Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
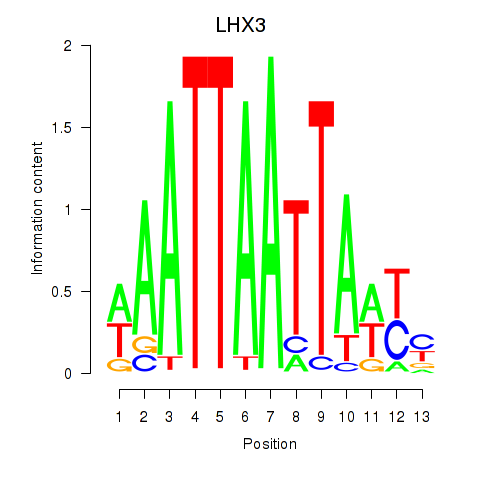
Results for LHX3
Z-value: 0.47
Transcription factors associated with LHX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
LHX3
|
ENSG00000107187.11 | LIM homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| LHX3 | hg19_v2_chr9_-_139094988_139095012, hg19_v2_chr9_-_139096955_139096979 | -0.42 | 6.8e-11 | Click! |
Activity profile of LHX3 motif
Sorted Z-values of LHX3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_24126023 | 6.16 |
ENST00000429356.1
|
GALE
|
UDP-galactose-4-epimerase |
| chr1_-_150208320 | 6.10 |
ENST00000534220.1
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr1_-_150208412 | 5.87 |
ENST00000532744.1
ENST00000369114.5 ENST00000369115.2 ENST00000369116.4 |
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr1_-_150208363 | 5.83 |
ENST00000436748.2
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr6_+_26104104 | 5.34 |
ENST00000377803.2
|
HIST1H4C
|
histone cluster 1, H4c |
| chr3_-_141747950 | 5.31 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chrX_+_107288197 | 5.13 |
ENST00000415430.3
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr1_-_150208291 | 4.79 |
ENST00000533654.1
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr4_+_113568207 | 4.60 |
ENST00000511529.1
|
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chrX_+_107288239 | 4.56 |
ENST00000217957.5
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr15_+_65843130 | 3.95 |
ENST00000569894.1
|
PTPLAD1
|
protein tyrosine phosphatase-like A domain containing 1 |
| chr17_-_60142609 | 3.29 |
ENST00000397786.2
|
MED13
|
mediator complex subunit 13 |
| chr5_-_43557791 | 3.22 |
ENST00000338972.4
ENST00000511321.1 ENST00000515338.1 |
PAIP1
|
poly(A) binding protein interacting protein 1 |
| chr9_-_128246769 | 3.15 |
ENST00000444226.1
|
MAPKAP1
|
mitogen-activated protein kinase associated protein 1 |
| chr3_+_141106643 | 3.01 |
ENST00000514251.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr14_+_39583427 | 2.98 |
ENST00000308317.6
ENST00000396249.2 ENST00000250379.8 ENST00000534684.2 ENST00000527381.1 |
GEMIN2
|
gem (nuclear organelle) associated protein 2 |
| chrX_-_55057403 | 2.93 |
ENST00000396198.3
ENST00000335854.4 ENST00000455688.1 ENST00000330807.5 |
ALAS2
|
aminolevulinate, delta-, synthase 2 |
| chr3_-_141719195 | 2.90 |
ENST00000397991.4
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr15_+_64680003 | 2.88 |
ENST00000261884.3
|
TRIP4
|
thyroid hormone receptor interactor 4 |
| chr17_+_35851570 | 2.79 |
ENST00000394386.1
|
DUSP14
|
dual specificity phosphatase 14 |
| chr12_-_118628350 | 2.78 |
ENST00000537952.1
ENST00000537822.1 |
TAOK3
|
TAO kinase 3 |
| chr1_-_45988542 | 2.60 |
ENST00000424390.1
|
PRDX1
|
peroxiredoxin 1 |
| chr3_+_138340049 | 2.48 |
ENST00000464668.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr17_-_27418537 | 2.42 |
ENST00000408971.2
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1 |
| chr3_+_69812877 | 2.38 |
ENST00000457080.1
ENST00000328528.6 |
MITF
|
microphthalmia-associated transcription factor |
| chr4_+_41937131 | 2.31 |
ENST00000504986.1
ENST00000508448.1 ENST00000513702.1 ENST00000325094.5 |
TMEM33
|
transmembrane protein 33 |
| chrX_-_106243451 | 2.29 |
ENST00000355610.4
ENST00000535534.1 |
MORC4
|
MORC family CW-type zinc finger 4 |
| chr16_+_15489603 | 2.25 |
ENST00000568766.1
ENST00000287594.7 |
RP11-1021N1.1
MPV17L
|
Uncharacterized protein MPV17 mitochondrial membrane protein-like |
| chr3_+_157827841 | 2.18 |
ENST00000295930.3
ENST00000471994.1 ENST00000464171.1 ENST00000312179.6 ENST00000475278.2 |
RSRC1
|
arginine/serine-rich coiled-coil 1 |
| chr16_-_28634874 | 2.13 |
ENST00000395609.1
ENST00000350842.4 |
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr11_-_67980744 | 1.85 |
ENST00000401547.2
ENST00000453170.1 ENST00000304363.4 |
SUV420H1
|
suppressor of variegation 4-20 homolog 1 (Drosophila) |
| chr5_+_67586465 | 1.81 |
ENST00000336483.5
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr3_+_138340067 | 1.77 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr7_+_107224364 | 1.74 |
ENST00000491150.1
|
BCAP29
|
B-cell receptor-associated protein 29 |
| chr5_-_78809950 | 1.63 |
ENST00000334082.6
|
HOMER1
|
homer homolog 1 (Drosophila) |
| chr12_+_64798095 | 1.58 |
ENST00000332707.5
|
XPOT
|
exportin, tRNA |
| chr12_+_4385230 | 1.47 |
ENST00000536537.1
|
CCND2
|
cyclin D2 |
| chr14_-_21566731 | 1.39 |
ENST00000360947.3
|
ZNF219
|
zinc finger protein 219 |
| chr9_-_75567962 | 1.36 |
ENST00000297785.3
ENST00000376939.1 |
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr15_-_20193370 | 1.21 |
ENST00000558565.2
|
IGHV3OR15-7
|
immunoglobulin heavy variable 3/OR15-7 (pseudogene) |
| chr7_-_22862406 | 1.16 |
ENST00000372879.4
|
TOMM7
|
translocase of outer mitochondrial membrane 7 homolog (yeast) |
| chr2_-_99279928 | 1.16 |
ENST00000414521.2
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr2_-_145278475 | 1.15 |
ENST00000558170.2
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr19_+_48949030 | 1.14 |
ENST00000253237.5
|
GRWD1
|
glutamate-rich WD repeat containing 1 |
| chr11_+_59824127 | 1.12 |
ENST00000278865.3
|
MS4A3
|
membrane-spanning 4-domains, subfamily A, member 3 (hematopoietic cell-specific) |
| chr4_-_41884620 | 1.11 |
ENST00000504870.1
|
LINC00682
|
long intergenic non-protein coding RNA 682 |
| chr5_-_88119580 | 1.11 |
ENST00000539796.1
|
MEF2C
|
myocyte enhancer factor 2C |
| chr11_+_59824060 | 1.10 |
ENST00000395032.2
ENST00000358152.2 |
MS4A3
|
membrane-spanning 4-domains, subfamily A, member 3 (hematopoietic cell-specific) |
| chr12_-_7656357 | 1.05 |
ENST00000396620.3
ENST00000432237.2 ENST00000359156.4 |
CD163
|
CD163 molecule |
| chrX_-_13835147 | 1.05 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr18_+_72201829 | 1.02 |
ENST00000582365.1
|
CNDP1
|
carnosine dipeptidase 1 (metallopeptidase M20 family) |
| chr6_+_83072923 | 0.91 |
ENST00000535040.1
|
TPBG
|
trophoblast glycoprotein |
| chrX_+_9431324 | 0.89 |
ENST00000407597.2
ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X
|
transducin (beta)-like 1X-linked |
| chr7_-_36764142 | 0.86 |
ENST00000258749.5
ENST00000535891.1 |
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chr3_+_63953415 | 0.85 |
ENST00000484332.1
|
ATXN7
|
ataxin 7 |
| chr12_+_59989918 | 0.82 |
ENST00000547379.1
ENST00000549465.1 |
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr4_-_185275104 | 0.82 |
ENST00000317596.3
|
RP11-290F5.2
|
RP11-290F5.2 |
| chr19_-_49149553 | 0.81 |
ENST00000084798.4
|
CA11
|
carbonic anhydrase XI |
| chrX_+_108779004 | 0.80 |
ENST00000218004.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr12_-_10282836 | 0.78 |
ENST00000304084.8
ENST00000353231.5 ENST00000525605.1 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr18_-_21891460 | 0.72 |
ENST00000357041.4
|
OSBPL1A
|
oxysterol binding protein-like 1A |
| chr14_+_22963806 | 0.69 |
ENST00000390493.1
|
TRAJ44
|
T cell receptor alpha joining 44 |
| chr4_+_158142750 | 0.68 |
ENST00000505888.1
ENST00000449365.1 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr3_-_47517302 | 0.63 |
ENST00000441517.2
ENST00000545718.1 |
SCAP
|
SREBF chaperone |
| chr5_-_16916624 | 0.62 |
ENST00000513882.1
|
MYO10
|
myosin X |
| chr4_+_158141899 | 0.58 |
ENST00000264426.9
ENST00000506284.1 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr12_-_86650077 | 0.57 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr4_+_155484103 | 0.55 |
ENST00000302068.4
|
FGB
|
fibrinogen beta chain |
| chr7_-_122840015 | 0.54 |
ENST00000194130.2
|
SLC13A1
|
solute carrier family 13 (sodium/sulfate symporter), member 1 |
| chr1_+_81771806 | 0.52 |
ENST00000370721.1
ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2
|
latrophilin 2 |
| chr1_+_168250194 | 0.52 |
ENST00000367821.3
|
TBX19
|
T-box 19 |
| chr12_-_53994805 | 0.45 |
ENST00000328463.7
|
ATF7
|
activating transcription factor 7 |
| chr4_-_23891693 | 0.41 |
ENST00000264867.2
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
| chr18_-_21852143 | 0.40 |
ENST00000399443.3
|
OSBPL1A
|
oxysterol binding protein-like 1A |
| chr9_-_215744 | 0.39 |
ENST00000382387.2
|
C9orf66
|
chromosome 9 open reading frame 66 |
| chr9_+_125132803 | 0.38 |
ENST00000540753.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr9_-_100000957 | 0.31 |
ENST00000366109.2
ENST00000607322.1 |
RP11-498P14.5
|
RP11-498P14.5 |
| chr19_+_49199209 | 0.27 |
ENST00000522966.1
ENST00000425340.2 ENST00000391876.4 |
FUT2
|
fucosyltransferase 2 (secretor status included) |
| chr12_-_10282742 | 0.27 |
ENST00000298523.5
ENST00000396484.2 ENST00000310002.4 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr14_-_107049312 | 0.23 |
ENST00000390627.2
|
IGHV3-53
|
immunoglobulin heavy variable 3-53 |
| chr12_-_42631529 | 0.23 |
ENST00000548917.1
|
YAF2
|
YY1 associated factor 2 |
| chr4_+_74301880 | 0.22 |
ENST00000395792.2
ENST00000226359.2 |
AFP
|
alpha-fetoprotein |
| chr18_+_616672 | 0.21 |
ENST00000338387.7
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr9_+_125133315 | 0.21 |
ENST00000223423.4
ENST00000362012.2 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr17_-_72772462 | 0.17 |
ENST00000582870.1
ENST00000581136.1 ENST00000357814.3 ENST00000579218.1 ENST00000583476.1 ENST00000580301.1 ENST00000583757.1 ENST00000582524.1 |
NAT9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr1_+_115572415 | 0.16 |
ENST00000256592.1
|
TSHB
|
thyroid stimulating hormone, beta |
| chr4_+_158141843 | 0.12 |
ENST00000509417.1
ENST00000296526.7 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr8_+_119294456 | 0.12 |
ENST00000366457.2
|
AC023590.1
|
Uncharacterized protein |
| chr1_+_244214577 | 0.11 |
ENST00000358704.4
|
ZBTB18
|
zinc finger and BTB domain containing 18 |
| chr17_-_10372875 | 0.11 |
ENST00000255381.2
|
MYH4
|
myosin, heavy chain 4, skeletal muscle |
| chr5_+_140529630 | 0.10 |
ENST00000543635.1
|
PCDHB6
|
protocadherin beta 6 |
| chr6_+_153019023 | 0.07 |
ENST00000367245.5
ENST00000529453.1 |
MYCT1
|
myc target 1 |
| chr19_-_43969796 | 0.07 |
ENST00000244333.3
|
LYPD3
|
LY6/PLAUR domain containing 3 |
| chr12_+_49740700 | 0.06 |
ENST00000549441.2
ENST00000395069.3 |
DNAJC22
|
DnaJ (Hsp40) homolog, subfamily C, member 22 |
| chr12_-_86650045 | 0.03 |
ENST00000604798.1
|
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr8_-_57123815 | 0.02 |
ENST00000316981.3
ENST00000423799.2 ENST00000429357.2 |
PLAG1
|
pleiomorphic adenoma gene 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of LHX3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.6 | 2.2 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.5 | 1.6 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.5 | 3.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.5 | 9.7 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.5 | 6.2 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.5 | 1.4 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.4 | 22.6 | GO:0043486 | histone exchange(GO:0043486) |
| 0.3 | 1.1 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.3 | 1.1 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.2 | 2.9 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.2 | 1.7 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.2 | 7.3 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 0.4 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) regulation of progesterone biosynthetic process(GO:2000182) |
| 0.1 | 0.6 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 | 2.8 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 1.8 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.1 | 1.2 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.1 | 1.0 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 0.8 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 1.5 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.1 | 2.1 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 2.6 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.1 | 3.3 | GO:0070328 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.1 | 0.9 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 2.8 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 3.0 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 3.2 | GO:0048255 | mRNA stabilization(GO:0048255) |
| 0.1 | 2.9 | GO:0045661 | regulation of myoblast differentiation(GO:0045661) |
| 0.1 | 1.1 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.1 | 1.1 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 2.4 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.1 | 0.5 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.0 | 1.2 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 1.4 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.6 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 1.6 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.3 | GO:0019317 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 2.2 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 1.1 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.2 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 1.1 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 3.0 | GO:0006275 | regulation of DNA replication(GO:0006275) |
| 0.0 | 0.9 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.2 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.0 | 0.6 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 22.6 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.5 | 1.8 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.4 | 3.0 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.3 | 1.4 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.2 | 1.5 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.2 | 1.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.2 | 3.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 3.3 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 8.2 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 1.6 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 3.9 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.6 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 2.9 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 2.2 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.4 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 3.8 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 2.2 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 0.8 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.9 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 2.6 | GO:0043209 | myelin sheath(GO:0043209) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.2 | GO:0003974 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 1.0 | 2.9 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.4 | 22.6 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.3 | 2.6 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.3 | 2.1 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.3 | 0.9 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.3 | 1.4 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.3 | 1.8 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.2 | 1.8 | GO:0043559 | insulin binding(GO:0043559) |
| 0.2 | 0.8 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.2 | 3.3 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.2 | 3.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.2 | 2.8 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.6 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 3.1 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 3.0 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 3.3 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 1.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 1.4 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 1.6 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 1.2 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.1 | 2.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 0.6 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.1 | 0.5 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.1 | 1.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 1.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.8 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 1.0 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 1.1 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 1.6 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 1.1 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 1.1 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 2.8 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 1.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 1.1 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.6 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.3 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 8.7 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.4 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.6 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.9 | GO:0008013 | beta-catenin binding(GO:0008013) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 2.8 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 8.2 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 5.4 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 3.1 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 2.6 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 3.4 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.4 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.6 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 2.1 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.6 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 5.0 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.1 | 2.9 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 5.3 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 3.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 1.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 2.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 1.8 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 1.4 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 1.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 3.0 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 1.5 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.8 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 6.2 | REACTOME METABOLISM OF CARBOHYDRATES | Genes involved in Metabolism of carbohydrates |
| 0.0 | 1.2 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.6 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |