1.2 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.3 | 1.5 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.3 | 1.5 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.3 | 3.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.3 | 0.8 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.3 | 1.8 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.2 | 1.2 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.2 | 0.9 | GO:0035276 | aldehyde oxidase activity(GO:0004031) ethanol binding(GO:0035276) |
| 0.2 | 6.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.2 | 1.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.2 | 3.7 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.2 | 4.5 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.2 | 1.8 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.2 | 3.3 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.2 | 1.3 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.2 | 4.3 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.2 | 3.0 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.2 | 4.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.2 | 4.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 4.0 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 2.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 3.1 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 1.7 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.1 | 0.5 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.1 | 0.7 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.1 | 0.9 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 5.0 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 0.6 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 4.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 2.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.9 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 2.1 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 3.9 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 0.6 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.1 | 0.7 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 1.0 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 2.7 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 1.9 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 1.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 2.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.5 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 3.9 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 2.5 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 1.5 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 5.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.2 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 10.1 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.5 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 3.0 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.6 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 2.1 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 2.9 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 2.8 | GO:0004857 | enzyme inhibitor activity(GO:0004857) |
| 0.0 | 2.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.3 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 1.0 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 0.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.4 | GO:0031593 | polyubiquitin binding(GO:0031593) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 20.7 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.3 | 22.9 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.2 | 6.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.2 | 4.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.2 | 4.5 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.2 | 3.9 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 2.0 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.1 | 3.1 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 13.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 2.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 2.7 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 3.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 5.7 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 2.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 2.7 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 2.6 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 3.0 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 2.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.6 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 1.3 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 2.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.0 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.3 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.5 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 35.9 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.6 | 8.7 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.3 | 3.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.3 | 20.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.3 | 3.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.3 | 3.7 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.3 | 4.5 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.2 | 6.5 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.2 | 3.9 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.2 | 8.1 | REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | Genes involved in Integrin alphaIIb beta3 signaling |
| 0.1 | 29.5 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 6.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 2.1 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 3.9 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.1 | 2.0 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 0.9 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 3.1 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 18.3 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 0.8 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 10.8 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 2.1 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 2.0 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 2.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 2.5 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 1.0 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 2.6 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 1.8 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 1.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.3 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |