Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
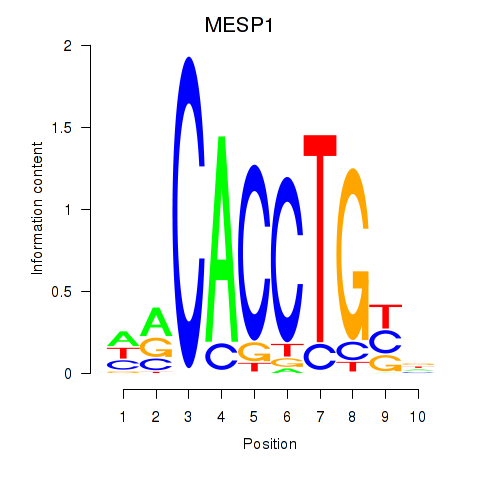
Results for MESP1
Z-value: 1.09
Transcription factors associated with MESP1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MESP1
|
ENSG00000166823.5 | mesoderm posterior bHLH transcription factor 1 |
Activity profile of MESP1 motif
Sorted Z-values of MESP1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MESP1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 16.9 | 50.6 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 5.3 | 26.3 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 5.2 | 31.2 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 5.0 | 19.8 | GO:0031179 | peptide amidation(GO:0001519) protein amidation(GO:0018032) peptide modification(GO:0031179) |
| 4.8 | 19.3 | GO:0021564 | vagus nerve development(GO:0021564) |
| 4.5 | 26.9 | GO:0007296 | vitellogenesis(GO:0007296) |
| 4.4 | 13.3 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 4.3 | 12.8 | GO:0021678 | fourth ventricle development(GO:0021592) third ventricle development(GO:0021678) |
| 3.8 | 11.4 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 3.8 | 15.2 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 3.6 | 10.8 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 3.6 | 3.6 | GO:0060920 | cardiac pacemaker cell differentiation(GO:0060920) cardiac pacemaker cell development(GO:0060926) |
| 3.5 | 21.1 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 3.3 | 26.8 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 3.3 | 10.0 | GO:0044501 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 3.1 | 9.3 | GO:1904693 | midbrain morphogenesis(GO:1904693) |
| 3.1 | 15.3 | GO:0003164 | His-Purkinje system development(GO:0003164) |
| 2.9 | 26.4 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 2.9 | 8.8 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 2.9 | 11.5 | GO:0046833 | snRNA export from nucleus(GO:0006408) positive regulation of RNA export from nucleus(GO:0046833) |
| 2.8 | 8.5 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 2.8 | 33.6 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 2.8 | 11.2 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 2.7 | 21.3 | GO:0010994 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) positive regulation of exit from mitosis(GO:0031536) |
| 2.5 | 7.5 | GO:1900060 | negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) |
| 2.5 | 12.3 | GO:2000542 | negative regulation of gastrulation(GO:2000542) |
| 2.4 | 31.4 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 2.3 | 9.4 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 2.3 | 13.7 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 2.3 | 13.6 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 2.2 | 8.9 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 2.2 | 8.9 | GO:0044571 | [2Fe-2S] cluster assembly(GO:0044571) |
| 2.2 | 11.0 | GO:2000784 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) regulation of barbed-end actin filament capping(GO:2000812) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 2.1 | 12.5 | GO:0061441 | renal artery morphogenesis(GO:0061441) |
| 2.1 | 6.2 | GO:0098968 | neurotransmitter receptor transport postsynaptic membrane to endosome(GO:0098968) |
| 2.0 | 16.3 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 2.0 | 6.1 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 2.0 | 7.9 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 1.9 | 30.7 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 1.9 | 5.7 | GO:0036333 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 1.9 | 28.3 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 1.9 | 5.6 | GO:1904351 | negative regulation of protein catabolic process in the vacuole(GO:1904351) negative regulation of lysosomal protein catabolic process(GO:1905166) |
| 1.9 | 7.4 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 1.8 | 16.5 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 1.7 | 14.0 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 1.7 | 5.2 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 1.7 | 3.5 | GO:0019087 | transformation of host cell by virus(GO:0019087) |
| 1.7 | 5.2 | GO:0050894 | determination of affect(GO:0050894) |
| 1.6 | 4.9 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 1.6 | 3.2 | GO:1990637 | response to prolactin(GO:1990637) |
| 1.6 | 10.9 | GO:0046836 | glycolipid transport(GO:0046836) |
| 1.5 | 6.1 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 1.5 | 24.3 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 1.5 | 7.5 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 1.5 | 16.5 | GO:0009414 | response to water deprivation(GO:0009414) |
| 1.5 | 5.9 | GO:0006050 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 1.5 | 8.8 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 1.4 | 4.1 | GO:0032773 | regulation of monophenol monooxygenase activity(GO:0032771) positive regulation of monophenol monooxygenase activity(GO:0032773) negative regulation of transforming growth factor beta2 production(GO:0032912) negative regulation of catagen(GO:0051796) regulation of hair cycle by canonical Wnt signaling pathway(GO:0060901) positive regulation of melanosome transport(GO:1902910) |
| 1.4 | 9.6 | GO:0090647 | modulation of age-related behavioral decline(GO:0090647) |
| 1.3 | 40.5 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 1.3 | 6.6 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 1.3 | 9.1 | GO:0021903 | rostrocaudal neural tube patterning(GO:0021903) |
| 1.3 | 6.4 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 1.2 | 10.5 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 1.2 | 13.9 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 1.1 | 3.4 | GO:1901624 | negative regulation of lymphocyte chemotaxis(GO:1901624) |
| 1.1 | 4.6 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 1.1 | 4.5 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 1.1 | 3.3 | GO:0070902 | mitochondrial tRNA pseudouridine synthesis(GO:0070902) |
| 1.1 | 18.7 | GO:0034063 | stress granule assembly(GO:0034063) |
| 1.1 | 5.4 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 1.0 | 13.6 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 1.0 | 5.2 | GO:0015862 | uridine transport(GO:0015862) |
| 1.0 | 4.1 | GO:0032954 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 1.0 | 16.9 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 1.0 | 5.9 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 1.0 | 4.9 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 1.0 | 12.7 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 1.0 | 5.9 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.9 | 9.4 | GO:0009415 | response to water(GO:0009415) |
| 0.9 | 4.7 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.9 | 2.8 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.9 | 3.7 | GO:1904386 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.9 | 6.5 | GO:0009597 | detection of virus(GO:0009597) |
| 0.9 | 4.6 | GO:0006041 | glucosamine metabolic process(GO:0006041) glucose 1-phosphate metabolic process(GO:0019255) |
| 0.9 | 5.4 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.9 | 8.9 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.9 | 3.6 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.9 | 15.6 | GO:1904816 | positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.9 | 7.7 | GO:1902669 | positive regulation of axon guidance(GO:1902669) |
| 0.9 | 9.5 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.8 | 2.5 | GO:1903925 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.8 | 3.4 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.8 | 10.1 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.8 | 34.0 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.8 | 2.5 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.8 | 2.4 | GO:2000106 | regulation of leukocyte apoptotic process(GO:2000106) |
| 0.8 | 9.5 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.8 | 8.6 | GO:0015868 | purine nucleotide transport(GO:0015865) purine ribonucleotide transport(GO:0015868) |
| 0.8 | 1.6 | GO:0007497 | posterior midgut development(GO:0007497) |
| 0.8 | 5.3 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.8 | 17.5 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.8 | 6.0 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.7 | 4.5 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 0.7 | 9.7 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.7 | 12.0 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.7 | 0.7 | GO:1902908 | regulation of melanosome transport(GO:1902908) |
| 0.7 | 17.7 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.7 | 8.8 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.7 | 5.1 | GO:1900045 | negative regulation of histone ubiquitination(GO:0033183) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.7 | 2.1 | GO:0015783 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.7 | 2.9 | GO:0072423 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) |
| 0.7 | 7.6 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.7 | 2.7 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.7 | 8.1 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.7 | 49.3 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.6 | 3.9 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.6 | 3.2 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.6 | 7.0 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.6 | 16.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.6 | 3.1 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.6 | 3.1 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.6 | 2.4 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.6 | 24.3 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.6 | 6.0 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.6 | 2.4 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.6 | 2.3 | GO:2001270 | regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.6 | 5.3 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.6 | 2.9 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.6 | 1.7 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.6 | 15.1 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.6 | 2.9 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.6 | 2.2 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.5 | 3.3 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.5 | 2.2 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.5 | 13.1 | GO:1903077 | negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.5 | 1.6 | GO:0033005 | positive regulation of mast cell activation(GO:0033005) |
| 0.5 | 1.6 | GO:0002232 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.5 | 2.1 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.5 | 25.2 | GO:0048255 | mRNA stabilization(GO:0048255) |
| 0.5 | 2.0 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.5 | 4.5 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.5 | 5.0 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.5 | 2.9 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.5 | 8.6 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.5 | 4.8 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.5 | 2.9 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.5 | 1.4 | GO:1901420 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) negative regulation of response to alcohol(GO:1901420) |
| 0.5 | 48.7 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.4 | 5.7 | GO:0070234 | positive regulation of T cell apoptotic process(GO:0070234) |
| 0.4 | 4.8 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.4 | 7.5 | GO:2000637 | positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.4 | 9.1 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.4 | 2.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.4 | 0.4 | GO:0098917 | retrograde trans-synaptic signaling(GO:0098917) |
| 0.4 | 2.9 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.4 | 5.3 | GO:2000680 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.4 | 2.4 | GO:2000567 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.4 | 2.8 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.4 | 2.0 | GO:0072244 | metanephric glomerular epithelium development(GO:0072244) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.4 | 5.2 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) |
| 0.4 | 10.3 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.4 | 1.2 | GO:0002566 | somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.4 | 17.1 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.4 | 9.6 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.4 | 10.4 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.4 | 1.9 | GO:0010728 | regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 0.4 | 1.9 | GO:0042695 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.4 | 0.4 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) |
| 0.4 | 7.3 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.3 | 5.7 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) |
| 0.3 | 2.3 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.3 | 1.6 | GO:0009439 | cyanate metabolic process(GO:0009439) cyanate catabolic process(GO:0009440) |
| 0.3 | 6.8 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.3 | 1.3 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.3 | 7.6 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.3 | 1.2 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.3 | 1.9 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.3 | 5.8 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.3 | 2.1 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.3 | 0.9 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.3 | 0.9 | GO:0006833 | water transport(GO:0006833) |
| 0.3 | 3.2 | GO:0048711 | positive regulation of astrocyte differentiation(GO:0048711) |
| 0.3 | 0.6 | GO:0061743 | motor learning(GO:0061743) |
| 0.3 | 4.0 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.3 | 4.0 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.3 | 0.8 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.3 | 2.4 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.3 | 6.4 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.3 | 1.1 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.3 | 1.3 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.3 | 2.1 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.3 | 5.5 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.2 | 7.5 | GO:0008334 | histone mRNA metabolic process(GO:0008334) |
| 0.2 | 1.2 | GO:1904073 | trophectodermal cell proliferation(GO:0001834) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.2 | 1.9 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.2 | 13.1 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.2 | 0.7 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.2 | 2.1 | GO:0070424 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) |
| 0.2 | 2.1 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.2 | 0.5 | GO:0060467 | negative regulation of fertilization(GO:0060467) |
| 0.2 | 1.8 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.2 | 2.6 | GO:0031113 | regulation of microtubule polymerization(GO:0031113) |
| 0.2 | 4.8 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.2 | 7.4 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.2 | 1.9 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.2 | 5.7 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.2 | 0.8 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.2 | 2.7 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.2 | 3.9 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.2 | 24.6 | GO:0031424 | keratinization(GO:0031424) |
| 0.2 | 13.8 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.2 | 3.8 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.2 | 3.7 | GO:0006415 | translational termination(GO:0006415) |
| 0.2 | 0.6 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.2 | 0.9 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.2 | 7.5 | GO:0001913 | T cell mediated cytotoxicity(GO:0001913) |
| 0.2 | 3.3 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.2 | 5.7 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.2 | 1.3 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.2 | 2.6 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.2 | 0.5 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.2 | 1.5 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.2 | 4.3 | GO:0019370 | leukotriene biosynthetic process(GO:0019370) |
| 0.2 | 1.1 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.2 | 2.7 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.2 | 9.8 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.2 | 3.3 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.2 | 7.6 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.2 | 5.2 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.2 | 61.2 | GO:0043687 | post-translational protein modification(GO:0043687) |
| 0.1 | 3.4 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 7.3 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.1 | 1.2 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 2.4 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.1 | 5.5 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 2.0 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.1 | 0.5 | GO:0070997 | neuron death(GO:0070997) |
| 0.1 | 1.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.5 | GO:0035814 | negative regulation of urine volume(GO:0035811) negative regulation of renal sodium excretion(GO:0035814) |
| 0.1 | 2.4 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.1 | 1.1 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 10.5 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.1 | 5.8 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 1.5 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.1 | 2.1 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.1 | 4.4 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 0.1 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.1 | 0.4 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.1 | 1.5 | GO:0008105 | asymmetric protein localization(GO:0008105) |
| 0.1 | 15.0 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.1 | 0.8 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.1 | 0.6 | GO:0043523 | regulation of neuron apoptotic process(GO:0043523) |
| 0.1 | 2.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 4.4 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.1 | 0.4 | GO:0070255 | regulation of mucus secretion(GO:0070255) positive regulation of mucus secretion(GO:0070257) |
| 0.1 | 3.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.9 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 | 0.5 | GO:0010644 | cell communication by electrical coupling(GO:0010644) |
| 0.1 | 2.6 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.1 | 0.4 | GO:0010666 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.1 | 3.4 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 2.5 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.1 | 6.9 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.1 | 2.8 | GO:0060071 | Wnt signaling pathway, planar cell polarity pathway(GO:0060071) regulation of establishment of planar polarity(GO:0090175) |
| 0.1 | 0.1 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.1 | 3.6 | GO:0071349 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.1 | 4.3 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.1 | 0.7 | GO:0019317 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.1 | 0.8 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 2.1 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 2.6 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 2.8 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.1 | 1.6 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 3.0 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.1 | 1.0 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 4.7 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.1 | 11.9 | GO:0001649 | osteoblast differentiation(GO:0001649) |
| 0.1 | 1.8 | GO:0006310 | DNA recombination(GO:0006310) |
| 0.1 | 14.6 | GO:0016042 | lipid catabolic process(GO:0016042) |
| 0.1 | 0.6 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.1 | 0.3 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.1 | 2.4 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.1 | 0.2 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 2.2 | GO:0021766 | hippocampus development(GO:0021766) |
| 0.1 | 1.5 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 3.1 | GO:0098869 | cellular oxidant detoxification(GO:0098869) |
| 0.0 | 0.3 | GO:0014896 | cardiac muscle hypertrophy(GO:0003300) muscle hypertrophy(GO:0014896) striated muscle hypertrophy(GO:0014897) |
| 0.0 | 0.5 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.0 | 0.7 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.0 | 1.6 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.7 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) |
| 0.0 | 2.2 | GO:0002220 | innate immune response activating cell surface receptor signaling pathway(GO:0002220) stimulatory C-type lectin receptor signaling pathway(GO:0002223) |
| 0.0 | 0.8 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.9 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.0 | 0.9 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 1.3 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 1.2 | GO:0032508 | DNA duplex unwinding(GO:0032508) |
| 0.0 | 0.5 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.3 | GO:0007628 | adult walking behavior(GO:0007628) |
| 0.0 | 1.8 | GO:0021543 | pallium development(GO:0021543) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.5 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.2 | GO:0043388 | positive regulation of DNA binding(GO:0043388) |
| 0.0 | 0.6 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.5 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 1.9 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.0 | 0.2 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.1 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.7 | 28.3 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 5.4 | 16.3 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 5.3 | 26.4 | GO:1905202 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 4.0 | 28.1 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 3.9 | 11.6 | GO:0034676 | integrin alpha6-beta4 complex(GO:0034676) |
| 3.6 | 50.6 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 3.5 | 10.4 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 3.3 | 13.1 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 3.1 | 9.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 2.9 | 11.5 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 2.6 | 12.9 | GO:0097513 | myosin II filament(GO:0097513) |
| 2.5 | 2.5 | GO:0070695 | FHF complex(GO:0070695) |
| 2.4 | 26.7 | GO:0005614 | interstitial matrix(GO:0005614) |
| 2.1 | 6.4 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 2.0 | 24.0 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 1.9 | 9.5 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 1.9 | 7.5 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 1.8 | 34.9 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 1.6 | 4.7 | GO:1990393 | 3M complex(GO:1990393) |
| 1.4 | 8.6 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 1.4 | 5.6 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) |
| 1.4 | 16.3 | GO:0005915 | zonula adherens(GO:0005915) |
| 1.3 | 6.3 | GO:0071986 | Ragulator complex(GO:0071986) |
| 1.3 | 8.8 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 1.3 | 11.3 | GO:1990812 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 1.2 | 6.2 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 1.2 | 11.0 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 1.2 | 53.9 | GO:0045095 | keratin filament(GO:0045095) |
| 1.2 | 35.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 1.1 | 5.7 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 1.1 | 3.4 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 1.1 | 4.5 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 1.1 | 7.6 | GO:0070852 | cell body fiber(GO:0070852) |
| 1.1 | 3.2 | GO:1990015 | mesaxon(GO:0097453) ensheathing process(GO:1990015) |
| 1.0 | 2.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 1.0 | 2.0 | GO:0031143 | pseudopodium(GO:0031143) |
| 1.0 | 13.7 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 1.0 | 14.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 1.0 | 5.8 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.9 | 7.5 | GO:0005683 | U7 snRNP(GO:0005683) pICln-Sm protein complex(GO:0034715) |
| 0.9 | 11.4 | GO:0033643 | host cell part(GO:0033643) |
| 0.9 | 5.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.8 | 4.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.8 | 2.5 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.8 | 10.5 | GO:0042555 | MCM complex(GO:0042555) |
| 0.8 | 8.9 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.8 | 4.0 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.8 | 3.2 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.8 | 4.7 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.8 | 2.3 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.8 | 3.8 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.7 | 4.5 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.7 | 7.8 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.7 | 23.8 | GO:0032420 | stereocilium(GO:0032420) |
| 0.7 | 5.4 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.6 | 8.0 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.6 | 3.9 | GO:0005818 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.5 | 4.8 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.5 | 2.6 | GO:0030891 | VCB complex(GO:0030891) |
| 0.5 | 3.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.5 | 28.3 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.5 | 14.6 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.5 | 7.5 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.5 | 2.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.5 | 11.5 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.4 | 8.5 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) cytosolic proteasome complex(GO:0031597) |
| 0.4 | 7.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.4 | 3.4 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.4 | 10.6 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.4 | 12.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.4 | 10.9 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.4 | 5.9 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.4 | 21.7 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.4 | 2.6 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.4 | 5.0 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.4 | 2.8 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.3 | 26.3 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.3 | 4.1 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.3 | 0.7 | GO:0032039 | integrator complex(GO:0032039) |
| 0.3 | 4.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.3 | 5.4 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.3 | 1.2 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.3 | 2.3 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.3 | 4.9 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.3 | 5.5 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.3 | 25.2 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.3 | 11.2 | GO:0000313 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.3 | 5.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.3 | 7.4 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.3 | 2.8 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.3 | 8.0 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.2 | 3.0 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.2 | 5.3 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.2 | 2.1 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.2 | 2.8 | GO:0060293 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.2 | 2.3 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.2 | 5.2 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.2 | 6.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.2 | 2.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.2 | 20.4 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.2 | 10.5 | GO:0002102 | podosome(GO:0002102) |
| 0.2 | 7.4 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.2 | 3.8 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.2 | 17.5 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.2 | 1.2 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.2 | 5.7 | GO:0005921 | gap junction(GO:0005921) |
| 0.2 | 1.6 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.2 | 4.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.2 | 9.8 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.2 | 6.9 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.2 | 11.2 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.2 | 2.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.2 | 1.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.2 | 1.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.2 | 4.4 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.2 | 1.6 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.2 | 8.5 | GO:0005844 | polysome(GO:0005844) |
| 0.2 | 8.3 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.2 | 4.2 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 23.8 | GO:1904813 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.1 | 19.2 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 0.1 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.1 | 11.8 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.1 | 1.8 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 6.9 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.1 | 10.6 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 3.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 2.9 | GO:0034385 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.1 | 2.1 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 0.5 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 1.5 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.1 | 5.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 2.6 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 0.4 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 14.3 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.1 | 2.3 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 4.5 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 10.5 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 2.8 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 2.7 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 2.5 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 0.7 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 2.2 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 3.5 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 6.9 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 4.7 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.1 | 7.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 1.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 1.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 3.1 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.1 | 4.7 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.1 | 8.9 | GO:0005819 | spindle(GO:0005819) |
| 0.1 | 1.9 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 0.6 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.9 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.4 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 33.9 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 1.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 8.9 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 3.2 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 3.3 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 96.3 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 4.5 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 2.5 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 0.3 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 2.4 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.7 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.8 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 0.1 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 1.2 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 1.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 16.9 | 50.6 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 10.5 | 31.4 | GO:0003974 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 6.8 | 34.0 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 5.3 | 26.4 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 5.0 | 19.8 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 4.9 | 24.3 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 3.8 | 11.4 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 3.3 | 16.3 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 3.0 | 12.1 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 2.7 | 10.9 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 2.7 | 10.8 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 2.6 | 7.8 | GO:0004853 | uroporphyrinogen decarboxylase activity(GO:0004853) |
| 2.2 | 15.2 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 2.1 | 6.4 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 2.0 | 8.1 | GO:0033981 | D-dopachrome decarboxylase activity(GO:0033981) |
| 2.0 | 13.9 | GO:0004459 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 2.0 | 5.9 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 1.9 | 5.7 | GO:0048030 | disaccharide binding(GO:0048030) |
| 1.7 | 3.5 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 1.7 | 5.2 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 1.7 | 15.0 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 1.6 | 4.7 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 1.5 | 15.3 | GO:0051425 | PTB domain binding(GO:0051425) |
| 1.5 | 4.5 | GO:0002134 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 1.5 | 25.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 1.5 | 7.3 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 1.4 | 11.5 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 1.3 | 5.4 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 1.2 | 5.0 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 1.2 | 3.6 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 1.2 | 6.0 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 1.2 | 32.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 1.2 | 4.7 | GO:0048763 | HLH domain binding(GO:0043398) calcium-induced calcium release activity(GO:0048763) |
| 1.2 | 3.5 | GO:0032427 | GBD domain binding(GO:0032427) |
| 1.1 | 3.4 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 1.1 | 3.4 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 1.1 | 3.3 | GO:0004730 | pseudouridylate synthase activity(GO:0004730) |
| 1.1 | 43.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 1.1 | 22.8 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 1.1 | 6.3 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 1.1 | 48.4 | GO:0061650 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 1.0 | 5.2 | GO:0070404 | NADH binding(GO:0070404) |
| 1.0 | 6.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 1.0 | 12.8 | GO:0038132 | neuregulin binding(GO:0038132) |
| 1.0 | 16.5 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.9 | 2.8 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.9 | 5.6 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.9 | 14.9 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.9 | 7.4 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.9 | 2.8 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.9 | 3.5 | GO:0033906 | protein tyrosine kinase inhibitor activity(GO:0030292) hyaluronoglucuronidase activity(GO:0033906) |
| 0.9 | 7.8 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.9 | 12.0 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.9 | 8.5 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.8 | 5.8 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.8 | 4.1 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.8 | 9.1 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) |
| 0.8 | 2.5 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.8 | 10.7 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.8 | 5.7 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.8 | 4.0 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.8 | 24.0 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.8 | 3.1 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.8 | 4.6 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.8 | 6.1 | GO:0015216 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine ribonucleotide transmembrane transporter activity(GO:0005346) ATP transmembrane transporter activity(GO:0005347) purine nucleotide transmembrane transporter activity(GO:0015216) ADP transmembrane transporter activity(GO:0015217) |
| 0.8 | 17.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.7 | 4.5 | GO:0044594 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.7 | 7.2 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.7 | 2.1 | GO:0005457 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.7 | 8.6 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.7 | 2.9 | GO:0038025 | glycoprotein transporter activity(GO:0034437) reelin receptor activity(GO:0038025) |
| 0.7 | 20.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.7 | 9.5 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.7 | 4.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.7 | 5.4 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.7 | 9.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.6 | 5.7 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.6 | 32.1 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.6 | 8.9 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.6 | 2.3 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 0.6 | 2.3 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.6 | 2.3 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.6 | 4.0 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.6 | 2.9 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 0.6 | 2.2 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.5 | 6.5 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.5 | 10.6 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.5 | 1.6 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.5 | 5.6 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.5 | 11.5 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.5 | 1.9 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.5 | 7.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.4 | 1.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.4 | 3.6 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.4 | 7.1 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.4 | 20.0 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.4 | 37.6 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.4 | 3.8 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.4 | 5.0 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.4 | 1.2 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.4 | 2.8 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.4 | 2.4 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.4 | 5.2 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.4 | 3.8 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.4 | 7.4 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.4 | 5.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.4 | 5.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.4 | 5.5 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.4 | 7.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.4 | 5.4 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.4 | 7.5 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.3 | 8.0 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.3 | 2.6 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.3 | 12.2 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.3 | 6.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.3 | 2.5 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.3 | 8.5 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.3 | 3.3 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.3 | 3.0 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.3 | 3.3 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.3 | 6.4 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.3 | 0.8 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.3 | 7.5 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.3 | 1.9 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.3 | 4.8 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.3 | 1.1 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.3 | 13.1 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.3 | 1.6 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.3 | 6.1 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.3 | 1.3 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.3 | 4.5 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) dynein intermediate chain binding(GO:0045505) |
| 0.3 | 2.9 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.3 | 6.1 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.2 | 2.7 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) NFAT protein binding(GO:0051525) |
| 0.2 | 7.9 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.2 | 1.2 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.2 | 3.3 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.2 | 4.0 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.2 | 10.0 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.2 | 3.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.2 | 5.0 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.2 | 0.9 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.2 | 0.4 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.2 | 2.0 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.2 | 11.2 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.2 | 5.8 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.2 | 17.5 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.2 | 8.9 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.2 | 17.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.2 | 12.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.2 | 3.1 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.2 | 2.0 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.2 | 0.8 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.2 | 4.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 6.0 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.2 | 0.6 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.2 | 1.1 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) oxaloacetate decarboxylase activity(GO:0008948) |
| 0.2 | 2.9 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.2 | 5.7 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.2 | 2.9 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.2 | 11.3 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.2 | 2.4 | GO:0016918 | retinal binding(GO:0016918) |
| 0.2 | 0.5 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.2 | 5.6 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.2 | 3.5 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.2 | 3.3 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.9 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 4.9 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 5.7 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 2.0 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.9 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 4.9 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.1 | 2.0 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 5.0 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 8.8 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 1.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.5 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.1 | 2.1 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 2.0 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 3.4 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 1.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 1.2 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.1 | 1.6 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 6.4 | GO:1902936 | phosphatidylinositol bisphosphate binding(GO:1902936) |
| 0.1 | 3.8 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 3.6 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 2.6 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 3.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 3.6 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 31.2 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 2.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 4.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 1.3 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 4.6 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.4 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 10.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 10.1 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 10.7 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 13.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 1.9 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 8.1 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 8.3 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 0.6 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.1 | 0.7 | GO:0035240 | dopamine binding(GO:0035240) |
| 0.1 | 1.6 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 0.7 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.1 | 4.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.6 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 2.0 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.5 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 2.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 1.6 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 2.4 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 2.7 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 3.5 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.3 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.4 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.9 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.2 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.0 | 2.4 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.7 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 1.1 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.7 | GO:0004950 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.0 | 1.3 | GO:0005525 | GTP binding(GO:0005525) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 50.6 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 1.2 | 43.0 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 1.0 | 23.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 1.0 | 41.3 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.9 | 51.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.8 | 0.8 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.7 | 24.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.7 | 8.9 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.6 | 24.0 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.6 | 4.0 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.6 | 15.9 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.5 | 19.9 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.5 | 31.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.5 | 28.3 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.5 | 10.4 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.4 | 8.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.4 | 6.5 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.4 | 24.2 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.4 | 11.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.3 | 4.0 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.3 | 5.5 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.3 | 19.2 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.3 | 42.4 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.3 | 5.3 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.3 | 3.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.3 | 23.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.3 | 7.9 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.3 | 10.1 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.3 | 6.2 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.2 | 8.0 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.2 | 6.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.2 | 8.5 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.2 | 8.5 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.2 | 5.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 4.1 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.2 | 2.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.2 | 6.5 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.2 | 10.5 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 3.6 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.1 | 1.8 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 5.7 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 8.3 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 4.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 6.0 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 1.9 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 3.2 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 4.8 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 4.5 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 2.5 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 3.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 1.5 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 3.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 2.1 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 0.8 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.4 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.2 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.7 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 9.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.9 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 3.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 66.5 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 1.7 | 31.5 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 1.3 | 35.4 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 1.2 | 18.9 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 1.0 | 19.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 1.0 | 8.9 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 1.0 | 41.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 1.0 | 26.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 1.0 | 27.0 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.9 | 37.9 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.9 | 17.1 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.8 | 28.0 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.8 | 16.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.7 | 12.7 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.7 | 26.4 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.7 | 36.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.7 | 26.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.7 | 19.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.7 | 10.7 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.7 | 9.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.7 | 51.7 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.7 | 11.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.7 | 20.4 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.7 | 10.5 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.6 | 17.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.6 | 12.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.6 | 12.8 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.5 | 11.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.5 | 10.6 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.5 | 11.5 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.5 | 5.7 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.5 | 12.7 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.4 | 8.9 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.4 | 6.4 | REACTOME ORC1 REMOVAL FROM CHROMATIN | Genes involved in Orc1 removal from chromatin |
| 0.4 | 32.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.4 | 16.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.3 | 24.4 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.3 | 14.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.3 | 2.5 | REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | Genes involved in p53-Independent G1/S DNA damage checkpoint |
| 0.3 | 8.8 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.3 | 3.3 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.3 | 13.0 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.2 | 1.7 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.2 | 6.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.2 | 4.5 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.2 | 6.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.2 | 5.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.2 | 8.0 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.2 | 6.1 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.2 | 5.7 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.2 | 1.6 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.2 | 5.9 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.2 | 2.9 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.2 | 21.0 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.2 | 4.0 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.2 | 4.5 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.2 | 3.7 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.2 | 2.3 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.2 | 5.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 2.0 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.2 | 17.4 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 4.4 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.1 | 10.8 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.1 | 2.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 3.4 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 9.5 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 16.6 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.1 | 2.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 2.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 4.9 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 2.0 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 4.1 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.1 | 6.0 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 2.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 1.2 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 3.3 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 4.9 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 0.2 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 8.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 2.1 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.1 | 7.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 4.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 1.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 8.7 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 1.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.9 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 1.5 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 1.3 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.0 | 1.0 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 8.6 | REACTOME METABOLISM OF CARBOHYDRATES | Genes involved in Metabolism of carbohydrates |
| 0.0 | 2.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.8 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.5 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.4 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 1.1 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.0 | 1.1 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |