Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
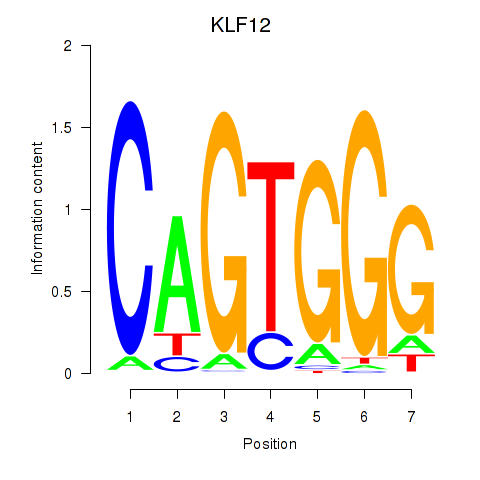
Results for KLF12
Z-value: 0.84
Transcription factors associated with KLF12
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF12
|
ENSG00000118922.12 | Kruppel like factor 12 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| KLF12 | hg19_v2_chr13_-_74708372_74708409 | -0.34 | 2.5e-07 | Click! |
Activity profile of KLF12 motif
Sorted Z-values of KLF12 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_89402148 | 23.04 |
ENST00000560601.1
|
ACAN
|
aggrecan |
| chrX_+_23682379 | 16.47 |
ENST00000379349.1
|
PRDX4
|
peroxiredoxin 4 |
| chrX_+_49028265 | 11.17 |
ENST00000376322.3
ENST00000376327.5 |
PLP2
|
proteolipid protein 2 (colonic epithelium-enriched) |
| chr12_-_56120838 | 10.75 |
ENST00000548160.1
|
CD63
|
CD63 molecule |
| chr1_+_155178481 | 10.68 |
ENST00000368376.3
|
MTX1
|
metaxin 1 |
| chr12_-_53012343 | 10.19 |
ENST00000305748.3
|
KRT73
|
keratin 73 |
| chr11_-_64013288 | 9.88 |
ENST00000542235.1
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr19_+_13051206 | 9.51 |
ENST00000586760.1
|
CALR
|
calreticulin |
| chr15_+_64428529 | 9.39 |
ENST00000560861.1
|
SNX1
|
sorting nexin 1 |
| chr12_+_53848549 | 9.10 |
ENST00000439930.3
ENST00000548933.1 ENST00000562264.1 |
PCBP2
|
poly(rC) binding protein 2 |
| chr8_-_71519889 | 8.72 |
ENST00000521425.1
|
TRAM1
|
translocation associated membrane protein 1 |
| chr1_+_155178518 | 8.19 |
ENST00000316721.4
|
MTX1
|
metaxin 1 |
| chr15_-_34394119 | 8.17 |
ENST00000256545.4
|
EMC7
|
ER membrane protein complex subunit 7 |
| chr12_-_109125285 | 7.94 |
ENST00000552871.1
ENST00000261401.3 |
CORO1C
|
coronin, actin binding protein, 1C |
| chr12_-_120907374 | 7.85 |
ENST00000550458.1
|
SRSF9
|
serine/arginine-rich splicing factor 9 |
| chr7_+_65552756 | 7.66 |
ENST00000450043.1
|
AC068533.7
|
AC068533.7 |
| chr10_+_51371390 | 7.64 |
ENST00000478381.1
ENST00000451577.2 ENST00000374098.2 ENST00000374097.2 |
TIMM23B
|
translocase of inner mitochondrial membrane 23 homolog B (yeast) |
| chr17_+_27071002 | 7.48 |
ENST00000262395.5
ENST00000422344.1 ENST00000444415.3 ENST00000262396.6 |
TRAF4
|
TNF receptor-associated factor 4 |
| chr17_-_38978847 | 7.46 |
ENST00000269576.5
|
KRT10
|
keratin 10 |
| chr17_-_79827808 | 7.38 |
ENST00000580685.1
|
ARHGDIA
|
Rho GDP dissociation inhibitor (GDI) alpha |
| chr1_+_153750622 | 7.34 |
ENST00000532853.1
|
SLC27A3
|
solute carrier family 27 (fatty acid transporter), member 3 |
| chr12_-_56120865 | 7.25 |
ENST00000548898.1
ENST00000552067.1 |
CD63
|
CD63 molecule |
| chr2_-_32236002 | 7.17 |
ENST00000404530.1
|
MEMO1
|
mediator of cell motility 1 |
| chr15_-_64673630 | 7.14 |
ENST00000558008.1
ENST00000559519.1 ENST00000380258.2 |
KIAA0101
|
KIAA0101 |
| chr15_-_34394008 | 6.88 |
ENST00000527822.1
ENST00000528949.1 |
EMC7
|
ER membrane protein complex subunit 7 |
| chr15_-_75230368 | 6.61 |
ENST00000564811.1
ENST00000562233.1 ENST00000567270.1 ENST00000568783.1 |
COX5A
|
cytochrome c oxidase subunit Va |
| chr12_-_120907459 | 6.57 |
ENST00000229390.3
|
SRSF9
|
serine/arginine-rich splicing factor 9 |
| chr9_-_127952032 | 6.51 |
ENST00000456642.1
ENST00000373546.3 ENST00000373547.4 |
PPP6C
|
protein phosphatase 6, catalytic subunit |
| chr1_+_32687971 | 6.41 |
ENST00000373586.1
|
EIF3I
|
eukaryotic translation initiation factor 3, subunit I |
| chr10_+_81272287 | 6.33 |
ENST00000520547.2
|
EIF5AL1
|
eukaryotic translation initiation factor 5A-like 1 |
| chr14_+_24605361 | 6.21 |
ENST00000206451.6
ENST00000559123.1 |
PSME1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr14_-_106174960 | 6.10 |
ENST00000390547.2
|
IGHA1
|
immunoglobulin heavy constant alpha 1 |
| chr11_-_46142948 | 6.09 |
ENST00000257821.4
|
PHF21A
|
PHD finger protein 21A |
| chr16_+_29819096 | 5.93 |
ENST00000568411.1
ENST00000563012.1 ENST00000562557.1 |
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr13_-_46716969 | 5.91 |
ENST00000435666.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr2_-_89459813 | 5.90 |
ENST00000390256.2
|
IGKV6-21
|
immunoglobulin kappa variable 6-21 (non-functional) |
| chr17_+_7210898 | 5.90 |
ENST00000572815.1
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr9_+_131445928 | 5.86 |
ENST00000372692.4
|
SET
|
SET nuclear oncogene |
| chr5_-_179227540 | 5.70 |
ENST00000520875.1
|
MGAT4B
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme B |
| chr8_-_101962777 | 5.64 |
ENST00000395951.3
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr1_+_24117693 | 5.62 |
ENST00000374503.3
ENST00000374502.3 |
LYPLA2
|
lysophospholipase II |
| chr20_-_49547731 | 5.62 |
ENST00000396029.3
|
ADNP
|
activity-dependent neuroprotector homeobox |
| chr9_-_127952187 | 5.46 |
ENST00000451402.1
ENST00000415905.1 |
PPP6C
|
protein phosphatase 6, catalytic subunit |
| chr9_-_139371533 | 5.43 |
ENST00000290037.6
ENST00000431893.2 ENST00000371706.3 |
SEC16A
|
SEC16 homolog A (S. cerevisiae) |
| chr1_+_26798955 | 5.37 |
ENST00000361427.5
|
HMGN2
|
high mobility group nucleosomal binding domain 2 |
| chr12_-_56122220 | 5.28 |
ENST00000552692.1
|
CD63
|
CD63 molecule |
| chr2_-_153573887 | 5.27 |
ENST00000493468.2
ENST00000545856.1 |
PRPF40A
|
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr17_-_79818354 | 5.27 |
ENST00000576541.1
ENST00000576380.1 ENST00000571617.1 ENST00000576052.1 ENST00000576390.1 ENST00000573778.2 ENST00000439918.2 ENST00000574914.1 ENST00000331483.4 |
P4HB
|
prolyl 4-hydroxylase, beta polypeptide |
| chr19_-_47349395 | 5.16 |
ENST00000597020.1
|
AP2S1
|
adaptor-related protein complex 2, sigma 1 subunit |
| chr2_-_220435963 | 5.14 |
ENST00000373876.1
ENST00000404537.1 ENST00000603926.1 ENST00000373873.4 ENST00000289656.3 |
OBSL1
|
obscurin-like 1 |
| chr7_-_140179276 | 5.10 |
ENST00000443720.2
ENST00000255977.2 |
MKRN1
|
makorin ring finger protein 1 |
| chr10_-_51623203 | 5.09 |
ENST00000444743.1
ENST00000374065.3 ENST00000374064.3 ENST00000260867.4 |
TIMM23
|
translocase of inner mitochondrial membrane 23 homolog (yeast) |
| chr12_-_56122124 | 5.04 |
ENST00000552754.1
|
CD63
|
CD63 molecule |
| chr15_-_75230478 | 5.02 |
ENST00000322347.6
|
COX5A
|
cytochrome c oxidase subunit Va |
| chr9_-_34637718 | 4.99 |
ENST00000378892.1
ENST00000277010.4 |
SIGMAR1
|
sigma non-opioid intracellular receptor 1 |
| chrX_-_107334750 | 4.97 |
ENST00000340200.5
ENST00000372296.1 ENST00000372295.1 ENST00000361815.5 |
PSMD10
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 10 |
| chr7_-_65447192 | 4.88 |
ENST00000421103.1
ENST00000345660.6 ENST00000304895.4 |
GUSB
|
glucuronidase, beta |
| chr16_+_103816 | 4.87 |
ENST00000383018.3
ENST00000417493.1 |
SNRNP25
|
small nuclear ribonucleoprotein 25kDa (U11/U12) |
| chr19_+_39214797 | 4.83 |
ENST00000440400.1
|
ACTN4
|
actinin, alpha 4 |
| chr2_+_90139056 | 4.80 |
ENST00000492446.1
|
IGKV1D-16
|
immunoglobulin kappa variable 1D-16 |
| chr12_-_56121580 | 4.77 |
ENST00000550776.1
|
CD63
|
CD63 molecule |
| chrX_-_107334790 | 4.70 |
ENST00000217958.3
|
PSMD10
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 10 |
| chr12_+_53848505 | 4.64 |
ENST00000552819.1
ENST00000455667.3 |
PCBP2
|
poly(rC) binding protein 2 |
| chr17_-_47841485 | 4.61 |
ENST00000506156.1
ENST00000240364.2 |
FAM117A
|
family with sequence similarity 117, member A |
| chr16_+_29818857 | 4.59 |
ENST00000567444.1
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr16_+_29819372 | 4.58 |
ENST00000568544.1
ENST00000569978.1 |
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr16_-_103572 | 4.58 |
ENST00000293860.5
|
POLR3K
|
polymerase (RNA) III (DNA directed) polypeptide K, 12.3 kDa |
| chrX_-_129299638 | 4.51 |
ENST00000535724.1
ENST00000346424.2 |
AIFM1
|
apoptosis-inducing factor, mitochondrion-associated, 1 |
| chr2_-_89568263 | 4.51 |
ENST00000473726.1
|
IGKV1-33
|
immunoglobulin kappa variable 1-33 |
| chr14_-_106054659 | 4.51 |
ENST00000390539.2
|
IGHA2
|
immunoglobulin heavy constant alpha 2 (A2m marker) |
| chrX_-_129299847 | 4.50 |
ENST00000319908.3
ENST00000287295.3 |
AIFM1
|
apoptosis-inducing factor, mitochondrion-associated, 1 |
| chr5_+_34656331 | 4.40 |
ENST00000265109.3
|
RAI14
|
retinoic acid induced 14 |
| chr11_-_1643368 | 4.38 |
ENST00000399682.1
|
KRTAP5-4
|
keratin associated protein 5-4 |
| chr1_+_156084461 | 4.37 |
ENST00000347559.2
ENST00000361308.4 ENST00000368300.4 ENST00000368299.3 |
LMNA
|
lamin A/C |
| chr1_-_244615425 | 4.35 |
ENST00000366535.3
|
ADSS
|
adenylosuccinate synthase |
| chr16_+_29819446 | 4.35 |
ENST00000568282.1
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr14_+_75348592 | 4.32 |
ENST00000334220.4
|
DLST
|
dihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) |
| chr17_+_8191815 | 4.31 |
ENST00000226105.6
ENST00000407006.4 ENST00000580434.1 ENST00000439238.3 |
RANGRF
|
RAN guanine nucleotide release factor |
| chr10_-_103880209 | 4.29 |
ENST00000425280.1
|
LDB1
|
LIM domain binding 1 |
| chr7_+_107220660 | 4.28 |
ENST00000465919.1
ENST00000445771.2 ENST00000479917.1 ENST00000421217.1 ENST00000457837.1 |
BCAP29
|
B-cell receptor-associated protein 29 |
| chr1_-_153538011 | 4.27 |
ENST00000368707.4
|
S100A2
|
S100 calcium binding protein A2 |
| chr7_+_107220899 | 4.26 |
ENST00000379117.2
ENST00000473124.1 |
BCAP29
|
B-cell receptor-associated protein 29 |
| chr2_-_89399845 | 4.26 |
ENST00000479981.1
|
IGKV1-16
|
immunoglobulin kappa variable 1-16 |
| chr2_+_89952792 | 4.25 |
ENST00000390265.2
|
IGKV1D-33
|
immunoglobulin kappa variable 1D-33 |
| chr19_+_19303720 | 4.24 |
ENST00000392324.4
|
RFXANK
|
regulatory factor X-associated ankyrin-containing protein |
| chr4_+_184426147 | 4.23 |
ENST00000302327.3
|
ING2
|
inhibitor of growth family, member 2 |
| chr19_-_39330818 | 4.17 |
ENST00000594769.1
ENST00000602021.1 |
AC104534.3
|
Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, mitochondrial |
| chr17_+_57784997 | 4.16 |
ENST00000537567.1
ENST00000539763.1 ENST00000587945.1 ENST00000536180.1 ENST00000589823.2 ENST00000592106.1 ENST00000591315.1 ENST00000545362.1 |
VMP1
|
vacuole membrane protein 1 |
| chr17_-_76899275 | 4.01 |
ENST00000322630.2
ENST00000586713.1 |
DDC8
|
Protein DDC8 homolog |
| chr16_-_20911641 | 3.96 |
ENST00000564349.1
ENST00000324344.4 |
ERI2
DCUN1D3
|
ERI1 exoribonuclease family member 2 DCN1, defective in cullin neddylation 1, domain containing 3 |
| chrX_+_47082408 | 3.95 |
ENST00000518022.1
ENST00000276052.6 |
CDK16
|
cyclin-dependent kinase 16 |
| chr5_+_34656569 | 3.92 |
ENST00000428746.2
|
RAI14
|
retinoic acid induced 14 |
| chr12_+_7060432 | 3.91 |
ENST00000318974.9
ENST00000456013.1 |
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr17_+_7155343 | 3.89 |
ENST00000573513.1
ENST00000354429.2 ENST00000574255.1 ENST00000396627.2 ENST00000356683.2 |
ELP5
|
elongator acetyltransferase complex subunit 5 |
| chr11_-_73689037 | 3.89 |
ENST00000544615.1
|
UCP2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr7_+_2394445 | 3.82 |
ENST00000360876.4
ENST00000413917.1 ENST00000397011.2 |
EIF3B
|
eukaryotic translation initiation factor 3, subunit B |
| chr15_+_74218787 | 3.81 |
ENST00000261921.7
|
LOXL1
|
lysyl oxidase-like 1 |
| chr2_+_99953816 | 3.79 |
ENST00000289371.6
|
EIF5B
|
eukaryotic translation initiation factor 5B |
| chr19_+_19303572 | 3.74 |
ENST00000407360.3
ENST00000540981.1 |
RFXANK
|
regulatory factor X-associated ankyrin-containing protein |
| chr2_-_153573965 | 3.71 |
ENST00000448428.1
|
PRPF40A
|
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr17_-_57784755 | 3.70 |
ENST00000537860.1
ENST00000393038.2 ENST00000409433.2 |
PTRH2
|
peptidyl-tRNA hydrolase 2 |
| chr12_-_46766577 | 3.67 |
ENST00000256689.5
|
SLC38A2
|
solute carrier family 38, member 2 |
| chr1_-_32403903 | 3.61 |
ENST00000344035.6
ENST00000356536.3 |
PTP4A2
|
protein tyrosine phosphatase type IVA, member 2 |
| chr20_-_49547910 | 3.56 |
ENST00000396032.3
|
ADNP
|
activity-dependent neuroprotector homeobox |
| chr21_-_38445011 | 3.54 |
ENST00000464265.1
ENST00000399102.1 |
PIGP
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr12_+_32832134 | 3.47 |
ENST00000452533.2
|
DNM1L
|
dynamin 1-like |
| chr7_-_140178726 | 3.47 |
ENST00000480552.1
|
MKRN1
|
makorin ring finger protein 1 |
| chr3_+_184037466 | 3.45 |
ENST00000441154.1
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr17_+_57784826 | 3.41 |
ENST00000262291.4
|
VMP1
|
vacuole membrane protein 1 |
| chr19_-_2050852 | 3.38 |
ENST00000541165.1
ENST00000591601.1 |
MKNK2
|
MAP kinase interacting serine/threonine kinase 2 |
| chr22_+_47016277 | 3.35 |
ENST00000406902.1
|
GRAMD4
|
GRAM domain containing 4 |
| chr1_+_24117627 | 3.34 |
ENST00000400061.1
|
LYPLA2
|
lysophospholipase II |
| chr3_-_52713729 | 3.34 |
ENST00000296302.7
ENST00000356770.4 ENST00000337303.4 ENST00000409057.1 ENST00000410007.1 ENST00000409114.3 ENST00000409767.1 ENST00000423351.1 |
PBRM1
|
polybromo 1 |
| chr17_+_7210921 | 3.28 |
ENST00000573542.1
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr21_-_38445297 | 3.26 |
ENST00000430792.1
ENST00000399103.1 |
PIGP
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr12_+_120907622 | 3.24 |
ENST00000392509.2
ENST00000549649.1 ENST00000548342.1 |
DYNLL1
|
dynein, light chain, LC8-type 1 |
| chr16_-_67969888 | 3.23 |
ENST00000574576.2
|
PSMB10
|
proteasome (prosome, macropain) subunit, beta type, 10 |
| chr20_-_1373682 | 3.23 |
ENST00000381724.3
|
FKBP1A
|
FK506 binding protein 1A, 12kDa |
| chr10_-_16859361 | 3.22 |
ENST00000377921.3
|
RSU1
|
Ras suppressor protein 1 |
| chr2_-_111291587 | 3.20 |
ENST00000437167.1
|
RGPD6
|
RANBP2-like and GRIP domain containing 6 |
| chr3_+_49058444 | 3.19 |
ENST00000326925.6
ENST00000395458.2 |
NDUFAF3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr6_-_114292284 | 3.16 |
ENST00000520895.1
ENST00000521163.1 ENST00000524334.1 ENST00000368632.2 ENST00000398283.2 |
HDAC2
|
histone deacetylase 2 |
| chr11_-_67120974 | 3.14 |
ENST00000539074.1
ENST00000312419.3 |
POLD4
|
polymerase (DNA-directed), delta 4, accessory subunit |
| chr16_+_75256507 | 3.13 |
ENST00000495583.1
|
CTRB1
|
chymotrypsinogen B1 |
| chr10_+_21823079 | 3.13 |
ENST00000377100.3
ENST00000377072.3 ENST00000446906.2 |
MLLT10
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
| chr11_+_118889456 | 3.11 |
ENST00000528230.1
ENST00000525303.1 ENST00000434101.2 ENST00000359005.4 ENST00000533058.1 |
TRAPPC4
|
trafficking protein particle complex 4 |
| chr2_-_27593180 | 3.09 |
ENST00000493344.2
ENST00000445933.2 |
EIF2B4
|
eukaryotic translation initiation factor 2B, subunit 4 delta, 67kDa |
| chr6_-_114292449 | 3.08 |
ENST00000519065.1
|
HDAC2
|
histone deacetylase 2 |
| chr17_-_36891830 | 3.06 |
ENST00000578487.1
|
PCGF2
|
polycomb group ring finger 2 |
| chr12_+_32832203 | 3.02 |
ENST00000553257.1
ENST00000549701.1 ENST00000358214.5 ENST00000266481.6 ENST00000551476.1 ENST00000550154.1 ENST00000547312.1 ENST00000414834.2 ENST00000381000.4 ENST00000548750.1 |
DNM1L
|
dynamin 1-like |
| chr3_-_52567792 | 2.99 |
ENST00000307092.4
ENST00000422318.2 ENST00000459839.1 |
NT5DC2
|
5'-nucleotidase domain containing 2 |
| chr3_+_148508845 | 2.94 |
ENST00000491148.1
|
CPB1
|
carboxypeptidase B1 (tissue) |
| chr9_-_34637806 | 2.92 |
ENST00000477726.1
|
SIGMAR1
|
sigma non-opioid intracellular receptor 1 |
| chr5_+_137688285 | 2.91 |
ENST00000314358.5
|
KDM3B
|
lysine (K)-specific demethylase 3B |
| chr14_+_102276209 | 2.86 |
ENST00000445439.3
ENST00000334743.5 ENST00000557095.1 |
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr1_-_202777535 | 2.86 |
ENST00000367264.2
|
KDM5B
|
lysine (K)-specific demethylase 5B |
| chr6_+_52535878 | 2.84 |
ENST00000211314.4
|
TMEM14A
|
transmembrane protein 14A |
| chrX_+_152953505 | 2.84 |
ENST00000253122.5
|
SLC6A8
|
solute carrier family 6 (neurotransmitter transporter), member 8 |
| chr16_+_85646891 | 2.76 |
ENST00000393243.1
|
GSE1
|
Gse1 coiled-coil protein |
| chr22_-_39268308 | 2.72 |
ENST00000407418.3
|
CBX6
|
chromobox homolog 6 |
| chr6_-_111804393 | 2.69 |
ENST00000368802.3
ENST00000368805.1 |
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr22_+_38093005 | 2.69 |
ENST00000406386.3
|
TRIOBP
|
TRIO and F-actin binding protein |
| chr15_-_64673665 | 2.65 |
ENST00000300035.4
|
KIAA0101
|
KIAA0101 |
| chr6_+_150070857 | 2.60 |
ENST00000544496.1
|
PCMT1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr17_-_7154984 | 2.59 |
ENST00000574322.1
|
CTDNEP1
|
CTD nuclear envelope phosphatase 1 |
| chr2_+_89901292 | 2.58 |
ENST00000448155.2
|
IGKV1D-39
|
immunoglobulin kappa variable 1D-39 |
| chr2_-_89340242 | 2.58 |
ENST00000480492.1
|
IGKV1-12
|
immunoglobulin kappa variable 1-12 |
| chr17_-_72864739 | 2.56 |
ENST00000579893.1
ENST00000544854.1 |
FDXR
|
ferredoxin reductase |
| chr3_-_148804275 | 2.49 |
ENST00000392912.2
ENST00000465259.1 ENST00000310053.5 ENST00000494055.1 |
HLTF
|
helicase-like transcription factor |
| chr18_+_19749386 | 2.47 |
ENST00000269216.3
|
GATA6
|
GATA binding protein 6 |
| chr2_+_90458201 | 2.47 |
ENST00000603238.1
|
CH17-132F21.1
|
Uncharacterized protein |
| chr1_-_31538517 | 2.47 |
ENST00000440538.2
ENST00000423018.2 ENST00000424085.2 ENST00000426105.2 ENST00000257075.5 ENST00000373747.3 ENST00000525843.1 ENST00000373742.2 |
PUM1
|
pumilio RNA-binding family member 1 |
| chr11_+_66360665 | 2.46 |
ENST00000310190.4
|
CCS
|
copper chaperone for superoxide dismutase |
| chrX_+_48644962 | 2.45 |
ENST00000376670.3
ENST00000376665.3 |
GATA1
|
GATA binding protein 1 (globin transcription factor 1) |
| chr16_-_70729496 | 2.41 |
ENST00000567648.1
|
VAC14
|
Vac14 homolog (S. cerevisiae) |
| chr19_-_8386238 | 2.38 |
ENST00000301457.2
|
NDUFA7
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kDa |
| chr1_-_212965104 | 2.35 |
ENST00000422588.2
ENST00000366975.6 ENST00000366977.3 ENST00000366976.1 |
NSL1
|
NSL1, MIS12 kinetochore complex component |
| chr2_+_89999259 | 2.32 |
ENST00000558026.1
|
IGKV2D-28
|
immunoglobulin kappa variable 2D-28 |
| chr16_+_31044413 | 2.30 |
ENST00000394998.1
|
STX4
|
syntaxin 4 |
| chr1_+_116915855 | 2.30 |
ENST00000295598.5
|
ATP1A1
|
ATPase, Na+/K+ transporting, alpha 1 polypeptide |
| chr1_-_6259641 | 2.29 |
ENST00000234875.4
|
RPL22
|
ribosomal protein L22 |
| chr7_+_142458507 | 2.28 |
ENST00000492062.1
|
PRSS1
|
protease, serine, 1 (trypsin 1) |
| chr2_-_89247338 | 2.27 |
ENST00000496168.1
|
IGKV1-5
|
immunoglobulin kappa variable 1-5 |
| chr16_+_30194916 | 2.26 |
ENST00000570045.1
ENST00000565497.1 ENST00000570244.1 |
CORO1A
|
coronin, actin binding protein, 1A |
| chr5_+_74633036 | 2.25 |
ENST00000343975.5
|
HMGCR
|
3-hydroxy-3-methylglutaryl-CoA reductase |
| chr7_+_100273736 | 2.25 |
ENST00000412215.1
ENST00000393924.1 |
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr17_-_8534031 | 2.21 |
ENST00000411957.1
ENST00000396239.1 ENST00000379980.4 |
MYH10
|
myosin, heavy chain 10, non-muscle |
| chr16_-_67978016 | 2.19 |
ENST00000264005.5
|
LCAT
|
lecithin-cholesterol acyltransferase |
| chr3_+_130569429 | 2.14 |
ENST00000505330.1
ENST00000504381.1 ENST00000507488.2 ENST00000393221.4 |
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr10_-_27149792 | 2.12 |
ENST00000376140.3
ENST00000376170.4 |
ABI1
|
abl-interactor 1 |
| chr3_+_33840050 | 2.10 |
ENST00000457054.2
ENST00000413073.1 |
PDCD6IP
|
programmed cell death 6 interacting protein |
| chr17_+_80477571 | 2.10 |
ENST00000335255.5
|
FOXK2
|
forkhead box K2 |
| chr17_-_8534067 | 2.07 |
ENST00000360416.3
ENST00000269243.4 |
MYH10
|
myosin, heavy chain 10, non-muscle |
| chr16_-_29875057 | 2.06 |
ENST00000219789.6
|
CDIPT
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase |
| chr3_+_193853927 | 2.05 |
ENST00000232424.3
|
HES1
|
hes family bHLH transcription factor 1 |
| chr14_-_75530693 | 1.98 |
ENST00000555135.1
ENST00000357971.3 ENST00000553302.1 ENST00000555694.1 ENST00000238618.3 |
ACYP1
|
acylphosphatase 1, erythrocyte (common) type |
| chr2_-_47168906 | 1.98 |
ENST00000444761.2
ENST00000409147.1 |
MCFD2
|
multiple coagulation factor deficiency 2 |
| chr1_-_155211017 | 1.97 |
ENST00000536770.1
ENST00000368373.3 |
GBA
|
glucosidase, beta, acid |
| chr17_+_7487146 | 1.95 |
ENST00000396501.4
ENST00000584378.1 ENST00000423172.2 ENST00000579445.1 ENST00000585217.1 ENST00000581380.1 |
MPDU1
|
mannose-P-dolichol utilization defect 1 |
| chr1_-_47655686 | 1.95 |
ENST00000294338.2
|
PDZK1IP1
|
PDZK1 interacting protein 1 |
| chrX_-_10544942 | 1.92 |
ENST00000380779.1
|
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr15_-_23034322 | 1.90 |
ENST00000539711.2
ENST00000560039.1 ENST00000398013.3 ENST00000337451.3 ENST00000359727.4 ENST00000398014.2 |
NIPA2
|
non imprinted in Prader-Willi/Angelman syndrome 2 |
| chr10_-_27149851 | 1.87 |
ENST00000376142.2
ENST00000359188.4 ENST00000376139.2 ENST00000376160.1 |
ABI1
|
abl-interactor 1 |
| chr22_-_39268192 | 1.87 |
ENST00000216083.6
|
CBX6
|
chromobox homolog 6 |
| chr9_-_117853297 | 1.87 |
ENST00000542877.1
ENST00000537320.1 ENST00000341037.4 |
TNC
|
tenascin C |
| chr5_+_52776228 | 1.79 |
ENST00000256759.3
|
FST
|
follistatin |
| chr6_-_30640811 | 1.78 |
ENST00000376442.3
|
DHX16
|
DEAH (Asp-Glu-Ala-His) box polypeptide 16 |
| chr1_-_114355083 | 1.78 |
ENST00000261441.5
|
RSBN1
|
round spermatid basic protein 1 |
| chr2_+_234104079 | 1.78 |
ENST00000417661.1
|
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa |
| chr16_+_85646763 | 1.77 |
ENST00000411612.1
ENST00000253458.7 |
GSE1
|
Gse1 coiled-coil protein |
| chr6_+_35995531 | 1.76 |
ENST00000229794.4
|
MAPK14
|
mitogen-activated protein kinase 14 |
| chr17_+_7211280 | 1.75 |
ENST00000419711.2
ENST00000571955.1 ENST00000573714.1 |
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr17_-_27418537 | 1.74 |
ENST00000408971.2
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1 |
| chr10_-_27149904 | 1.73 |
ENST00000376166.1
ENST00000376138.3 ENST00000355394.4 ENST00000346832.5 ENST00000376134.3 ENST00000376137.4 ENST00000536334.1 ENST00000490841.2 |
ABI1
|
abl-interactor 1 |
| chr17_+_4618734 | 1.68 |
ENST00000571206.1
|
ARRB2
|
arrestin, beta 2 |
| chr11_-_102401469 | 1.67 |
ENST00000260227.4
|
MMP7
|
matrix metallopeptidase 7 (matrilysin, uterine) |
| chr2_+_46769798 | 1.65 |
ENST00000238738.4
|
RHOQ
|
ras homolog family member Q |
| chr5_+_74632993 | 1.65 |
ENST00000287936.4
|
HMGCR
|
3-hydroxy-3-methylglutaryl-CoA reductase |
| chr1_+_214161272 | 1.65 |
ENST00000498508.2
ENST00000366958.4 |
PROX1
|
prospero homeobox 1 |
| chr4_-_185570590 | 1.65 |
ENST00000517513.1
ENST00000447121.2 ENST00000393588.4 ENST00000308394.4 ENST00000523916.1 |
CASP3
|
caspase 3, apoptosis-related cysteine peptidase |
Network of associatons between targets according to the STRING database.
First level regulatory network of KLF12
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 16.5 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 2.5 | 33.1 | GO:0035826 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 2.3 | 18.8 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 2.2 | 15.5 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 1.9 | 9.7 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 1.8 | 9.0 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 1.6 | 4.8 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 1.6 | 9.5 | GO:0002501 | peptide antigen assembly with MHC protein complex(GO:0002501) |
| 1.5 | 4.4 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 1.4 | 4.3 | GO:0019474 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 1.4 | 4.3 | GO:0021678 | fourth ventricle development(GO:0021592) third ventricle development(GO:0021678) |
| 1.3 | 9.2 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 1.2 | 6.2 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 1.2 | 4.9 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 1.2 | 3.6 | GO:1902568 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 1.2 | 23.0 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 1.1 | 4.3 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 1.1 | 2.1 | GO:2000977 | regulation of forebrain neuron differentiation(GO:2000977) |
| 1.1 | 10.6 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 1.0 | 17.6 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 1.0 | 2.0 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 1.0 | 2.9 | GO:2000861 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 0.9 | 4.4 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.9 | 8.5 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.8 | 12.7 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.8 | 4.2 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.8 | 2.5 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.8 | 2.5 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.8 | 2.5 | GO:0010725 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) |
| 0.8 | 7.9 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.7 | 4.3 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.7 | 2.1 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.7 | 9.4 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.7 | 1.3 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.7 | 5.3 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.6 | 7.5 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.6 | 1.9 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.6 | 3.7 | GO:0032328 | alanine transport(GO:0032328) |
| 0.6 | 1.8 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.6 | 2.3 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.6 | 11.6 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.5 | 2.2 | GO:0090107 | regulation of high-density lipoprotein particle assembly(GO:0090107) |
| 0.5 | 2.7 | GO:0030047 | actin modification(GO:0030047) |
| 0.5 | 2.6 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.5 | 2.0 | GO:1904457 | beta-glucoside metabolic process(GO:1901804) beta-glucoside catabolic process(GO:1901805) positive regulation of neuronal action potential(GO:1904457) |
| 0.5 | 3.9 | GO:0010666 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.5 | 3.9 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.5 | 14.4 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.5 | 4.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.4 | 2.6 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.4 | 1.2 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.4 | 5.8 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.4 | 5.5 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.4 | 3.5 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.4 | 5.6 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.4 | 5.9 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.4 | 2.9 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.3 | 1.4 | GO:0060263 | regulation of respiratory burst(GO:0060263) |
| 0.3 | 6.8 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.3 | 3.7 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.3 | 1.7 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.3 | 1.3 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.3 | 2.3 | GO:0031947 | negative regulation of glucocorticoid metabolic process(GO:0031944) negative regulation of glucocorticoid biosynthetic process(GO:0031947) negative regulation of steroid hormone biosynthetic process(GO:0090032) response to glycoside(GO:1903416) |
| 0.3 | 5.7 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.3 | 1.6 | GO:0003150 | muscular septum morphogenesis(GO:0003150) |
| 0.3 | 3.2 | GO:0035721 | intraciliary retrograde transport(GO:0035721) positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.3 | 1.5 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) |
| 0.3 | 5.2 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 0.3 | 7.2 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.3 | 1.6 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.3 | 1.1 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.3 | 13.0 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.3 | 2.8 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.2 | 19.4 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.2 | 4.0 | GO:0045116 | response to UV-C(GO:0010225) protein neddylation(GO:0045116) |
| 0.2 | 1.9 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.2 | 4.4 | GO:0060347 | heart trabecula formation(GO:0060347) |
| 0.2 | 4.6 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.2 | 1.5 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.2 | 1.5 | GO:0014066 | regulation of phosphatidylinositol 3-kinase signaling(GO:0014066) |
| 0.2 | 1.5 | GO:2001268 | positive regulation of keratinocyte migration(GO:0051549) negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.2 | 7.4 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.2 | 29.7 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.2 | 5.4 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.2 | 1.6 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.2 | 17.1 | GO:0048207 | vesicle coating(GO:0006901) vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.2 | 1.4 | GO:0002679 | respiratory burst involved in defense response(GO:0002679) |
| 0.2 | 5.7 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.2 | 4.9 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.2 | 2.9 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.2 | 2.9 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.2 | 1.3 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.2 | 1.3 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.2 | 3.4 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.2 | 1.0 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.2 | 0.8 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.2 | 1.3 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.2 | 23.4 | GO:0031424 | keratinization(GO:0031424) |
| 0.2 | 0.6 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.1 | 1.9 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 3.9 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.1 | 0.5 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 | 0.4 | GO:0035989 | tendon development(GO:0035989) |
| 0.1 | 0.6 | GO:0021562 | vestibulocochlear nerve development(GO:0021562) |
| 0.1 | 8.3 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.1 | 1.2 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.1 | 1.8 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 0.1 | 2.0 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 2.1 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 0.2 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.1 | 1.7 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.1 | 8.0 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.1 | 1.1 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.1 | 5.4 | GO:0031640 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) |
| 0.1 | 11.7 | GO:0006413 | translational initiation(GO:0006413) |
| 0.1 | 1.1 | GO:0006939 | smooth muscle contraction(GO:0006939) |
| 0.1 | 9.4 | GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) |
| 0.1 | 0.3 | GO:0035993 | subthalamic nucleus development(GO:0021763) deltoid tuberosity development(GO:0035993) prolactin secreting cell differentiation(GO:0060127) left lung development(GO:0060459) left lung morphogenesis(GO:0060460) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) cell proliferation involved in outflow tract morphogenesis(GO:0061325) |
| 0.1 | 0.3 | GO:0035565 | regulation of pronephros size(GO:0035565) renal glucose absorption(GO:0035623) |
| 0.1 | 0.5 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) basic amino acid transmembrane transport(GO:1990822) |
| 0.1 | 11.5 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.1 | 0.7 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.1 | 5.6 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.7 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 1.4 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 0.1 | GO:0070272 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.1 | 0.8 | GO:0060346 | bone trabecula formation(GO:0060346) bone trabecula morphogenesis(GO:0061430) |
| 0.1 | 0.8 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 0.6 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.1 | 9.3 | GO:0006626 | protein targeting to mitochondrion(GO:0006626) |
| 0.1 | 2.1 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.1 | 0.9 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.1 | 7.3 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.1 | 0.3 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 2.5 | GO:0035904 | aorta development(GO:0035904) |
| 0.0 | 1.7 | GO:0007249 | I-kappaB kinase/NF-kappaB signaling(GO:0007249) |
| 0.0 | 0.8 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 0.9 | GO:0002717 | positive regulation of natural killer cell mediated immunity(GO:0002717) positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.0 | 5.9 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.3 | GO:0070535 | negative regulation of histone ubiquitination(GO:0033183) histone H2A K63-linked ubiquitination(GO:0070535) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.6 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 1.3 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.8 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 2.7 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 1.7 | GO:0060135 | maternal process involved in female pregnancy(GO:0060135) |
| 0.0 | 0.5 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 1.1 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.8 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 2.7 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.2 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.9 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 1.3 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.0 | 1.9 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 1.1 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 2.3 | GO:0032886 | regulation of microtubule-based process(GO:0032886) |
| 0.0 | 0.3 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 0.4 | GO:0051930 | regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.0 | 0.8 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.1 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.5 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.0 | 0.1 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.1 | GO:2000121 | regulation of removal of superoxide radicals(GO:2000121) |
| 0.0 | 0.0 | GO:0031446 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) maintenance of mitochondrion location(GO:0051659) relaxation of skeletal muscle(GO:0090076) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 3.9 | GO:0030335 | positive regulation of cell migration(GO:0030335) |
| 0.0 | 0.3 | GO:0016311 | dephosphorylation(GO:0016311) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 10.6 | GO:0071746 | IgA immunoglobulin complex(GO:0071745) IgA immunoglobulin complex, circulating(GO:0071746) monomeric IgA immunoglobulin complex(GO:0071748) polymeric IgA immunoglobulin complex(GO:0071749) secretory IgA immunoglobulin complex(GO:0071751) |
| 2.8 | 33.1 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 1.8 | 5.3 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 1.7 | 15.1 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 1.6 | 9.4 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 1.3 | 6.4 | GO:1990393 | 3M complex(GO:1990393) |
| 1.2 | 9.5 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 1.1 | 10.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 1.1 | 12.7 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 1.0 | 12.5 | GO:0005642 | annulate lamellae(GO:0005642) |
| 1.0 | 6.2 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 1.0 | 2.9 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.9 | 4.4 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.9 | 4.3 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.8 | 11.6 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.8 | 3.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.7 | 6.8 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.7 | 5.4 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.6 | 4.3 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.6 | 7.9 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.5 | 9.7 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.5 | 4.4 | GO:0005638 | lamin filament(GO:0005638) |
| 0.5 | 2.4 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.5 | 3.3 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.5 | 3.6 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.4 | 4.9 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.4 | 1.3 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.4 | 10.5 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.4 | 3.9 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.4 | 8.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.4 | 5.7 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.4 | 3.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.4 | 16.5 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.4 | 9.0 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.4 | 1.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.3 | 15.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.3 | 5.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) endolysosome membrane(GO:0036020) |
| 0.3 | 2.7 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.3 | 3.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.2 | 7.9 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.2 | 4.9 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.2 | 7.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.2 | 29.3 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.2 | 4.6 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.2 | 2.1 | GO:0090543 | Flemming body(GO:0090543) |
| 0.2 | 1.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 3.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 7.5 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 0.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 1.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 2.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 24.6 | GO:0031968 | mitochondrial outer membrane(GO:0005741) organelle outer membrane(GO:0031968) |
| 0.1 | 22.8 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 1.3 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 3.3 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.1 | 0.9 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 1.6 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.1 | 4.4 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 2.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 4.2 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 3.2 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 5.7 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 0.4 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 6.3 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 0.8 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 3.9 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 7.2 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 3.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 2.8 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 0.9 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 2.4 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.8 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.8 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.5 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 1.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 2.7 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 2.0 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 1.4 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 3.5 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.7 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 10.9 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 1.2 | GO:0030532 | small nuclear ribonucleoprotein complex(GO:0030532) |
| 0.0 | 2.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 3.0 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 1.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.5 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 1.4 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 16.4 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.4 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.1 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 6.3 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 1.8 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 1.9 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 13.7 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 2.1 | 16.5 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 1.9 | 9.4 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 1.4 | 4.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 1.3 | 12.7 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 1.2 | 3.7 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 1.2 | 4.9 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 1.2 | 9.5 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 1.1 | 9.0 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 1.1 | 4.3 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 1.1 | 5.3 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.9 | 3.6 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.9 | 3.6 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.9 | 6.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.9 | 2.6 | GO:0016730 | ferredoxin-NADP+ reductase activity(GO:0004324) NADPH-adrenodoxin reductase activity(GO:0015039) oxidoreductase activity, acting on iron-sulfur proteins as donors(GO:0016730) oxidoreductase activity, acting on iron-sulfur proteins as donors, NAD or NADP as acceptor(GO:0016731) |
| 0.8 | 5.7 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.8 | 23.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.7 | 20.1 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.7 | 7.4 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.7 | 2.9 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.7 | 2.1 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.7 | 6.8 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.6 | 2.5 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.6 | 4.3 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.6 | 8.6 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.6 | 9.0 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.6 | 1.8 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.6 | 1.7 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.6 | 3.9 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.6 | 1.7 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.5 | 2.2 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.5 | 7.5 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.5 | 2.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.5 | 7.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.5 | 2.0 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.4 | 6.2 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.4 | 5.8 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.4 | 11.6 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.4 | 4.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.4 | 4.4 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.3 | 6.1 | GO:0048185 | activin binding(GO:0048185) |
| 0.3 | 15.7 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.3 | 3.6 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.3 | 2.0 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.3 | 6.5 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.3 | 22.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.3 | 10.6 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.3 | 9.0 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.3 | 3.0 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.3 | 4.4 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.3 | 1.9 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.3 | 2.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.2 | 5.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 5.2 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.2 | 3.6 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.2 | 2.7 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.2 | 23.2 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.2 | 4.0 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.2 | 0.6 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.2 | 4.6 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.2 | 3.2 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.2 | 2.3 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.2 | 4.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 43.5 | GO:0003823 | antigen binding(GO:0003823) |
| 0.2 | 3.4 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.2 | 2.3 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.2 | 8.2 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.2 | 0.7 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.2 | 0.8 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.2 | 3.9 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.2 | 3.9 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 0.8 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 1.9 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 11.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 8.9 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 0.8 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 1.1 | GO:0015278 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.1 | 1.6 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.1 | 4.8 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.1 | 0.7 | GO:0038049 | transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) |
| 0.1 | 2.4 | GO:0072349 | modified amino acid transmembrane transporter activity(GO:0072349) |
| 0.1 | 3.0 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 0.6 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 3.2 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.6 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 2.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 2.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 4.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 2.1 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 1.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 3.0 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 1.2 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.1 | 2.1 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 3.9 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 1.3 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 1.3 | GO:0046961 | hydrogen-exporting ATPase activity(GO:0036442) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 1.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 15.1 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 1.2 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.9 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.5 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.4 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 2.4 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.8 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.4 | GO:0016773 | phosphotransferase activity, alcohol group as acceptor(GO:0016773) |
| 0.0 | 0.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 2.2 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 1.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 1.5 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 3.2 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 5.4 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 5.2 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 1.8 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 3.6 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 1.6 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.4 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.5 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 1.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 4.5 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 21.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.3 | 10.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.3 | 3.6 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.2 | 15.4 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.2 | 4.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.2 | 10.4 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.2 | 6.5 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.2 | 14.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 3.0 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 6.7 | PID EPO PATHWAY | EPO signaling pathway |
| 0.1 | 5.7 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 2.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 9.0 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 8.6 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 3.4 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 0.9 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 11.9 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 3.3 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 4.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 5.9 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 1.5 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.1 | 3.1 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 5.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 1.9 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 1.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 1.4 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 1.6 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.4 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.7 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 1.3 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 2.5 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.6 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 1.1 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.6 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 1.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.9 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.5 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.4 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.9 | PID P73PATHWAY | p73 transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 23.0 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.7 | 9.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.4 | 23.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.3 | 4.9 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.3 | 14.4 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.3 | 12.0 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.3 | 8.2 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.3 | 5.7 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.3 | 6.8 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.3 | 41.6 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.3 | 3.1 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.3 | 19.1 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.3 | 4.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.3 | 5.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 12.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.2 | 2.4 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.2 | 5.7 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.2 | 5.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.2 | 4.4 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.2 | 4.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.2 | 6.9 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 4.6 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.2 | 0.7 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 0.2 | 14.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.2 | 3.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.2 | 3.5 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.2 | 3.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.2 | 16.2 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.2 | 2.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.2 | 6.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 10.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 21.5 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.1 | 3.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 2.1 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 11.6 | REACTOME APOPTOTIC EXECUTION PHASE | Genes involved in Apoptotic execution phase |
| 0.1 | 1.7 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 4.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 2.1 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 2.0 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 2.0 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.9 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 6.4 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.9 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.4 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 2.5 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.7 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 3.7 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 1.1 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 1.0 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 1.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.6 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 1.2 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |