Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for KLF7
Z-value: 0.41
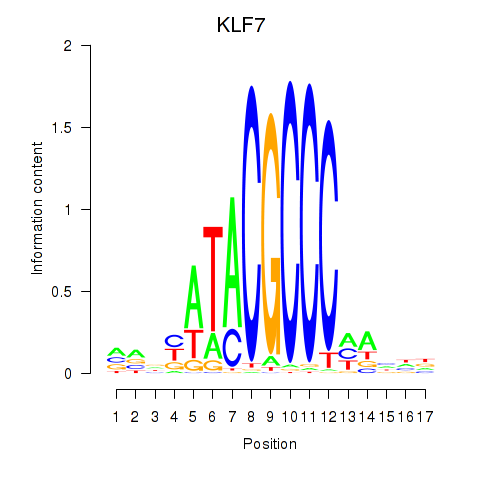
Transcription factors associated with KLF7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF7
|
ENSG00000118263.10 | Kruppel like factor 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| KLF7 | hg19_v2_chr2_-_208030647_208030689, hg19_v2_chr2_-_208031943_208031991 | 0.32 | 2.0e-06 | Click! |
Activity profile of KLF7 motif
Sorted Z-values of KLF7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_39390440 | 7.22 |
ENST00000249396.7
ENST00000414941.1 ENST00000392081.2 |
SIRT2
|
sirtuin 2 |
| chr1_-_19229014 | 6.74 |
ENST00000538839.1
ENST00000290597.5 |
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr16_+_31539197 | 6.70 |
ENST00000564707.1
|
AHSP
|
alpha hemoglobin stabilizing protein |
| chr1_-_19229248 | 6.69 |
ENST00000375341.3
|
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr19_-_39390350 | 6.60 |
ENST00000447739.1
ENST00000358931.5 ENST00000407552.1 |
SIRT2
|
sirtuin 2 |
| chr17_-_18218237 | 6.28 |
ENST00000542570.1
|
TOP3A
|
topoisomerase (DNA) III alpha |
| chr15_+_74422585 | 5.93 |
ENST00000561740.1
ENST00000435464.1 ENST00000453268.2 |
ISLR2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr1_-_21948906 | 5.61 |
ENST00000374761.2
ENST00000599760.1 |
RAP1GAP
|
RAP1 GTPase activating protein |
| chr16_+_31539183 | 5.54 |
ENST00000302312.4
|
AHSP
|
alpha hemoglobin stabilizing protein |
| chr1_-_157108266 | 5.15 |
ENST00000326786.4
|
ETV3
|
ets variant 3 |
| chr8_-_6837602 | 4.60 |
ENST00000382692.2
|
DEFA1
|
defensin, alpha 1 |
| chr6_-_33239712 | 4.50 |
ENST00000436044.2
|
VPS52
|
vacuolar protein sorting 52 homolog (S. cerevisiae) |
| chr8_-_6875778 | 4.07 |
ENST00000535841.1
ENST00000327857.2 |
DEFA1B
DEFA3
|
defensin, alpha 1B defensin, alpha 3, neutrophil-specific |
| chr6_+_31865552 | 3.78 |
ENST00000469372.1
ENST00000497706.1 |
C2
|
complement component 2 |
| chrX_-_92928557 | 3.71 |
ENST00000373079.3
ENST00000475430.2 |
NAP1L3
|
nucleosome assembly protein 1-like 3 |
| chr2_+_67624430 | 3.69 |
ENST00000272342.5
|
ETAA1
|
Ewing tumor-associated antigen 1 |
| chr15_+_92937144 | 3.39 |
ENST00000539113.1
ENST00000555434.1 |
ST8SIA2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
| chr17_-_18218270 | 3.23 |
ENST00000321105.5
|
TOP3A
|
topoisomerase (DNA) III alpha |
| chrX_+_102631844 | 2.91 |
ENST00000372634.1
ENST00000299872.7 |
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1 |
| chr5_+_176560007 | 2.77 |
ENST00000510954.1
ENST00000354179.4 |
NSD1
|
nuclear receptor binding SET domain protein 1 |
| chr15_+_92937058 | 2.56 |
ENST00000268164.3
|
ST8SIA2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
| chr14_+_29234870 | 2.49 |
ENST00000382535.3
|
FOXG1
|
forkhead box G1 |
| chr6_-_33239612 | 2.31 |
ENST00000482399.1
ENST00000445902.2 |
VPS52
|
vacuolar protein sorting 52 homolog (S. cerevisiae) |
| chr1_-_161102421 | 2.19 |
ENST00000490843.2
ENST00000368006.3 ENST00000392188.1 ENST00000545495.1 |
DEDD
|
death effector domain containing |
| chr1_-_161102367 | 2.17 |
ENST00000464113.1
|
DEDD
|
death effector domain containing |
| chr2_-_188430478 | 2.10 |
ENST00000421427.1
|
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr17_-_19281203 | 1.78 |
ENST00000487415.2
|
B9D1
|
B9 protein domain 1 |
| chr19_+_39390320 | 1.61 |
ENST00000576510.1
|
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr7_+_49813255 | 1.61 |
ENST00000340652.4
|
VWC2
|
von Willebrand factor C domain containing 2 |
| chr19_+_39390587 | 1.50 |
ENST00000572515.1
ENST00000392079.3 ENST00000575359.1 ENST00000313582.5 |
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr3_-_105587879 | 1.47 |
ENST00000264122.4
ENST00000403724.1 ENST00000405772.1 |
CBLB
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr2_-_74710078 | 1.25 |
ENST00000290418.4
|
CCDC142
|
coiled-coil domain containing 142 |
| chr6_-_30585009 | 1.24 |
ENST00000376511.2
|
PPP1R10
|
protein phosphatase 1, regulatory subunit 10 |
| chr5_+_141348598 | 1.02 |
ENST00000394520.2
ENST00000347642.3 |
RNF14
|
ring finger protein 14 |
| chr19_-_7694417 | 0.82 |
ENST00000358368.4
ENST00000534844.1 |
XAB2
|
XPA binding protein 2 |
| chr17_+_41476327 | 0.78 |
ENST00000320033.4
|
ARL4D
|
ADP-ribosylation factor-like 4D |
| chr5_+_141348721 | 0.54 |
ENST00000507163.1
ENST00000394519.1 |
RNF14
|
ring finger protein 14 |
| chr20_+_2821340 | 0.52 |
ENST00000380445.3
ENST00000380469.3 |
VPS16
|
vacuolar protein sorting 16 homolog (S. cerevisiae) |
| chr19_-_14629224 | 0.35 |
ENST00000254322.2
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr9_+_130565147 | 0.02 |
ENST00000373247.2
ENST00000373245.1 ENST00000393706.2 ENST00000373228.1 |
FPGS
|
folylpolyglutamate synthase |
| chr13_-_108867101 | 0.01 |
ENST00000356922.4
|
LIG4
|
ligase IV, DNA, ATP-dependent |
Network of associatons between targets according to the STRING database.
First level regulatory network of KLF7
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 13.8 | GO:2000777 | positive regulation of oocyte maturation(GO:1900195) negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 3.4 | 13.4 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 2.3 | 6.8 | GO:0048611 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 1.9 | 5.6 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 1.5 | 4.6 | GO:0035915 | pore formation in membrane of other organism(GO:0035915) |
| 0.7 | 6.0 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.6 | 12.2 | GO:0020027 | hemoglobin metabolic process(GO:0020027) |
| 0.6 | 9.5 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.4 | 2.8 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.4 | 5.2 | GO:0097012 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.3 | 3.8 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.3 | 2.1 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.2 | 4.1 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.2 | 3.7 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 0.8 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.1 | 5.9 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 0.5 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 1.6 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 | 1.6 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 4.4 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 0.3 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 3.7 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 1.8 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.0 | 0.4 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 3.1 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 6.8 | GO:1990745 | EARP complex(GO:1990745) |
| 1.5 | 13.8 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.9 | 12.2 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.2 | 0.8 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.2 | 5.2 | GO:0090568 | nuclear transcriptional repressor complex(GO:0090568) |
| 0.1 | 1.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 1.8 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 1.2 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 3.7 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.1 | 9.5 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 8.7 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.5 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 10.9 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 3.0 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 2.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 13.8 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 2.0 | 12.2 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 1.6 | 9.5 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.9 | 2.8 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.9 | 6.0 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.6 | 13.4 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.3 | 5.2 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.2 | 1.8 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.1 | 3.7 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 6.8 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 1.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 2.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 5.6 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 3.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 13.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.2 | 9.5 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 7.5 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 2.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 2.7 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 2.5 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.3 | PID CD40 PATHWAY | CD40/CD40L signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 8.7 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.2 | 6.0 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.2 | 3.8 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 5.6 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 9.5 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 3.1 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 2.1 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 13.4 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 2.9 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.0 | 0.8 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |