Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
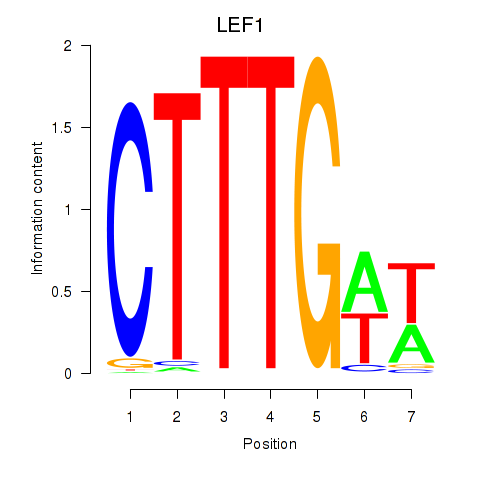
Results for LEF1
Z-value: 0.89
Transcription factors associated with LEF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
LEF1
|
ENSG00000138795.5 | lymphoid enhancer binding factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| LEF1 | hg19_v2_chr4_-_109089573_109089585 | -0.22 | 9.4e-04 | Click! |
Activity profile of LEF1 motif
Sorted Z-values of LEF1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_54074033 | 18.25 |
ENST00000373970.3
|
DKK1
|
dickkopf WNT signaling pathway inhibitor 1 |
| chr15_+_96869165 | 14.15 |
ENST00000421109.2
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr20_+_11898507 | 12.12 |
ENST00000378226.2
|
BTBD3
|
BTB (POZ) domain containing 3 |
| chr11_+_69455855 | 10.57 |
ENST00000227507.2
ENST00000536559.1 |
CCND1
|
cyclin D1 |
| chr3_-_134093395 | 9.84 |
ENST00000249883.5
|
AMOTL2
|
angiomotin like 2 |
| chr15_-_37392086 | 9.77 |
ENST00000561208.1
|
MEIS2
|
Meis homeobox 2 |
| chr16_+_89989687 | 8.92 |
ENST00000315491.7
ENST00000555576.1 ENST00000554336.1 ENST00000553967.1 |
TUBB3
|
Tubulin beta-3 chain |
| chr14_+_54863739 | 8.80 |
ENST00000541304.1
|
CDKN3
|
cyclin-dependent kinase inhibitor 3 |
| chr14_+_54863667 | 8.55 |
ENST00000335183.6
|
CDKN3
|
cyclin-dependent kinase inhibitor 3 |
| chr14_+_54863682 | 8.23 |
ENST00000543789.2
ENST00000442975.2 ENST00000458126.2 ENST00000556102.2 |
CDKN3
|
cyclin-dependent kinase inhibitor 3 |
| chr5_+_137514834 | 8.16 |
ENST00000508792.1
ENST00000504621.1 |
KIF20A
|
kinesin family member 20A |
| chr13_+_76334567 | 8.02 |
ENST00000321797.8
|
LMO7
|
LIM domain 7 |
| chr5_+_137514687 | 8.00 |
ENST00000394894.3
|
KIF20A
|
kinesin family member 20A |
| chr13_+_76334795 | 7.98 |
ENST00000526202.1
ENST00000465261.2 |
LMO7
|
LIM domain 7 |
| chr2_+_181845843 | 7.91 |
ENST00000602710.1
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr13_-_45150392 | 7.76 |
ENST00000501704.2
|
TSC22D1
|
TSC22 domain family, member 1 |
| chr3_-_134093275 | 7.58 |
ENST00000513145.1
ENST00000422605.2 |
AMOTL2
|
angiomotin like 2 |
| chr12_-_118498958 | 7.44 |
ENST00000315436.3
|
WSB2
|
WD repeat and SOCS box containing 2 |
| chr7_+_16793160 | 7.34 |
ENST00000262067.4
|
TSPAN13
|
tetraspanin 13 |
| chr3_-_149375783 | 7.25 |
ENST00000467467.1
ENST00000460517.1 ENST00000360632.3 |
WWTR1
|
WW domain containing transcription regulator 1 |
| chr2_+_201170596 | 6.93 |
ENST00000439084.1
ENST00000409718.1 |
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr22_-_36357671 | 6.91 |
ENST00000408983.2
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr5_-_16936340 | 6.80 |
ENST00000507288.1
ENST00000513610.1 |
MYO10
|
myosin X |
| chr2_+_201170770 | 6.76 |
ENST00000409988.3
ENST00000409385.1 |
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr11_-_62474803 | 6.59 |
ENST00000533982.1
ENST00000360796.5 |
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr5_+_34757309 | 6.38 |
ENST00000397449.1
|
RAI14
|
retinoic acid induced 14 |
| chrX_-_10588459 | 6.32 |
ENST00000380782.2
|
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr11_-_64013663 | 6.28 |
ENST00000392210.2
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr8_-_80993010 | 6.16 |
ENST00000537855.1
ENST00000520527.1 ENST00000517427.1 ENST00000448733.2 ENST00000379097.3 |
TPD52
|
tumor protein D52 |
| chr2_+_201171242 | 6.11 |
ENST00000360760.5
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr2_-_166060552 | 6.06 |
ENST00000283254.7
ENST00000453007.1 |
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit |
| chr1_+_93913713 | 6.06 |
ENST00000604705.1
ENST00000370253.2 |
FNBP1L
|
formin binding protein 1-like |
| chr11_-_64013288 | 5.97 |
ENST00000542235.1
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr4_-_157892498 | 5.95 |
ENST00000502773.1
|
PDGFC
|
platelet derived growth factor C |
| chr5_+_140864649 | 5.93 |
ENST00000306593.1
|
PCDHGC4
|
protocadherin gamma subfamily C, 4 |
| chr11_+_101983176 | 5.93 |
ENST00000524575.1
|
YAP1
|
Yes-associated protein 1 |
| chr9_+_71819927 | 5.86 |
ENST00000535702.1
|
TJP2
|
tight junction protein 2 |
| chr13_-_36429763 | 5.73 |
ENST00000379893.1
|
DCLK1
|
doublecortin-like kinase 1 |
| chrX_-_10588595 | 5.71 |
ENST00000423614.1
ENST00000317552.4 |
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr12_-_16759711 | 5.69 |
ENST00000447609.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr3_-_33700933 | 5.62 |
ENST00000480013.1
|
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr15_-_37393406 | 5.62 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr3_-_33686743 | 5.62 |
ENST00000333778.6
ENST00000539981.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chrX_+_102631844 | 5.60 |
ENST00000372634.1
ENST00000299872.7 |
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1 |
| chr12_+_6309963 | 5.46 |
ENST00000382515.2
|
CD9
|
CD9 molecule |
| chr2_-_216257849 | 5.45 |
ENST00000456923.1
|
FN1
|
fibronectin 1 |
| chr4_-_139163491 | 5.44 |
ENST00000280612.5
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
| chr1_-_95392635 | 5.38 |
ENST00000538964.1
ENST00000394202.4 ENST00000370206.4 |
CNN3
|
calponin 3, acidic |
| chr15_+_43985725 | 5.36 |
ENST00000413453.2
|
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr12_+_69979113 | 5.35 |
ENST00000299300.6
|
CCT2
|
chaperonin containing TCP1, subunit 2 (beta) |
| chr5_+_140797296 | 5.32 |
ENST00000398594.2
|
PCDHGB7
|
protocadherin gamma subfamily B, 7 |
| chr15_+_57511609 | 5.29 |
ENST00000543579.1
ENST00000537840.1 ENST00000343827.3 |
TCF12
|
transcription factor 12 |
| chr1_-_225840747 | 5.27 |
ENST00000366843.2
ENST00000366844.3 |
ENAH
|
enabled homolog (Drosophila) |
| chr4_-_111119804 | 5.18 |
ENST00000394607.3
ENST00000302274.3 |
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr20_+_10199468 | 5.18 |
ENST00000254976.2
ENST00000304886.2 |
SNAP25
|
synaptosomal-associated protein, 25kDa |
| chr1_-_32801825 | 5.14 |
ENST00000329421.7
|
MARCKSL1
|
MARCKS-like 1 |
| chr1_-_54303934 | 5.03 |
ENST00000537333.1
|
NDC1
|
NDC1 transmembrane nucleoporin |
| chr5_-_146833485 | 4.91 |
ENST00000398514.3
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr2_+_201171372 | 4.90 |
ENST00000409140.3
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr17_-_46682321 | 4.85 |
ENST00000225648.3
ENST00000484302.2 |
HOXB6
|
homeobox B6 |
| chr2_-_166060571 | 4.85 |
ENST00000360093.3
|
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit |
| chr8_+_80523321 | 4.80 |
ENST00000518111.1
|
STMN2
|
stathmin-like 2 |
| chr9_+_71820057 | 4.80 |
ENST00000539225.1
|
TJP2
|
tight junction protein 2 |
| chrX_+_38420783 | 4.79 |
ENST00000422612.2
ENST00000286824.6 ENST00000545599.1 |
TSPAN7
|
tetraspanin 7 |
| chr15_+_52311398 | 4.78 |
ENST00000261845.5
|
MAPK6
|
mitogen-activated protein kinase 6 |
| chr17_-_77924627 | 4.77 |
ENST00000572862.1
ENST00000573782.1 ENST00000574427.1 ENST00000570373.1 ENST00000340848.7 ENST00000576768.1 |
TBC1D16
|
TBC1 domain family, member 16 |
| chr3_-_141747439 | 4.69 |
ENST00000467667.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr10_-_95242044 | 4.68 |
ENST00000371501.4
ENST00000371502.4 ENST00000371489.1 |
MYOF
|
myoferlin |
| chr12_+_53491220 | 4.64 |
ENST00000548547.1
ENST00000301464.3 |
IGFBP6
|
insulin-like growth factor binding protein 6 |
| chr15_+_43885252 | 4.58 |
ENST00000453782.1
ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr1_-_54304212 | 4.58 |
ENST00000540001.1
|
NDC1
|
NDC1 transmembrane nucleoporin |
| chr10_-_95241951 | 4.57 |
ENST00000358334.5
ENST00000359263.4 ENST00000371488.3 |
MYOF
|
myoferlin |
| chr15_+_43985084 | 4.56 |
ENST00000434505.1
ENST00000411750.1 |
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr1_-_177133818 | 4.56 |
ENST00000424564.2
ENST00000361833.2 |
ASTN1
|
astrotactin 1 |
| chr4_-_74088800 | 4.56 |
ENST00000509867.2
|
ANKRD17
|
ankyrin repeat domain 17 |
| chr3_-_148804275 | 4.55 |
ENST00000392912.2
ENST00000465259.1 ENST00000310053.5 ENST00000494055.1 |
HLTF
|
helicase-like transcription factor |
| chr18_-_74728998 | 4.55 |
ENST00000359645.3
ENST00000397875.3 ENST00000397869.3 ENST00000578193.1 ENST00000578873.1 ENST00000397866.4 ENST00000528160.1 ENST00000527041.1 ENST00000526111.1 ENST00000397865.5 ENST00000382582.3 |
MBP
|
myelin basic protein |
| chr18_-_25616519 | 4.44 |
ENST00000399380.3
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr7_+_73868439 | 4.42 |
ENST00000424337.2
|
GTF2IRD1
|
GTF2I repeat domain containing 1 |
| chr2_-_86564776 | 4.41 |
ENST00000165698.5
ENST00000541910.1 ENST00000535845.1 |
REEP1
|
receptor accessory protein 1 |
| chr2_+_234621551 | 4.39 |
ENST00000608381.1
ENST00000373414.3 |
UGT1A1
UGT1A5
|
UDP glucuronosyltransferase 1 family, polypeptide A8 UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr15_-_37391614 | 4.38 |
ENST00000219869.9
|
MEIS2
|
Meis homeobox 2 |
| chr4_+_146403912 | 4.37 |
ENST00000507367.1
ENST00000394092.2 ENST00000515385.1 |
SMAD1
|
SMAD family member 1 |
| chr17_-_43025005 | 4.30 |
ENST00000587309.1
ENST00000593135.1 ENST00000339151.4 |
KIF18B
|
kinesin family member 18B |
| chr1_-_177134024 | 4.22 |
ENST00000367654.3
|
ASTN1
|
astrotactin 1 |
| chrX_-_80457385 | 4.21 |
ENST00000451455.1
ENST00000436386.1 ENST00000358130.2 |
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr1_+_166808692 | 4.21 |
ENST00000367876.4
|
POGK
|
pogo transposable element with KRAB domain |
| chr18_+_3449821 | 4.21 |
ENST00000407501.2
ENST00000405385.3 ENST00000546979.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr3_-_58563094 | 4.18 |
ENST00000464064.1
|
FAM107A
|
family with sequence similarity 107, member A |
| chr7_-_99698338 | 4.14 |
ENST00000354230.3
ENST00000425308.1 |
MCM7
|
minichromosome maintenance complex component 7 |
| chr12_+_79258547 | 4.13 |
ENST00000457153.2
|
SYT1
|
synaptotagmin I |
| chr14_+_29234870 | 4.12 |
ENST00000382535.3
|
FOXG1
|
forkhead box G1 |
| chr6_-_56707943 | 4.09 |
ENST00000370769.4
ENST00000421834.2 ENST00000312431.6 ENST00000361203.3 ENST00000523817.1 |
DST
|
dystonin |
| chr1_+_68150744 | 4.09 |
ENST00000370986.4
ENST00000370985.3 |
GADD45A
|
growth arrest and DNA-damage-inducible, alpha |
| chr1_+_82266053 | 4.00 |
ENST00000370715.1
ENST00000370713.1 ENST00000319517.6 ENST00000370717.2 ENST00000394879.1 ENST00000271029.4 ENST00000335786.5 |
LPHN2
|
latrophilin 2 |
| chr1_-_54303949 | 3.98 |
ENST00000234725.8
|
NDC1
|
NDC1 transmembrane nucleoporin |
| chrX_-_10645773 | 3.97 |
ENST00000453318.2
|
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr2_-_2334888 | 3.92 |
ENST00000428368.2
ENST00000399161.2 |
MYT1L
|
myelin transcription factor 1-like |
| chr15_+_63335899 | 3.89 |
ENST00000561266.1
|
TPM1
|
tropomyosin 1 (alpha) |
| chr8_-_22089845 | 3.89 |
ENST00000454243.2
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein |
| chr14_-_105444694 | 3.86 |
ENST00000333244.5
|
AHNAK2
|
AHNAK nucleoprotein 2 |
| chr7_-_80548667 | 3.86 |
ENST00000265361.3
|
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr8_-_22089533 | 3.86 |
ENST00000321613.3
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein |
| chr8_+_85095769 | 3.79 |
ENST00000518566.1
|
RALYL
|
RALY RNA binding protein-like |
| chr13_-_67802549 | 3.78 |
ENST00000328454.5
ENST00000377865.2 |
PCDH9
|
protocadherin 9 |
| chr1_+_87797351 | 3.75 |
ENST00000370542.1
|
LMO4
|
LIM domain only 4 |
| chr5_+_140749803 | 3.71 |
ENST00000576222.1
|
PCDHGB3
|
protocadherin gamma subfamily B, 3 |
| chr16_+_68678892 | 3.70 |
ENST00000429102.2
|
CDH3
|
cadherin 3, type 1, P-cadherin (placental) |
| chr3_-_79816965 | 3.67 |
ENST00000464233.1
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr1_+_164528866 | 3.65 |
ENST00000420696.2
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr1_-_38061522 | 3.62 |
ENST00000373062.3
|
GNL2
|
guanine nucleotide binding protein-like 2 (nucleolar) |
| chr8_-_145642267 | 3.56 |
ENST00000301305.3
|
SLC39A4
|
solute carrier family 39 (zinc transporter), member 4 |
| chr7_-_27213893 | 3.51 |
ENST00000283921.4
|
HOXA10
|
homeobox A10 |
| chr1_+_6845384 | 3.51 |
ENST00000303635.7
|
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr7_+_73868220 | 3.50 |
ENST00000455841.2
|
GTF2IRD1
|
GTF2I repeat domain containing 1 |
| chr7_-_27219849 | 3.44 |
ENST00000396344.4
|
HOXA10
|
homeobox A10 |
| chr10_+_114710211 | 3.43 |
ENST00000349937.2
ENST00000369397.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr2_+_201171064 | 3.42 |
ENST00000451764.2
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr16_+_6069586 | 3.41 |
ENST00000547372.1
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr9_+_504674 | 3.34 |
ENST00000382297.2
|
KANK1
|
KN motif and ankyrin repeat domains 1 |
| chr6_+_32146131 | 3.33 |
ENST00000375094.3
|
RNF5
|
ring finger protein 5, E3 ubiquitin protein ligase |
| chr1_-_115292591 | 3.22 |
ENST00000438362.2
|
CSDE1
|
cold shock domain containing E1, RNA-binding |
| chr14_+_101299520 | 3.19 |
ENST00000455531.1
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr16_+_68678739 | 3.16 |
ENST00000264012.4
|
CDH3
|
cadherin 3, type 1, P-cadherin (placental) |
| chr16_+_6069072 | 3.15 |
ENST00000547605.1
ENST00000550418.1 ENST00000553186.1 |
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr11_-_57298187 | 3.15 |
ENST00000525158.1
ENST00000257245.4 ENST00000525587.1 |
TIMM10
|
translocase of inner mitochondrial membrane 10 homolog (yeast) |
| chr1_-_211752073 | 3.10 |
ENST00000367001.4
|
SLC30A1
|
solute carrier family 30 (zinc transporter), member 1 |
| chr5_+_112074029 | 3.09 |
ENST00000512211.2
|
APC
|
adenomatous polyposis coli |
| chr6_+_114178512 | 3.08 |
ENST00000368635.4
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate |
| chr18_+_55888767 | 3.05 |
ENST00000431212.2
ENST00000586268.1 ENST00000587190.1 |
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr14_+_56078695 | 3.01 |
ENST00000416613.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr5_+_140729649 | 3.01 |
ENST00000523390.1
|
PCDHGB1
|
protocadherin gamma subfamily B, 1 |
| chrX_+_119737806 | 3.01 |
ENST00000371317.5
|
MCTS1
|
malignant T cell amplified sequence 1 |
| chr11_-_111794446 | 3.00 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr4_+_83351791 | 2.99 |
ENST00000509635.1
|
ENOPH1
|
enolase-phosphatase 1 |
| chr2_-_170681324 | 2.97 |
ENST00000409340.1
|
METTL5
|
methyltransferase like 5 |
| chr14_+_61201445 | 2.96 |
ENST00000261245.4
ENST00000539616.2 |
MNAT1
|
MNAT CDK-activating kinase assembly factor 1 |
| chr7_+_56119323 | 2.96 |
ENST00000275603.4
ENST00000335503.3 ENST00000540286.1 |
CCT6A
|
chaperonin containing TCP1, subunit 6A (zeta 1) |
| chr6_-_153304148 | 2.94 |
ENST00000229758.3
|
FBXO5
|
F-box protein 5 |
| chr10_+_114709999 | 2.94 |
ENST00000355995.4
ENST00000545257.1 ENST00000543371.1 ENST00000536810.1 ENST00000355717.4 ENST00000538897.1 ENST00000534894.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr2_-_9143786 | 2.93 |
ENST00000462696.1
ENST00000305997.3 |
MBOAT2
|
membrane bound O-acyltransferase domain containing 2 |
| chr11_+_57529234 | 2.92 |
ENST00000360682.6
ENST00000361796.4 ENST00000529526.1 ENST00000426142.2 ENST00000399050.4 ENST00000361391.6 ENST00000361332.4 ENST00000532463.1 ENST00000529986.1 ENST00000358694.6 ENST00000532787.1 ENST00000533667.1 ENST00000532649.1 ENST00000528621.1 ENST00000530748.1 ENST00000428599.2 ENST00000527467.1 ENST00000528232.1 ENST00000531014.1 ENST00000526772.1 ENST00000529873.1 ENST00000525902.1 ENST00000532844.1 ENST00000526357.1 ENST00000530094.1 ENST00000415361.2 ENST00000532245.1 ENST00000534579.1 ENST00000526938.1 |
CTNND1
|
catenin (cadherin-associated protein), delta 1 |
| chr14_-_71107921 | 2.86 |
ENST00000553982.1
ENST00000500016.1 |
CTD-2540L5.5
CTD-2540L5.6
|
CTD-2540L5.5 CTD-2540L5.6 |
| chr6_+_134274322 | 2.84 |
ENST00000367871.1
ENST00000237264.4 |
TBPL1
|
TBP-like 1 |
| chr4_+_83351715 | 2.83 |
ENST00000273920.3
|
ENOPH1
|
enolase-phosphatase 1 |
| chr15_+_80696666 | 2.83 |
ENST00000303329.4
|
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr2_-_211341411 | 2.82 |
ENST00000233714.4
ENST00000443314.1 ENST00000441020.3 ENST00000450366.2 ENST00000431941.2 |
LANCL1
|
LanC lantibiotic synthetase component C-like 1 (bacterial) |
| chr12_+_54422142 | 2.82 |
ENST00000243108.4
|
HOXC6
|
homeobox C6 |
| chr12_+_104359576 | 2.82 |
ENST00000392872.3
ENST00000436021.2 |
TDG
|
thymine-DNA glycosylase |
| chr3_-_141747950 | 2.81 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr4_+_20255123 | 2.78 |
ENST00000504154.1
ENST00000273739.5 |
SLIT2
|
slit homolog 2 (Drosophila) |
| chr11_+_120255997 | 2.77 |
ENST00000532993.1
|
ARHGEF12
|
Rho guanine nucleotide exchange factor (GEF) 12 |
| chr1_+_155036204 | 2.76 |
ENST00000368409.3
ENST00000359751.4 ENST00000427683.2 ENST00000556931.1 ENST00000505139.1 |
EFNA4
EFNA3
EFNA3
|
ephrin-A4 ephrin-A3 Ephrin-A3; Uncharacterized protein; cDNA FLJ57652, highly similar to Ephrin-A3 |
| chr17_-_685559 | 2.76 |
ENST00000301329.6
|
GLOD4
|
glyoxalase domain containing 4 |
| chr3_+_173302222 | 2.74 |
ENST00000361589.4
|
NLGN1
|
neuroligin 1 |
| chr2_-_176046391 | 2.74 |
ENST00000392541.3
ENST00000409194.1 |
ATP5G3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr5_+_95066823 | 2.72 |
ENST00000506817.1
ENST00000379982.3 |
RHOBTB3
|
Rho-related BTB domain containing 3 |
| chr1_-_115259337 | 2.72 |
ENST00000369535.4
|
NRAS
|
neuroblastoma RAS viral (v-ras) oncogene homolog |
| chr10_+_123923205 | 2.68 |
ENST00000369004.3
ENST00000260733.3 |
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr13_+_37581115 | 2.68 |
ENST00000481013.1
|
EXOSC8
|
exosome component 8 |
| chr20_-_32891151 | 2.67 |
ENST00000217426.2
|
AHCY
|
adenosylhomocysteinase |
| chr5_-_176900610 | 2.67 |
ENST00000477391.2
ENST00000393565.1 ENST00000309007.5 |
DBN1
|
drebrin 1 |
| chr1_+_87794150 | 2.65 |
ENST00000370544.5
|
LMO4
|
LIM domain only 4 |
| chr7_-_47621736 | 2.64 |
ENST00000311160.9
|
TNS3
|
tensin 3 |
| chr14_-_60097297 | 2.63 |
ENST00000395090.1
|
RTN1
|
reticulon 1 |
| chr7_+_23146271 | 2.60 |
ENST00000545771.1
|
KLHL7
|
kelch-like family member 7 |
| chrX_-_109683446 | 2.58 |
ENST00000372057.1
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
| chr11_+_114128522 | 2.57 |
ENST00000535401.1
|
NNMT
|
nicotinamide N-methyltransferase |
| chr16_+_68679193 | 2.56 |
ENST00000581171.1
|
CDH3
|
cadherin 3, type 1, P-cadherin (placental) |
| chr5_-_146435501 | 2.54 |
ENST00000336640.6
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chrX_-_43832711 | 2.53 |
ENST00000378062.5
|
NDP
|
Norrie disease (pseudoglioma) |
| chr10_-_735553 | 2.53 |
ENST00000280886.6
ENST00000423550.1 |
DIP2C
|
DIP2 disco-interacting protein 2 homolog C (Drosophila) |
| chr17_-_685493 | 2.52 |
ENST00000536578.1
ENST00000301328.5 ENST00000576419.1 |
GLOD4
|
glyoxalase domain containing 4 |
| chr2_-_224467093 | 2.52 |
ENST00000305409.2
|
SCG2
|
secretogranin II |
| chr12_-_16761007 | 2.51 |
ENST00000354662.1
ENST00000441439.2 |
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr12_+_54378923 | 2.50 |
ENST00000303460.4
|
HOXC10
|
homeobox C10 |
| chr6_+_64282447 | 2.49 |
ENST00000370650.2
ENST00000578299.1 |
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr7_+_69064300 | 2.48 |
ENST00000342771.4
|
AUTS2
|
autism susceptibility candidate 2 |
| chr18_-_31802056 | 2.44 |
ENST00000538587.1
|
NOL4
|
nucleolar protein 4 |
| chr2_+_149632783 | 2.44 |
ENST00000435030.1
|
KIF5C
|
kinesin family member 5C |
| chr7_-_137028498 | 2.43 |
ENST00000393083.2
|
PTN
|
pleiotrophin |
| chr16_+_57673430 | 2.42 |
ENST00000540164.2
ENST00000568531.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr10_-_104192405 | 2.42 |
ENST00000369937.4
|
CUEDC2
|
CUE domain containing 2 |
| chr5_-_138210977 | 2.37 |
ENST00000274711.6
ENST00000521094.2 |
LRRTM2
|
leucine rich repeat transmembrane neuronal 2 |
| chr2_-_26205340 | 2.35 |
ENST00000264712.3
|
KIF3C
|
kinesin family member 3C |
| chr5_+_141346385 | 2.33 |
ENST00000513019.1
ENST00000356143.1 |
RNF14
|
ring finger protein 14 |
| chr13_+_52598827 | 2.32 |
ENST00000521776.2
|
UTP14C
|
UTP14, U3 small nucleolar ribonucleoprotein, homolog C (yeast) |
| chr3_+_123813509 | 2.31 |
ENST00000460856.1
ENST00000240874.3 |
KALRN
|
kalirin, RhoGEF kinase |
| chr20_+_60718785 | 2.31 |
ENST00000421564.1
ENST00000450482.1 ENST00000331758.3 |
SS18L1
|
synovial sarcoma translocation gene on chromosome 18-like 1 |
| chr18_-_53089723 | 2.30 |
ENST00000561992.1
ENST00000562512.2 |
TCF4
|
transcription factor 4 |
| chrX_+_54835493 | 2.29 |
ENST00000396224.1
|
MAGED2
|
melanoma antigen family D, 2 |
| chr5_+_92919043 | 2.28 |
ENST00000327111.3
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr15_+_50474385 | 2.25 |
ENST00000267842.5
|
SLC27A2
|
solute carrier family 27 (fatty acid transporter), member 2 |
| chr5_+_179921430 | 2.25 |
ENST00000393356.1
|
CNOT6
|
CCR4-NOT transcription complex, subunit 6 |
| chr5_-_177580777 | 2.23 |
ENST00000314397.4
|
NHP2
|
NHP2 ribonucleoprotein |
| chr17_-_37934466 | 2.22 |
ENST00000583368.1
|
IKZF3
|
IKAROS family zinc finger 3 (Aiolos) |
| chr7_+_116312411 | 2.21 |
ENST00000456159.1
ENST00000397752.3 ENST00000318493.6 |
MET
|
met proto-oncogene |
| chr11_-_62432641 | 2.21 |
ENST00000528405.1
ENST00000524958.1 ENST00000525675.1 |
RP11-831H9.11
C11orf48
|
Uncharacterized protein chromosome 11 open reading frame 48 |
| chr5_+_179921344 | 2.21 |
ENST00000261951.4
|
CNOT6
|
CCR4-NOT transcription complex, subunit 6 |
| chr1_-_94079648 | 2.20 |
ENST00000370247.3
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of LEF1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.7 | 20.2 | GO:0009996 | negative regulation of cell fate specification(GO:0009996) |
| 4.5 | 13.6 | GO:0030474 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 4.0 | 16.2 | GO:0009956 | radial pattern formation(GO:0009956) |
| 3.1 | 9.4 | GO:0060901 | regulation of monophenol monooxygenase activity(GO:0032771) positive regulation of monophenol monooxygenase activity(GO:0032773) negative regulation of catagen(GO:0051796) regulation of hair cycle by canonical Wnt signaling pathway(GO:0060901) positive regulation of melanosome transport(GO:1902910) |
| 2.2 | 6.5 | GO:0050923 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 1.8 | 5.4 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 1.8 | 5.4 | GO:0051086 | chaperone mediated protein folding independent of cofactor(GO:0051086) |
| 1.7 | 5.1 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 1.6 | 7.9 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 1.5 | 4.6 | GO:1904395 | retinal rod cell differentiation(GO:0060221) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 1.5 | 6.1 | GO:1902544 | oxidative DNA demethylation(GO:0035511) regulation of DNA N-glycosylase activity(GO:1902544) |
| 1.4 | 7.2 | GO:1901631 | positive regulation of presynaptic membrane organization(GO:1901631) positive regulation of synaptic vesicle clustering(GO:2000809) |
| 1.4 | 11.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 1.4 | 4.1 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 1.4 | 4.1 | GO:0036333 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 1.4 | 8.2 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 1.3 | 7.7 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 1.3 | 6.3 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 1.2 | 16.0 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 1.2 | 4.8 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 1.2 | 6.9 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 1.1 | 4.6 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 1.1 | 3.3 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 1.1 | 13.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 1.1 | 10.7 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 1.1 | 10.6 | GO:0070141 | response to UV-A(GO:0070141) |
| 1.0 | 5.2 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 1.0 | 4.0 | GO:1904529 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 1.0 | 3.0 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.9 | 13.2 | GO:0072257 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.9 | 4.6 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.9 | 9.9 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.9 | 16.2 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.9 | 4.4 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.9 | 5.2 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.8 | 3.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.7 | 2.2 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.7 | 2.8 | GO:0006235 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.7 | 2.7 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.7 | 2.7 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 0.7 | 6.7 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.6 | 3.2 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.6 | 4.5 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.6 | 5.8 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.6 | 3.9 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.6 | 6.4 | GO:0042659 | regulation of cell fate specification(GO:0042659) |
| 0.6 | 2.5 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.6 | 2.5 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.6 | 1.8 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.6 | 2.3 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.5 | 4.4 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.5 | 3.3 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.5 | 2.5 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.5 | 3.9 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.5 | 2.4 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.5 | 1.9 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.5 | 6.6 | GO:0030913 | paranodal junction assembly(GO:0030913) |
| 0.5 | 3.2 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.5 | 4.1 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.4 | 0.9 | GO:1904849 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.4 | 2.7 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.4 | 2.2 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.4 | 2.2 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.4 | 2.1 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.4 | 4.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.4 | 1.2 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.4 | 2.0 | GO:0007068 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) |
| 0.4 | 10.2 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.4 | 1.9 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.4 | 8.0 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.4 | 1.1 | GO:0002678 | positive regulation of chronic inflammatory response(GO:0002678) |
| 0.4 | 12.0 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.4 | 4.8 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.4 | 1.4 | GO:2001270 | regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.3 | 3.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.3 | 1.4 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.3 | 13.5 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.3 | 1.3 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.3 | 0.7 | GO:0051572 | negative regulation of histone H3-K4 methylation(GO:0051572) |
| 0.3 | 1.3 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.3 | 0.9 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.3 | 30.2 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) |
| 0.3 | 3.0 | GO:1904871 | regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.3 | 8.2 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.3 | 4.4 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.3 | 1.5 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.3 | 6.6 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.3 | 6.1 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.3 | 1.3 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.3 | 19.0 | GO:0008542 | visual learning(GO:0008542) |
| 0.3 | 6.0 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.3 | 1.0 | GO:2000320 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.3 | 1.8 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.3 | 2.8 | GO:1902961 | positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.2 | 4.8 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.2 | 14.1 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.2 | 0.7 | GO:0032899 | regulation of nucleoside transport(GO:0032242) regulation of neurotrophin production(GO:0032899) negative regulation of neurotrophin production(GO:0032900) |
| 0.2 | 11.2 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.2 | 2.4 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.2 | 2.6 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.2 | 4.2 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.2 | 2.5 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.2 | 0.8 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.2 | 2.0 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.2 | 1.8 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.2 | 0.6 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.2 | 0.8 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.2 | 3.0 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.2 | 0.4 | GO:2000015 | regulation of determination of dorsal identity(GO:2000015) |
| 0.2 | 10.7 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.2 | 2.2 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.2 | 0.9 | GO:1904816 | positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.2 | 7.1 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.2 | 0.5 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.2 | 0.7 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.2 | 0.8 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.2 | 2.1 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.2 | 1.1 | GO:0060339 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.2 | 4.0 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.2 | 1.7 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.2 | 1.1 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 4.2 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.1 | 1.4 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.6 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.1 | 0.6 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.1 | 18.4 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.1 | 0.8 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 1.1 | GO:0090240 | positive regulation of histone H4 acetylation(GO:0090240) |
| 0.1 | 2.0 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.1 | 2.1 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 0.9 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 1.4 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.1 | 1.8 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.8 | GO:1990564 | protein ufmylation(GO:0071569) protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 | 0.6 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 3.2 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.1 | 0.8 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 1.4 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.1 | 0.2 | GO:0048880 | sensory system development(GO:0048880) |
| 0.1 | 0.4 | GO:0035983 | response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.1 | 0.8 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.1 | 2.2 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.1 | 0.5 | GO:0030879 | mammary gland development(GO:0030879) |
| 0.1 | 3.9 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 1.5 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 0.5 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.1 | 3.5 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.1 | 0.9 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.1 | 0.6 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 0.1 | 5.7 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 3.1 | GO:0006833 | water transport(GO:0006833) |
| 0.1 | 1.0 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.1 | 0.4 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.1 | 1.0 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 0.2 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.1 | 0.2 | GO:0021553 | olfactory nerve development(GO:0021553) ureteric bud invasion(GO:0072092) |
| 0.1 | 1.2 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.1 | 1.3 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 2.7 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 0.7 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.1 | 2.4 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 2.7 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.1 | 0.6 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.1 | 0.1 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.1 | 10.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 2.1 | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 0.1 | 4.2 | GO:0001558 | regulation of cell growth(GO:0001558) |
| 0.1 | 1.3 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.1 | 2.3 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.1 | 0.9 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.1 | 3.4 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.1 | 2.0 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.1 | 0.1 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.1 | 1.5 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.1 | 0.9 | GO:0035246 | peptidyl-arginine N-methylation(GO:0035246) |
| 0.1 | 0.8 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.1 | 0.3 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 1.2 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 | 0.4 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.1 | 2.3 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 2.1 | GO:0043403 | skeletal muscle tissue regeneration(GO:0043403) |
| 0.1 | 1.7 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 1.6 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 2.8 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 2.8 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.8 | GO:0019054 | modulation by virus of host process(GO:0019054) |
| 0.0 | 1.3 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.4 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.0 | 1.2 | GO:0050850 | positive regulation of calcium-mediated signaling(GO:0050850) |
| 0.0 | 0.8 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.0 | 0.8 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 3.8 | GO:1903169 | regulation of calcium ion transmembrane transport(GO:1903169) |
| 0.0 | 2.9 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 1.0 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.0 | 14.3 | GO:0043687 | post-translational protein modification(GO:0043687) |
| 0.0 | 0.3 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.7 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 1.4 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.3 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.0 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 1.7 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 1.7 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.2 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.7 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 1.9 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.8 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.8 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 1.3 | GO:0006497 | protein lipidation(GO:0006497) |
| 0.0 | 0.4 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 2.3 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.4 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 2.3 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 4.6 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.0 | 0.1 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.0 | 1.4 | GO:0034101 | erythrocyte homeostasis(GO:0034101) |
| 0.0 | 1.1 | GO:0097581 | lamellipodium organization(GO:0097581) |
| 0.0 | 0.3 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.1 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.1 | GO:0033198 | response to ATP(GO:0033198) |
| 0.0 | 0.5 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 0.4 | GO:0030850 | prostate gland development(GO:0030850) |
| 0.0 | 0.3 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.3 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 13.6 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 1.3 | 5.2 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 1.2 | 5.9 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 1.1 | 11.2 | GO:0045180 | basal cortex(GO:0045180) |
| 1.1 | 6.3 | GO:0090661 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 1.0 | 4.1 | GO:0031673 | H zone(GO:0031673) |
| 0.8 | 4.1 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.7 | 2.0 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.7 | 11.1 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.7 | 4.6 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.6 | 7.5 | GO:0016342 | catenin complex(GO:0016342) |
| 0.6 | 1.8 | GO:1903349 | omegasome membrane(GO:1903349) |
| 0.5 | 1.1 | GO:0042025 | host cell nucleus(GO:0042025) host cell nuclear part(GO:0044094) |
| 0.5 | 4.3 | GO:0000235 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.5 | 1.6 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.5 | 9.5 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.5 | 2.3 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.5 | 4.5 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.4 | 8.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.4 | 5.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.4 | 1.2 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.4 | 3.2 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.4 | 3.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.4 | 4.6 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.3 | 2.9 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.3 | 4.1 | GO:0042555 | MCM complex(GO:0042555) |
| 0.3 | 5.3 | GO:0032059 | bleb(GO:0032059) |
| 0.3 | 3.3 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.3 | 3.0 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.3 | 7.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.3 | 1.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.3 | 2.8 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.3 | 4.5 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.3 | 2.5 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.3 | 13.5 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.3 | 0.8 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.2 | 1.2 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.2 | 2.7 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.2 | 1.9 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.2 | 36.2 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.2 | 4.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 12.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.2 | 4.4 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.2 | 1.7 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.2 | 0.8 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.2 | 3.1 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.2 | 4.6 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.2 | 3.0 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.2 | 6.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 0.9 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.2 | 1.4 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.2 | 2.5 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 20.0 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.2 | 2.7 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 1.3 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 0.6 | GO:0071664 | beta-catenin-TCF7L2 complex(GO:0070369) catenin-TCF7L2 complex(GO:0071664) |
| 0.1 | 2.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 14.7 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 2.7 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 5.5 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.1 | 0.8 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.1 | 5.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 17.2 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 2.6 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.1 | 2.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.6 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.1 | 8.0 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 15.9 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 8.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 8.1 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 0.9 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 0.8 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.3 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 0.8 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 5.3 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 0.7 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 0.7 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 2.3 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 2.7 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.1 | 1.9 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 0.9 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 14.5 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.1 | 1.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 3.1 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 1.3 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.1 | 0.3 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.1 | 22.5 | GO:0031252 | cell leading edge(GO:0031252) |
| 0.1 | 6.3 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 12.3 | GO:0005819 | spindle(GO:0005819) |
| 0.1 | 3.3 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 2.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 9.7 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.4 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 1.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 2.3 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.6 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 1.8 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 4.1 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.3 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.9 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 3.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.0 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 20.7 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 2.7 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 8.1 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 1.3 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.9 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.9 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 5.0 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 2.1 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.2 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 17.4 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.5 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.8 | GO:0018995 | host(GO:0018995) host cell(GO:0043657) |
| 0.0 | 0.1 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 0.6 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 2.6 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 3.5 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 5.2 | GO:0019866 | organelle inner membrane(GO:0019866) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 6.1 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 1.7 | 18.6 | GO:0039706 | co-receptor binding(GO:0039706) |
| 1.2 | 9.9 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 1.2 | 9.5 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 1.1 | 2.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 1.1 | 5.4 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 1.1 | 6.3 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.9 | 5.2 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.8 | 4.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.8 | 3.2 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.8 | 6.8 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.7 | 8.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.7 | 1.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.7 | 18.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.6 | 2.6 | GO:0008112 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.6 | 3.7 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.6 | 27.5 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.6 | 2.3 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.5 | 2.2 | GO:0004310 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.5 | 1.6 | GO:0004515 | nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.5 | 2.1 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.5 | 5.1 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.5 | 3.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.5 | 20.7 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.5 | 13.6 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.5 | 2.8 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.5 | 11.1 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.5 | 4.1 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.5 | 4.5 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.4 | 2.7 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.4 | 3.0 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.4 | 1.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.4 | 2.9 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.4 | 5.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.4 | 1.1 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.4 | 2.2 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.4 | 3.9 | GO:0008430 | selenium binding(GO:0008430) |
| 0.4 | 6.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.3 | 1.4 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.3 | 3.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.3 | 1.7 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.3 | 1.3 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.3 | 11.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.3 | 5.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.3 | 4.3 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.3 | 2.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.3 | 3.3 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.3 | 3.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.3 | 6.0 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.3 | 11.9 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.3 | 2.1 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.3 | 2.0 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.3 | 5.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.3 | 1.3 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.2 | 20.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.2 | 1.0 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.2 | 3.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.2 | 6.9 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.2 | 0.9 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.2 | 0.9 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.2 | 1.3 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.2 | 9.8 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.2 | 4.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.2 | 5.4 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.2 | 1.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.2 | 1.2 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.2 | 2.7 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.2 | 5.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.2 | 2.8 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.2 | 3.4 | GO:0031005 | filamin binding(GO:0031005) |
| 0.2 | 6.8 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.2 | 1.3 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.2 | 0.9 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.2 | 3.1 | GO:0015250 | water channel activity(GO:0015250) |
| 0.2 | 2.5 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.2 | 7.9 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.2 | 2.8 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.2 | 0.8 | GO:0070905 | serine binding(GO:0070905) |
| 0.2 | 2.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.2 | 1.2 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.2 | 2.0 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 5.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 2.7 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 2.0 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 4.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.7 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.1 | 0.8 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.1 | 1.7 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 1.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 0.7 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.1 | 0.6 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 5.4 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.1 | 3.1 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 4.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 1.3 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.1 | 1.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 1.4 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 2.7 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.6 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 3.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 2.5 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 6.1 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 1.8 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.1 | 4.3 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 4.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 1.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 2.3 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 2.5 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 29.8 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.1 | 3.5 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.1 | 3.0 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 3.3 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 11.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 19.0 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 0.8 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.1 | 0.4 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.1 | 0.9 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.1 | 1.0 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 0.2 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 0.3 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.9 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 1.0 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.6 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 4.0 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 2.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.4 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 1.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.3 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.7 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 1.2 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.8 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 1.6 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 4.9 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 4.1 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 9.2 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 1.3 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 1.3 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 1.8 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 4.7 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 1.9 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 1.3 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 4.1 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 2.2 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 0.1 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 2.5 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 0.9 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 0.5 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.1 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.8 | GO:0005179 | hormone activity(GO:0005179) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 18.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.6 | 10.5 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.4 | 23.9 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.2 | 4.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.2 | 10.2 | PID ATR PATHWAY | ATR signaling pathway |
| 0.2 | 2.8 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.2 | 4.5 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.2 | 3.0 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.2 | 6.1 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.2 | 4.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.2 | 12.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 4.6 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 6.8 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 5.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 4.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 4.2 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.1 | 4.1 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 8.9 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 4.5 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 9.6 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 4.1 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 7.5 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 4.1 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 8.2 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 10.7 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 5.7 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 3.9 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 1.8 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 7.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 8.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 0.8 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 5.9 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 3.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 2.0 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 1.1 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.7 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 1.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.1 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.8 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 1.4 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.1 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.9 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.7 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.9 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 41.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 1.3 | 16.2 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.8 | 20.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.6 | 17.2 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.5 | 16.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.5 | 9.3 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.4 | 6.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.4 | 9.2 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.3 | 6.1 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.3 | 6.0 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.3 | 2.7 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.3 | 3.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.3 | 5.6 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.3 | 4.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.2 | 3.2 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.2 | 5.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.2 | 5.5 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.2 | 5.8 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.2 | 5.1 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.2 | 11.8 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.2 | 2.7 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.2 | 2.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.2 | 3.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 4.5 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.2 | 1.7 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.2 | 6.3 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.2 | 4.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 4.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 6.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.2 | 5.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 2.9 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.1 | 2.7 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.1 | 8.6 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 5.1 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 5.0 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.1 | 1.6 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 1.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 3.2 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 2.0 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 1.4 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 2.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 3.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 17.2 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 2.5 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 3.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 1.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.6 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 1.9 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 1.1 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 1.0 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 2.5 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 1.7 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.4 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 4.7 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.1 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.1 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.0 | 1.3 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 1.0 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |