Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for MSX1
Z-value: 0.95
Transcription factors associated with MSX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MSX1
|
ENSG00000163132.6 | msh homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MSX1 | hg19_v2_chr4_+_4861385_4861398 | -0.08 | 2.6e-01 | Click! |
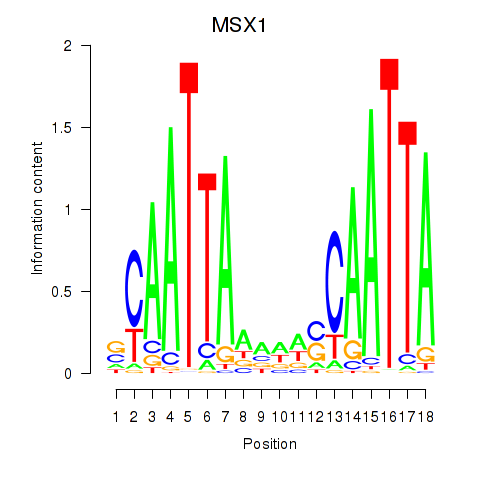
Activity profile of MSX1 motif
Sorted Z-values of MSX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_196313239 | 32.31 |
ENST00000413290.1
|
AC064834.1
|
AC064834.1 |
| chr13_-_88323218 | 26.68 |
ENST00000436290.2
ENST00000453832.2 ENST00000606590.1 |
MIR4500HG
|
MIR4500 host gene (non-protein coding) |
| chr5_+_150404904 | 17.51 |
ENST00000521632.1
|
GPX3
|
glutathione peroxidase 3 (plasma) |
| chrX_+_1710484 | 14.78 |
ENST00000313871.3
ENST00000381261.3 |
AKAP17A
|
A kinase (PRKA) anchor protein 17A |
| chr6_-_32095968 | 14.50 |
ENST00000375203.3
ENST00000375201.4 |
ATF6B
|
activating transcription factor 6 beta |
| chr2_-_136288113 | 13.48 |
ENST00000401392.1
|
ZRANB3
|
zinc finger, RAN-binding domain containing 3 |
| chrX_+_73164149 | 11.94 |
ENST00000602938.1
ENST00000602294.1 ENST00000602920.1 ENST00000602737.1 ENST00000602772.1 |
JPX
|
JPX transcript, XIST activator (non-protein coding) |
| chr6_-_32557610 | 11.88 |
ENST00000360004.5
|
HLA-DRB1
|
major histocompatibility complex, class II, DR beta 1 |
| chr18_-_5396271 | 10.68 |
ENST00000579951.1
|
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr6_-_84419101 | 10.60 |
ENST00000520302.1
ENST00000520213.1 ENST00000439399.2 ENST00000428679.2 ENST00000437520.1 |
SNAP91
|
synaptosomal-associated protein, 91kDa |
| chr13_-_38172863 | 10.56 |
ENST00000541481.1
ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN
|
periostin, osteoblast specific factor |
| chr22_+_51176624 | 10.54 |
ENST00000216139.5
ENST00000529621.1 |
ACR
|
acrosin |
| chr19_-_53757941 | 10.44 |
ENST00000594517.1
ENST00000601413.1 ENST00000594681.1 |
ZNF677
|
zinc finger protein 677 |
| chr1_-_160492994 | 10.42 |
ENST00000368055.1
ENST00000368057.3 ENST00000368059.3 |
SLAMF6
|
SLAM family member 6 |
| chr3_-_123339418 | 10.25 |
ENST00000583087.1
|
MYLK
|
myosin light chain kinase |
| chr5_-_11588907 | 10.18 |
ENST00000513598.1
ENST00000503622.1 |
CTNND2
|
catenin (cadherin-associated protein), delta 2 |
| chr11_-_85376121 | 10.04 |
ENST00000527447.1
|
CREBZF
|
CREB/ATF bZIP transcription factor |
| chr3_-_123339343 | 9.84 |
ENST00000578202.1
|
MYLK
|
myosin light chain kinase |
| chr11_+_92085262 | 9.83 |
ENST00000298047.6
ENST00000409404.2 ENST00000541502.1 |
FAT3
|
FAT atypical cadherin 3 |
| chr4_-_57547454 | 9.63 |
ENST00000556376.2
|
HOPX
|
HOP homeobox |
| chr6_-_52705641 | 9.34 |
ENST00000370989.2
|
GSTA5
|
glutathione S-transferase alpha 5 |
| chr19_-_53758094 | 9.11 |
ENST00000601828.1
ENST00000598513.1 ENST00000599012.1 ENST00000333952.4 ENST00000598806.1 |
ZNF677
|
zinc finger protein 677 |
| chr5_+_121647386 | 8.58 |
ENST00000542191.1
ENST00000506272.1 ENST00000508681.1 ENST00000509154.2 |
SNCAIP
|
synuclein, alpha interacting protein |
| chr13_-_52733980 | 8.43 |
ENST00000339406.3
|
NEK3
|
NIMA-related kinase 3 |
| chr6_-_32908765 | 8.12 |
ENST00000416244.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr3_-_21792838 | 7.86 |
ENST00000281523.2
|
ZNF385D
|
zinc finger protein 385D |
| chr7_-_36634181 | 7.76 |
ENST00000538464.1
|
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chr11_-_128894053 | 7.56 |
ENST00000392657.3
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr5_+_67586465 | 7.51 |
ENST00000336483.5
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr14_+_22964877 | 7.40 |
ENST00000390494.1
|
TRAJ43
|
T cell receptor alpha joining 43 |
| chr16_-_66584059 | 7.27 |
ENST00000417693.3
ENST00000544898.1 ENST00000569718.1 ENST00000527284.1 ENST00000299697.7 ENST00000451102.2 |
TK2
|
thymidine kinase 2, mitochondrial |
| chr10_-_101380121 | 7.14 |
ENST00000370495.4
|
SLC25A28
|
solute carrier family 25 (mitochondrial iron transporter), member 28 |
| chr14_-_75389975 | 6.93 |
ENST00000555647.1
ENST00000557413.1 |
RPS6KL1
|
ribosomal protein S6 kinase-like 1 |
| chr1_+_158985457 | 6.93 |
ENST00000567661.1
ENST00000474473.1 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr12_+_9980069 | 6.83 |
ENST00000354855.3
ENST00000324214.4 ENST00000279544.3 |
KLRF1
|
killer cell lectin-like receptor subfamily F, member 1 |
| chr3_-_123512688 | 6.79 |
ENST00000475616.1
|
MYLK
|
myosin light chain kinase |
| chr13_+_49551020 | 6.30 |
ENST00000541916.1
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr5_-_75919253 | 6.00 |
ENST00000296641.4
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2 |
| chr15_-_77712477 | 5.99 |
ENST00000560626.2
|
PEAK1
|
pseudopodium-enriched atypical kinase 1 |
| chr5_-_177207634 | 5.90 |
ENST00000513554.1
ENST00000440605.3 |
FAM153A
|
family with sequence similarity 153, member A |
| chr6_+_6588316 | 5.89 |
ENST00000379953.2
|
LY86
|
lymphocyte antigen 86 |
| chr6_+_134758827 | 5.88 |
ENST00000431422.1
|
LINC01010
|
long intergenic non-protein coding RNA 1010 |
| chr6_+_131894284 | 5.83 |
ENST00000368087.3
ENST00000356962.2 |
ARG1
|
arginase 1 |
| chr2_-_88927092 | 5.65 |
ENST00000303236.3
|
EIF2AK3
|
eukaryotic translation initiation factor 2-alpha kinase 3 |
| chr14_-_75389925 | 5.41 |
ENST00000556776.1
|
RPS6KL1
|
ribosomal protein S6 kinase-like 1 |
| chr11_+_12132117 | 5.34 |
ENST00000256194.4
|
MICAL2
|
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
| chr3_+_108308513 | 5.27 |
ENST00000361582.3
|
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr22_-_32766972 | 5.17 |
ENST00000382084.4
ENST00000382086.2 |
RFPL3S
|
RFPL3 antisense |
| chr2_-_158345462 | 5.13 |
ENST00000439355.1
ENST00000540637.1 |
CYTIP
|
cytohesin 1 interacting protein |
| chr11_+_101785727 | 4.98 |
ENST00000263468.8
|
KIAA1377
|
KIAA1377 |
| chr3_-_93781750 | 4.94 |
ENST00000314636.2
|
DHFRL1
|
dihydrofolate reductase-like 1 |
| chr5_+_175490540 | 4.94 |
ENST00000515817.1
|
FAM153B
|
family with sequence similarity 153, member B |
| chr3_+_171561127 | 4.90 |
ENST00000334567.5
ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr14_+_22293618 | 4.82 |
ENST00000390432.2
|
TRAV10
|
T cell receptor alpha variable 10 |
| chr1_+_158815588 | 4.77 |
ENST00000438394.1
|
MNDA
|
myeloid cell nuclear differentiation antigen |
| chr6_+_26020672 | 4.57 |
ENST00000357647.3
|
HIST1H3A
|
histone cluster 1, H3a |
| chr8_+_28748765 | 4.45 |
ENST00000355231.5
|
HMBOX1
|
homeobox containing 1 |
| chr22_-_32767017 | 4.39 |
ENST00000400234.1
|
RFPL3S
|
RFPL3 antisense |
| chr16_-_66583701 | 4.38 |
ENST00000527800.1
ENST00000525974.1 ENST00000563369.2 |
TK2
|
thymidine kinase 2, mitochondrial |
| chr18_+_61637159 | 4.36 |
ENST00000397985.2
ENST00000353706.2 ENST00000542677.1 ENST00000397988.3 ENST00000448851.1 |
SERPINB8
|
serpin peptidase inhibitor, clade B (ovalbumin), member 8 |
| chr14_+_50291993 | 4.22 |
ENST00000595378.1
|
AL627171.2
|
HCG1786899; PRO2610; Uncharacterized protein |
| chr2_+_32502952 | 4.08 |
ENST00000238831.4
|
YIPF4
|
Yip1 domain family, member 4 |
| chr2_-_209028300 | 4.05 |
ENST00000304502.4
|
CRYGA
|
crystallin, gamma A |
| chr4_-_120243545 | 4.02 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr10_+_13142225 | 3.89 |
ENST00000378747.3
|
OPTN
|
optineurin |
| chr12_+_56324756 | 3.80 |
ENST00000331886.5
ENST00000555090.1 |
DGKA
|
diacylglycerol kinase, alpha 80kDa |
| chr5_-_75919217 | 3.78 |
ENST00000504899.1
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2 |
| chr14_+_55595762 | 3.50 |
ENST00000254301.9
|
LGALS3
|
lectin, galactoside-binding, soluble, 3 |
| chr1_+_111415757 | 3.37 |
ENST00000429072.2
ENST00000271324.5 |
CD53
|
CD53 molecule |
| chr19_-_7812397 | 3.35 |
ENST00000593660.1
ENST00000354397.6 ENST00000593821.1 ENST00000602261.1 ENST00000315591.8 ENST00000394161.5 ENST00000204801.8 ENST00000601256.1 ENST00000601951.1 ENST00000315599.7 |
CD209
|
CD209 molecule |
| chr10_-_29923893 | 3.32 |
ENST00000355867.4
|
SVIL
|
supervillin |
| chr11_-_66964638 | 3.25 |
ENST00000444002.2
|
AP001885.1
|
AP001885.1 |
| chr3_-_190040223 | 3.22 |
ENST00000295522.3
|
CLDN1
|
claudin 1 |
| chr4_+_5527117 | 3.21 |
ENST00000505296.1
|
C4orf6
|
chromosome 4 open reading frame 6 |
| chr9_+_115913222 | 3.17 |
ENST00000259392.3
|
SLC31A2
|
solute carrier family 31 (copper transporter), member 2 |
| chrX_+_11311533 | 3.12 |
ENST00000380714.3
ENST00000380712.3 ENST00000348912.4 |
AMELX
|
amelogenin, X-linked |
| chr12_+_122688090 | 3.08 |
ENST00000324189.4
ENST00000546192.1 |
B3GNT4
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 4 |
| chr5_-_137475071 | 3.08 |
ENST00000265191.2
|
NME5
|
NME/NM23 family member 5 |
| chr14_-_101295407 | 3.06 |
ENST00000596284.1
|
AL117190.2
|
AL117190.2 |
| chr15_-_42500351 | 2.99 |
ENST00000348544.4
ENST00000318006.5 |
VPS39
|
vacuolar protein sorting 39 homolog (S. cerevisiae) |
| chr2_-_109605663 | 2.96 |
ENST00000409271.1
ENST00000258443.2 ENST00000376651.1 |
EDAR
|
ectodysplasin A receptor |
| chr3_+_93781728 | 2.93 |
ENST00000314622.4
|
NSUN3
|
NOP2/Sun domain family, member 3 |
| chr10_+_17270214 | 2.90 |
ENST00000544301.1
|
VIM
|
vimentin |
| chr5_+_82767487 | 2.89 |
ENST00000343200.5
ENST00000342785.4 |
VCAN
|
versican |
| chr5_+_140762268 | 2.89 |
ENST00000518325.1
|
PCDHGA7
|
protocadherin gamma subfamily A, 7 |
| chr19_-_7812446 | 2.87 |
ENST00000394173.4
ENST00000301357.8 |
CD209
|
CD209 molecule |
| chr5_+_140220769 | 2.86 |
ENST00000531613.1
ENST00000378123.3 |
PCDHA8
|
protocadherin alpha 8 |
| chr19_-_53662257 | 2.85 |
ENST00000599096.1
ENST00000334197.7 ENST00000597183.1 ENST00000601804.1 ENST00000601469.2 ENST00000452676.2 |
ZNF347
|
zinc finger protein 347 |
| chr7_+_2636522 | 2.81 |
ENST00000423196.1
|
IQCE
|
IQ motif containing E |
| chr21_+_43619796 | 2.79 |
ENST00000398457.2
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr10_+_135050908 | 2.78 |
ENST00000325980.9
|
VENTX
|
VENT homeobox |
| chr7_-_32338917 | 2.76 |
ENST00000396193.1
|
PDE1C
|
phosphodiesterase 1C, calmodulin-dependent 70kDa |
| chr6_+_118869452 | 2.71 |
ENST00000357525.5
|
PLN
|
phospholamban |
| chr5_+_82767583 | 2.62 |
ENST00000512590.2
ENST00000513960.1 ENST00000513984.1 ENST00000502527.2 |
VCAN
|
versican |
| chr8_+_1993152 | 2.54 |
ENST00000262113.4
|
MYOM2
|
myomesin 2 |
| chr8_-_121824374 | 2.53 |
ENST00000517992.1
|
SNTB1
|
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1) |
| chr11_+_62037622 | 2.47 |
ENST00000227918.2
ENST00000525380.1 |
SCGB2A2
|
secretoglobin, family 2A, member 2 |
| chr2_+_168675182 | 2.45 |
ENST00000305861.1
|
B3GALT1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr9_+_79074068 | 2.42 |
ENST00000444201.2
ENST00000376730.4 |
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2 |
| chr19_+_55043977 | 2.36 |
ENST00000335056.3
|
KIR3DX1
|
killer cell immunoglobulin-like receptor, three domains, X1 |
| chr4_+_5526883 | 2.34 |
ENST00000195455.2
|
C4orf6
|
chromosome 4 open reading frame 6 |
| chr1_+_160160283 | 2.31 |
ENST00000368079.3
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr18_+_29598335 | 2.23 |
ENST00000217740.3
|
RNF125
|
ring finger protein 125, E3 ubiquitin protein ligase |
| chr13_-_47471155 | 2.18 |
ENST00000543956.1
ENST00000542664.1 |
HTR2A
|
5-hydroxytryptamine (serotonin) receptor 2A, G protein-coupled |
| chr14_+_68086515 | 2.06 |
ENST00000261783.3
|
ARG2
|
arginase 2 |
| chr1_-_167487808 | 2.05 |
ENST00000392122.3
|
CD247
|
CD247 molecule |
| chr1_-_167487758 | 1.99 |
ENST00000362089.5
|
CD247
|
CD247 molecule |
| chr12_+_56324933 | 1.95 |
ENST00000549629.1
ENST00000555218.1 |
DGKA
|
diacylglycerol kinase, alpha 80kDa |
| chr7_+_99425633 | 1.88 |
ENST00000354829.2
ENST00000421837.2 ENST00000417625.1 ENST00000342499.4 ENST00000444905.1 ENST00000415413.1 ENST00000312017.5 ENST00000222382.5 |
CYP3A43
|
cytochrome P450, family 3, subfamily A, polypeptide 43 |
| chr8_-_72274095 | 1.84 |
ENST00000303824.7
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr4_-_100140331 | 1.81 |
ENST00000407820.2
ENST00000394897.1 ENST00000508558.1 ENST00000394899.2 |
ADH6
|
alcohol dehydrogenase 6 (class V) |
| chr12_+_106751436 | 1.72 |
ENST00000228347.4
|
POLR3B
|
polymerase (RNA) III (DNA directed) polypeptide B |
| chr6_+_136172820 | 1.69 |
ENST00000308191.6
|
PDE7B
|
phosphodiesterase 7B |
| chrY_-_2655644 | 1.69 |
ENST00000525526.2
ENST00000534739.2 ENST00000383070.1 |
SRY
|
sex determining region Y |
| chr9_+_90112117 | 1.57 |
ENST00000358077.5
|
DAPK1
|
death-associated protein kinase 1 |
| chr8_+_1993173 | 1.54 |
ENST00000523438.1
|
MYOM2
|
myomesin 2 |
| chr1_+_160160346 | 1.51 |
ENST00000368078.3
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr19_-_20150014 | 1.37 |
ENST00000358523.5
ENST00000397162.1 ENST00000601100.1 |
ZNF682
|
zinc finger protein 682 |
| chr6_-_32784687 | 1.35 |
ENST00000447394.1
ENST00000438763.2 |
HLA-DOB
|
major histocompatibility complex, class II, DO beta |
| chr10_+_32873190 | 1.30 |
ENST00000375025.4
|
C10orf68
|
Homo sapiens coiled-coil domain containing 7 (CCDC7), transcript variant 5, mRNA. |
| chr2_-_219031709 | 1.28 |
ENST00000295683.2
|
CXCR1
|
chemokine (C-X-C motif) receptor 1 |
| chr8_-_17555164 | 1.25 |
ENST00000297488.6
|
MTUS1
|
microtubule associated tumor suppressor 1 |
| chr2_-_77749474 | 1.24 |
ENST00000409093.1
ENST00000409088.3 |
LRRTM4
|
leucine rich repeat transmembrane neuronal 4 |
| chr10_+_13141585 | 1.20 |
ENST00000378764.2
|
OPTN
|
optineurin |
| chr9_+_133539981 | 1.16 |
ENST00000253008.2
|
PRDM12
|
PR domain containing 12 |
| chr13_+_50570019 | 1.14 |
ENST00000442421.1
|
TRIM13
|
tripartite motif containing 13 |
| chr11_-_102401469 | 1.06 |
ENST00000260227.4
|
MMP7
|
matrix metallopeptidase 7 (matrilysin, uterine) |
| chr7_-_20256965 | 1.03 |
ENST00000400331.5
ENST00000332878.4 |
MACC1
|
metastasis associated in colon cancer 1 |
| chr6_+_29141311 | 1.00 |
ENST00000377167.2
|
OR2J2
|
olfactory receptor, family 2, subfamily J, member 2 |
| chr5_+_82767284 | 0.96 |
ENST00000265077.3
|
VCAN
|
versican |
| chr14_+_55595960 | 0.92 |
ENST00000554715.1
|
LGALS3
|
lectin, galactoside-binding, soluble, 3 |
| chr6_+_28092338 | 0.91 |
ENST00000340487.4
|
ZSCAN16
|
zinc finger and SCAN domain containing 16 |
| chr6_-_109777128 | 0.91 |
ENST00000358807.3
ENST00000358577.3 |
MICAL1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr2_-_31361543 | 0.84 |
ENST00000349752.5
|
GALNT14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr22_+_41697520 | 0.69 |
ENST00000352645.4
|
ZC3H7B
|
zinc finger CCCH-type containing 7B |
| chr11_-_102496063 | 0.65 |
ENST00000260228.2
|
MMP20
|
matrix metallopeptidase 20 |
| chr12_+_9980113 | 0.54 |
ENST00000537723.1
|
KLRF1
|
killer cell lectin-like receptor subfamily F, member 1 |
| chr9_-_15510287 | 0.53 |
ENST00000397519.2
|
PSIP1
|
PC4 and SFRS1 interacting protein 1 |
| chr18_-_3874271 | 0.44 |
ENST00000400149.3
ENST00000400155.1 ENST00000400150.3 |
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr4_-_186696425 | 0.29 |
ENST00000430503.1
ENST00000319454.6 ENST00000450341.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr1_-_149908217 | 0.21 |
ENST00000369140.3
|
MTMR11
|
myotubularin related protein 11 |
| chr8_-_25281747 | 0.15 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr20_+_56964169 | 0.11 |
ENST00000475243.1
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr8_+_24241969 | 0.08 |
ENST00000522298.1
|
ADAMDEC1
|
ADAM-like, decysin 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of MSX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.8 | 17.5 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 4.0 | 20.0 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 3.5 | 10.6 | GO:1990523 | bone regeneration(GO:1990523) |
| 3.4 | 26.9 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 2.9 | 11.6 | GO:0046104 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 2.7 | 13.5 | GO:0048478 | replication fork protection(GO:0048478) |
| 2.4 | 7.1 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 2.1 | 6.2 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 1.9 | 5.7 | GO:1990737 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 1.5 | 10.7 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 1.5 | 10.5 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 1.5 | 4.4 | GO:2000521 | negative regulation of immunological synapse formation(GO:2000521) |
| 1.2 | 6.2 | GO:0046968 | peptide antigen transport(GO:0046968) |
| 1.2 | 14.5 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 1.2 | 5.8 | GO:0090467 | L-arginine import(GO:0043091) arginine import(GO:0090467) L-arginine transport(GO:1902023) |
| 1.1 | 3.2 | GO:1903348 | cellular response to lead ion(GO:0071284) positive regulation of bicellular tight junction assembly(GO:1903348) |
| 1.1 | 8.4 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 1.0 | 3.1 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 1.0 | 2.9 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.9 | 2.8 | GO:0009726 | detection of endogenous stimulus(GO:0009726) |
| 0.9 | 2.7 | GO:0086092 | regulation of the force of heart contraction by cardiac conduction(GO:0086092) regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.7 | 5.9 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.6 | 5.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.6 | 10.4 | GO:0001787 | natural killer cell proliferation(GO:0001787) |
| 0.6 | 3.0 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.5 | 3.1 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.5 | 4.1 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.5 | 4.6 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.4 | 1.3 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.4 | 9.8 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.4 | 7.5 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.4 | 2.2 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.4 | 1.3 | GO:0098759 | response to interleukin-8(GO:0098758) cellular response to interleukin-8(GO:0098759) |
| 0.4 | 9.6 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.4 | 3.8 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.4 | 10.6 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.4 | 3.2 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.3 | 2.4 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.3 | 1.2 | GO:1900109 | regulation of histone H3-K9 dimethylation(GO:1900109) |
| 0.3 | 6.3 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.3 | 4.4 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.3 | 3.0 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.3 | 2.9 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.3 | 8.6 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.3 | 6.5 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.3 | 3.1 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.3 | 5.8 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.2 | 2.1 | GO:0000050 | urea cycle(GO:0000050) |
| 0.2 | 1.8 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.2 | 2.2 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.2 | 4.5 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.2 | 2.4 | GO:0060352 | cell adhesion molecule production(GO:0060352) |
| 0.2 | 3.4 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.2 | 4.8 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.2 | 1.8 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.2 | 2.6 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.2 | 1.7 | GO:2000020 | positive regulation of male gonad development(GO:2000020) |
| 0.1 | 10.2 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.1 | 4.0 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 6.9 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 0.7 | GO:0070170 | regulation of tooth mineralization(GO:0070170) |
| 0.1 | 9.3 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 4.0 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 15.6 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 1.6 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.1 | 6.4 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.1 | 0.1 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.1 | 0.5 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 1.1 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.0 | 6.0 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 1.7 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 1.7 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 10.9 | GO:0042113 | B cell activation(GO:0042113) |
| 0.0 | 1.2 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 10.0 | GO:0009615 | response to virus(GO:0009615) |
| 0.0 | 3.3 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 4.1 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.8 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.7 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 10.5 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 1.9 | 7.5 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 1.1 | 5.3 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 1.0 | 21.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.6 | 10.7 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.5 | 3.8 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.5 | 3.3 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.4 | 4.0 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.4 | 26.9 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.3 | 4.6 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.3 | 2.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.3 | 13.5 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.3 | 3.0 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.2 | 4.1 | GO:0032982 | myosin filament(GO:0032982) |
| 0.2 | 2.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 20.2 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 7.8 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 10.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 4.5 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 2.7 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.1 | 10.6 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.1 | 1.1 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 5.1 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 2.5 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 8.1 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 8.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 6.3 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 10.3 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 2.9 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 1.7 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 6.2 | GO:0018995 | host(GO:0018995) host cell(GO:0043657) |
| 0.1 | 6.0 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 7.6 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.1 | 6.3 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 3.2 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 12.1 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 3.1 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 13.0 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 8.4 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 12.8 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.5 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 2.4 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 7.1 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 3.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 5.1 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 2.9 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.9 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 7.9 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 23.4 | GO:0005615 | extracellular space(GO:0005615) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.4 | 26.9 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 3.9 | 11.6 | GO:0004797 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 3.5 | 10.5 | GO:0004040 | amidase activity(GO:0004040) fucose binding(GO:0042806) |
| 2.6 | 7.8 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 1.9 | 5.7 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 1.6 | 9.8 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 1.6 | 17.5 | GO:0008430 | selenium binding(GO:0008430) |
| 1.3 | 13.5 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 1.0 | 3.1 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 1.0 | 3.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.9 | 7.5 | GO:0043559 | insulin binding(GO:0043559) |
| 0.9 | 2.8 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.8 | 2.4 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.8 | 2.4 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.7 | 14.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.7 | 21.4 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.7 | 2.7 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.6 | 3.2 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.6 | 2.9 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.6 | 7.4 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.6 | 6.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.6 | 7.9 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.6 | 4.4 | GO:0019863 | IgE binding(GO:0019863) |
| 0.5 | 6.4 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.5 | 2.9 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.5 | 10.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.4 | 7.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.4 | 4.5 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.4 | 1.3 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.3 | 5.8 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.3 | 1.8 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.3 | 2.8 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.3 | 5.1 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.2 | 6.2 | GO:0071949 | FAD binding(GO:0071949) |
| 0.2 | 6.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 9.3 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.2 | 2.2 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.2 | 4.1 | GO:0097493 | muscle alpha-actinin binding(GO:0051371) structural molecule activity conferring elasticity(GO:0097493) |
| 0.2 | 1.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.2 | 10.4 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.1 | 3.1 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 4.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 1.9 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 4.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 10.2 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 1.7 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.1 | 4.0 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 6.0 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 2.2 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 1.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 11.6 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 0.8 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 1.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.5 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.0 | 4.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 4.0 | GO:1990782 | protein tyrosine kinase binding(GO:1990782) |
| 0.0 | 6.9 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 6.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 3.2 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 2.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 7.6 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 3.3 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.8 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.7 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 10.0 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 1.2 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 9.8 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.5 | 26.9 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.3 | 7.5 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.3 | 15.9 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.2 | 6.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 4.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 3.2 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 7.6 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 5.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 2.7 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 5.8 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 1.3 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 1.6 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 13.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 2.7 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 3.3 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.7 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 5.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 15.9 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.4 | 26.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.4 | 11.6 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.4 | 7.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.3 | 9.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 6.5 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.2 | 1.8 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.2 | 9.8 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.2 | 2.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.2 | 4.4 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.2 | 5.7 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.1 | 6.3 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 2.8 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 5.8 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 1.6 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 2.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 1.7 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 9.5 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 0.5 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.1 | 2.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 2.2 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 5.9 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |
| 0.0 | 5.6 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 2.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 1.9 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 1.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.7 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |