Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
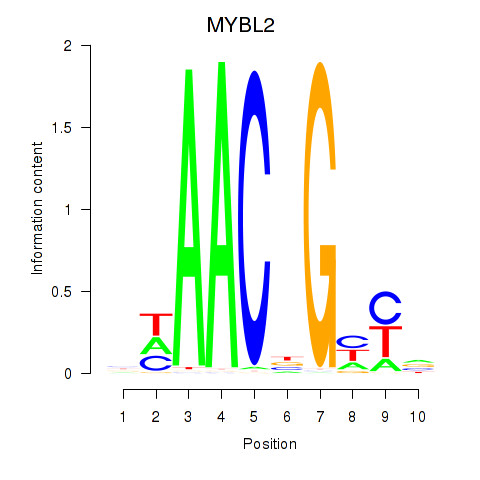
Results for MYBL2
Z-value: 2.21
Transcription factors associated with MYBL2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MYBL2
|
ENSG00000101057.11 | MYB proto-oncogene like 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MYBL2 | hg19_v2_chr20_+_42295745_42295797 | 0.56 | 3.7e-19 | Click! |
Activity profile of MYBL2 motif
Sorted Z-values of MYBL2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MYBL2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 16.3 | 65.3 | GO:0090579 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 15.8 | 47.3 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 13.6 | 40.9 | GO:1900195 | spindle assembly involved in female meiosis I(GO:0007057) positive regulation of oocyte maturation(GO:1900195) |
| 11.1 | 44.5 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 10.8 | 43.1 | GO:0046035 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 10.1 | 30.4 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 8.5 | 76.4 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 8.1 | 80.9 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 6.7 | 93.4 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 6.1 | 49.1 | GO:0060699 | regulation of endoribonuclease activity(GO:0060699) |
| 5.5 | 27.4 | GO:0090306 | spindle assembly involved in meiosis(GO:0090306) |
| 5.5 | 54.8 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 5.4 | 32.4 | GO:0019060 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 5.3 | 32.1 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 5.0 | 29.8 | GO:0019249 | lactate biosynthetic process(GO:0019249) |
| 4.9 | 44.5 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 4.7 | 37.6 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 4.6 | 18.4 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 4.5 | 13.4 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 4.4 | 8.9 | GO:0044211 | CTP salvage(GO:0044211) |
| 4.4 | 8.8 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 4.3 | 51.9 | GO:0015074 | DNA integration(GO:0015074) |
| 4.3 | 12.8 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 4.0 | 11.9 | GO:1901535 | regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) regulation of genetic imprinting(GO:2000653) |
| 3.9 | 11.7 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 3.9 | 7.7 | GO:0097695 | establishment of RNA localization to telomere(GO:0097694) establishment of macromolecular complex localization to telomere(GO:0097695) |
| 3.8 | 34.1 | GO:0030091 | protein repair(GO:0030091) |
| 3.7 | 11.2 | GO:0036333 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 3.7 | 63.5 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 3.5 | 56.5 | GO:0000338 | protein deneddylation(GO:0000338) |
| 3.5 | 65.8 | GO:0000022 | mitotic spindle elongation(GO:0000022) |
| 3.3 | 90.2 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 3.3 | 13.1 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 3.2 | 9.6 | GO:0044209 | AMP salvage(GO:0044209) |
| 3.2 | 373.7 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) |
| 3.2 | 15.8 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 3.0 | 12.2 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 3.0 | 14.8 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 2.9 | 11.6 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 2.9 | 8.6 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 2.7 | 21.7 | GO:0015866 | ADP transport(GO:0015866) |
| 2.7 | 16.2 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 2.7 | 24.0 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 2.5 | 27.8 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 2.5 | 4.9 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 2.5 | 12.3 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 2.5 | 12.3 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 2.4 | 94.6 | GO:2000816 | negative regulation of mitotic sister chromatid separation(GO:2000816) |
| 2.1 | 34.3 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 2.1 | 31.9 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 2.1 | 6.3 | GO:0060152 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 2.1 | 6.2 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 2.1 | 20.7 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 2.0 | 6.0 | GO:0071048 | generation of catalytic spliceosome for second transesterification step(GO:0000350) nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 2.0 | 6.0 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 2.0 | 13.8 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 1.9 | 13.5 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 1.9 | 7.6 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 1.9 | 17.0 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 1.9 | 7.5 | GO:0021564 | vagus nerve development(GO:0021564) |
| 1.9 | 97.0 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 1.9 | 5.6 | GO:0019482 | purine nucleobase catabolic process(GO:0006145) beta-alanine metabolic process(GO:0019482) |
| 1.9 | 46.4 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 1.8 | 7.4 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 1.8 | 35.0 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 1.8 | 7.3 | GO:0046833 | positive regulation of RNA export from nucleus(GO:0046833) |
| 1.8 | 38.3 | GO:0046931 | pore complex assembly(GO:0046931) |
| 1.8 | 32.6 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 1.8 | 23.4 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 1.8 | 14.4 | GO:1900004 | negative regulation of serine-type endopeptidase activity(GO:1900004) negative regulation of serine-type peptidase activity(GO:1902572) |
| 1.7 | 27.7 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 1.7 | 15.3 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 1.7 | 5.0 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 1.7 | 8.3 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 1.7 | 13.3 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 1.6 | 3.2 | GO:1903423 | positive regulation of synaptic vesicle endocytosis(GO:1900244) positive regulation of synaptic vesicle recycling(GO:1903423) |
| 1.6 | 11.1 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 1.6 | 44.1 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 1.6 | 9.4 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 1.6 | 17.2 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 1.5 | 4.6 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 1.5 | 21.4 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 1.5 | 22.4 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 1.5 | 7.4 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 1.4 | 24.2 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 1.4 | 21.1 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 1.4 | 2.8 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 1.4 | 30.2 | GO:0048255 | mRNA stabilization(GO:0048255) |
| 1.3 | 4.0 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 1.3 | 6.4 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 1.3 | 6.4 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 1.3 | 7.6 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) positive regulation of autophagosome maturation(GO:1901098) |
| 1.3 | 7.6 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 1.3 | 8.8 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 1.2 | 7.5 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 1.2 | 3.6 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 1.2 | 10.6 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 1.2 | 4.7 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 1.2 | 13.9 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 1.1 | 5.5 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) negative regulation by host of symbiont molecular function(GO:0052405) |
| 1.1 | 3.2 | GO:0046010 | positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) |
| 1.1 | 6.4 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 1.1 | 5.3 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 1.0 | 15.7 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 1.0 | 5.1 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 1.0 | 13.0 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 1.0 | 3.0 | GO:0060437 | lung growth(GO:0060437) |
| 1.0 | 7.9 | GO:0019695 | choline metabolic process(GO:0019695) |
| 1.0 | 95.1 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.9 | 2.8 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.9 | 36.5 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.9 | 3.7 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.9 | 3.6 | GO:1903436 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.9 | 16.9 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.9 | 19.3 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.9 | 5.3 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.9 | 8.7 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.8 | 9.8 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.8 | 18.8 | GO:0072663 | protein targeting to peroxisome(GO:0006625) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.8 | 22.1 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.8 | 5.6 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.8 | 8.7 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.8 | 15.5 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.8 | 5.4 | GO:0048511 | rhythmic process(GO:0048511) |
| 0.8 | 6.1 | GO:0010624 | regulation of Schwann cell proliferation(GO:0010624) negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.8 | 6.8 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.7 | 6.4 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.7 | 2.1 | GO:1904578 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) response to hypobaric hypoxia(GO:1990910) |
| 0.7 | 2.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.7 | 25.9 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.7 | 3.5 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.7 | 50.0 | GO:0070125 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 0.7 | 29.4 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.7 | 10.9 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.7 | 6.1 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.7 | 4.0 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.7 | 5.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.6 | 2.5 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.6 | 12.6 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.6 | 12.9 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.6 | 9.2 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.6 | 21.6 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.6 | 21.8 | GO:1902475 | neutral amino acid transport(GO:0015804) L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.6 | 1.1 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.5 | 8.1 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.5 | 4.2 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.5 | 14.3 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.5 | 13.3 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.5 | 4.9 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.5 | 1.9 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.5 | 26.8 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.5 | 2.7 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.4 | 0.9 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.4 | 2.2 | GO:0006581 | acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) |
| 0.4 | 1.8 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of nephron tubule epithelial cell differentiation(GO:0072183) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) negative regulation of epithelial cell differentiation involved in kidney development(GO:2000697) |
| 0.4 | 5.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.4 | 1.7 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.4 | 6.3 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.4 | 2.9 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.4 | 1.2 | GO:0070233 | regulation of T cell apoptotic process(GO:0070232) negative regulation of T cell apoptotic process(GO:0070233) regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.4 | 4.1 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.4 | 25.5 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.4 | 1.9 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.4 | 6.2 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.4 | 12.5 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.4 | 4.1 | GO:0030656 | regulation of vitamin metabolic process(GO:0030656) |
| 0.4 | 2.6 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.4 | 2.6 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.4 | 3.7 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.4 | 5.1 | GO:0006293 | nucleotide-excision repair, preincision complex stabilization(GO:0006293) nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.4 | 16.5 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.4 | 1.4 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.3 | 4.4 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.3 | 6.2 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.3 | 12.2 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.3 | 1.6 | GO:1901906 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.3 | 20.2 | GO:0001895 | retina homeostasis(GO:0001895) |
| 0.3 | 1.1 | GO:0010454 | negative regulation of cell fate commitment(GO:0010454) mammary gland bud morphogenesis(GO:0060648) regulation of determination of dorsal identity(GO:2000015) |
| 0.3 | 10.6 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.3 | 8.1 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.3 | 16.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.2 | 15.9 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.2 | 10.0 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.2 | 4.2 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.2 | 14.1 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.2 | 14.1 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.2 | 3.5 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.2 | 11.5 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.2 | 9.6 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.2 | 3.6 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.2 | 9.4 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.2 | 7.8 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.2 | 5.7 | GO:0016071 | mRNA metabolic process(GO:0016071) |
| 0.2 | 5.3 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.2 | 0.9 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) angiotensin maturation(GO:0002003) regulation of angiotensin metabolic process(GO:0060177) |
| 0.2 | 4.8 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.1 | 2.4 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.1 | 1.0 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 2.2 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.1 | 1.9 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 2.8 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.1 | 0.9 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.1 | 9.7 | GO:0035722 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.1 | 1.3 | GO:0017145 | stem cell division(GO:0017145) |
| 0.1 | 2.6 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 8.3 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.1 | 2.3 | GO:0045577 | regulation of B cell differentiation(GO:0045577) |
| 0.1 | 11.0 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 4.2 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.1 | 3.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 1.5 | GO:0046628 | positive regulation of insulin receptor signaling pathway(GO:0046628) |
| 0.1 | 3.2 | GO:0006333 | chromatin assembly or disassembly(GO:0006333) |
| 0.1 | 1.8 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.1 | 0.7 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.1 | 0.6 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 0.6 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 | 1.4 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.1 | 8.6 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.1 | 3.9 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.1 | 3.0 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.1 | 1.1 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.1 | 8.3 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.1 | 0.3 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.0 | 1.4 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 2.1 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 7.6 | GO:0070585 | protein localization to mitochondrion(GO:0070585) |
| 0.0 | 1.0 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 2.5 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.5 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.6 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) |
| 0.0 | 0.5 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.9 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 0.7 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.3 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.0 | 0.1 | GO:0035801 | adrenal cortex development(GO:0035801) adrenal cortex formation(GO:0035802) visceral serous pericardium development(GO:0061032) posterior mesonephric tubule development(GO:0072166) negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.0 | 0.6 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.4 | GO:0051353 | positive regulation of oxidoreductase activity(GO:0051353) |
| 0.0 | 2.2 | GO:0002576 | platelet degranulation(GO:0002576) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 19.1 | 76.4 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 14.8 | 44.5 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 9.2 | 73.5 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 8.1 | 32.4 | GO:0036502 | Derlin-1-VIMP complex(GO:0036502) |
| 7.4 | 66.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 6.9 | 69.4 | GO:0070938 | contractile ring(GO:0070938) |
| 6.5 | 65.3 | GO:0097255 | R2TP complex(GO:0097255) |
| 5.9 | 82.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 4.1 | 124.2 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 3.8 | 15.3 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 3.7 | 33.3 | GO:0000125 | PCAF complex(GO:0000125) |
| 3.3 | 117.2 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 3.0 | 27.0 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 2.9 | 34.9 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 2.7 | 21.6 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 2.7 | 10.7 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 2.6 | 71.1 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 2.4 | 40.6 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 2.3 | 13.9 | GO:0070545 | PeBoW complex(GO:0070545) |
| 2.3 | 13.8 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 2.2 | 15.5 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 2.2 | 10.9 | GO:0071986 | Ragulator complex(GO:0071986) |
| 2.1 | 12.5 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 2.0 | 6.0 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 2.0 | 11.8 | GO:0000796 | condensin complex(GO:0000796) |
| 1.9 | 29.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 1.9 | 5.8 | GO:1903349 | omegasome membrane(GO:1903349) |
| 1.9 | 61.8 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 1.9 | 11.5 | GO:1990037 | Lewy body core(GO:1990037) |
| 1.9 | 35.5 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 1.6 | 41.0 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 1.6 | 4.7 | GO:0031213 | RSF complex(GO:0031213) |
| 1.6 | 20.2 | GO:0097433 | dense body(GO:0097433) |
| 1.6 | 4.7 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 1.5 | 10.8 | GO:0070852 | cell body fiber(GO:0070852) |
| 1.5 | 7.4 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 1.5 | 33.7 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 1.4 | 25.9 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 1.4 | 15.8 | GO:0000243 | commitment complex(GO:0000243) |
| 1.4 | 70.0 | GO:0005876 | spindle microtubule(GO:0005876) |
| 1.4 | 10.8 | GO:0042382 | paraspeckles(GO:0042382) |
| 1.3 | 24.2 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 1.3 | 75.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 1.2 | 8.3 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) ERCC4-ERCC1 complex(GO:0070522) |
| 1.2 | 3.5 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 1.1 | 70.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 1.1 | 5.5 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 1.0 | 5.0 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 1.0 | 197.5 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 1.0 | 39.3 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.9 | 17.2 | GO:0031588 | cAMP-dependent protein kinase complex(GO:0005952) nucleotide-activated protein kinase complex(GO:0031588) |
| 0.9 | 41.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.8 | 21.4 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.8 | 12.3 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.7 | 8.9 | GO:0045120 | pronucleus(GO:0045120) |
| 0.7 | 2.9 | GO:0097413 | Lewy body(GO:0097413) |
| 0.7 | 7.6 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.7 | 7.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.7 | 4.0 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.7 | 5.3 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.6 | 38.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.6 | 5.1 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.6 | 8.7 | GO:0008091 | spectrin(GO:0008091) |
| 0.6 | 8.6 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.6 | 13.0 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.6 | 2.8 | GO:0032449 | CBM complex(GO:0032449) |
| 0.6 | 23.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.5 | 6.8 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.5 | 3.8 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.5 | 25.5 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.5 | 4.2 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.4 | 7.5 | GO:0031932 | TORC2 complex(GO:0031932) TOR complex(GO:0038201) |
| 0.4 | 10.7 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.4 | 4.2 | GO:0070187 | telosome(GO:0070187) |
| 0.4 | 2.8 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.4 | 15.5 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.3 | 35.9 | GO:0000776 | kinetochore(GO:0000776) |
| 0.3 | 4.0 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.3 | 11.0 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.3 | 8.4 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.3 | 49.9 | GO:0101002 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.3 | 1.5 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.3 | 1.8 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.3 | 12.3 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.3 | 26.9 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.3 | 6.0 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.3 | 5.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.3 | 16.2 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.3 | 8.8 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.3 | 3.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.3 | 1.6 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.3 | 214.3 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.2 | 12.5 | GO:0031672 | A band(GO:0031672) |
| 0.2 | 16.2 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.2 | 2.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.2 | 0.9 | GO:0032009 | early phagosome(GO:0032009) |
| 0.2 | 8.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.2 | 15.7 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.2 | 6.4 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 3.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 7.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 1.2 | GO:0016012 | dystroglycan complex(GO:0016011) sarcoglycan complex(GO:0016012) |
| 0.1 | 68.8 | GO:1990904 | ribonucleoprotein complex(GO:1990904) |
| 0.1 | 15.5 | GO:0031968 | organelle outer membrane(GO:0031968) |
| 0.1 | 12.2 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 22.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 13.2 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 8.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 1.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 1.0 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 3.6 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.1 | 3.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 10.0 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 2.2 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 4.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 1.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 2.4 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 4.9 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.1 | 1.0 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 2.9 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 4.7 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.7 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.2 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 2.4 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 1.4 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 79.4 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 0.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.3 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 3.2 | GO:0031966 | mitochondrial membrane(GO:0031966) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 21.5 | 86.0 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 19.1 | 76.4 | GO:0032143 | single thymine insertion binding(GO:0032143) |
| 14.8 | 44.5 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 10.9 | 65.3 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 10.6 | 63.5 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 10.1 | 80.9 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 8.4 | 59.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 7.4 | 51.9 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 6.6 | 32.9 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 6.3 | 31.5 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 6.2 | 43.5 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 6.0 | 18.0 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 5.6 | 16.8 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 5.4 | 244.2 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 5.3 | 15.8 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 4.5 | 40.9 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 4.5 | 27.0 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 4.3 | 12.8 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 4.2 | 21.2 | GO:0033265 | choline binding(GO:0033265) |
| 4.1 | 24.8 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 4.1 | 41.0 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 4.0 | 28.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 3.9 | 23.4 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 3.5 | 20.9 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 3.1 | 12.5 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 2.7 | 21.7 | GO:0005347 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine ribonucleotide transmembrane transporter activity(GO:0005346) ATP transmembrane transporter activity(GO:0005347) purine nucleotide transmembrane transporter activity(GO:0015216) ADP transmembrane transporter activity(GO:0015217) |
| 2.7 | 10.6 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 2.6 | 36.5 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 2.5 | 30.6 | GO:0031386 | protein tag(GO:0031386) |
| 2.5 | 17.3 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 2.2 | 8.8 | GO:0038025 | glycoprotein transporter activity(GO:0034437) reelin receptor activity(GO:0038025) |
| 2.2 | 15.3 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 2.0 | 6.0 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 1.9 | 7.6 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 1.9 | 7.5 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 1.8 | 11.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 1.8 | 30.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 1.7 | 8.7 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 1.6 | 51.1 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 1.6 | 23.8 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 1.5 | 20.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 1.5 | 12.3 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 1.5 | 67.1 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 1.5 | 11.9 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 1.5 | 43.0 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 1.5 | 20.7 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 1.4 | 25.9 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 1.4 | 40.6 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 1.4 | 4.2 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 1.4 | 4.1 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 1.3 | 10.7 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 1.3 | 104.1 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 1.3 | 24.2 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 1.3 | 17.8 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 1.3 | 20.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 1.2 | 11.2 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 1.2 | 63.8 | GO:0019894 | kinesin binding(GO:0019894) |
| 1.2 | 8.3 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 1.2 | 34.9 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 1.2 | 16.3 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 1.1 | 5.5 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 1.1 | 4.3 | GO:0047718 | enone reductase activity(GO:0035671) androsterone dehydrogenase activity(GO:0047023) indanol dehydrogenase activity(GO:0047718) |
| 1.1 | 9.7 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 1.1 | 4.3 | GO:0019003 | GDP binding(GO:0019003) |
| 1.1 | 10.6 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 1.0 | 6.2 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 1.0 | 20.7 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 1.0 | 6.2 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 1.0 | 13.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 1.0 | 6.8 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 1.0 | 9.6 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
| 0.9 | 19.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.9 | 6.4 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.9 | 8.7 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.9 | 32.0 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.9 | 55.2 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.9 | 17.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.8 | 21.7 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.8 | 23.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.8 | 3.2 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.8 | 13.7 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.8 | 15.9 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.7 | 3.7 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.7 | 13.8 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.7 | 10.8 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.7 | 12.9 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.7 | 15.7 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.7 | 3.6 | GO:0004306 | ethanolamine-phosphate cytidylyltransferase activity(GO:0004306) |
| 0.7 | 70.4 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.7 | 2.7 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.7 | 28.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.7 | 11.7 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.6 | 21.8 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.6 | 21.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.6 | 5.5 | GO:0008144 | drug binding(GO:0008144) |
| 0.6 | 36.7 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.6 | 3.4 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.5 | 18.3 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.5 | 4.2 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.5 | 5.2 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.5 | 3.5 | GO:0032405 | MutLalpha complex binding(GO:0032405) |
| 0.5 | 31.6 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.5 | 5.5 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.5 | 7.0 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.5 | 22.2 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.4 | 16.5 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.4 | 11.4 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.4 | 7.5 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.4 | 5.3 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.4 | 8.8 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.4 | 5.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.4 | 7.4 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.4 | 52.0 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.3 | 4.4 | GO:0031543 | peptidyl-proline dioxygenase activity(GO:0031543) |
| 0.3 | 1.9 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.3 | 3.6 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.3 | 7.6 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.3 | 1.6 | GO:0052843 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.3 | 1.8 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.3 | 15.5 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.3 | 1.7 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.3 | 7.4 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.3 | 6.3 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.2 | 5.6 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.2 | 2.5 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.2 | 7.3 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.2 | 48.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.2 | 3.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 5.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.2 | 74.0 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.2 | 9.5 | GO:0008135 | translation factor activity, RNA binding(GO:0008135) |
| 0.2 | 1.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.2 | 1.9 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.2 | 4.0 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.2 | 2.4 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.2 | 49.8 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.2 | 6.9 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 14.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 2.4 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 4.1 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 1.9 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 14.6 | GO:0005525 | GTP binding(GO:0005525) |
| 0.1 | 0.9 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 1.8 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.1 | 11.1 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.1 | 3.0 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 8.8 | GO:0004386 | helicase activity(GO:0004386) |
| 0.1 | 4.6 | GO:0016776 | phosphotransferase activity, phosphate group as acceptor(GO:0016776) |
| 0.1 | 0.9 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 6.5 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.1 | 2.9 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 1.1 | GO:0036122 | BMP binding(GO:0036122) |
| 0.1 | 6.9 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.1 | 2.8 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 1.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 4.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 5.1 | GO:0005496 | steroid binding(GO:0005496) |
| 0.1 | 9.7 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 2.3 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 75.2 | GO:0003723 | RNA binding(GO:0003723) |
| 0.0 | 1.1 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 1.7 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 2.6 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.2 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 1.3 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 1.0 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 216.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 2.3 | 140.4 | PID PLK1 PATHWAY | PLK1 signaling events |
| 2.0 | 102.3 | PID MYC PATHWAY | C-MYC pathway |
| 1.2 | 138.3 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 1.2 | 41.0 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.6 | 36.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.5 | 14.8 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.4 | 47.0 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.4 | 15.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.4 | 7.6 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.3 | 15.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.3 | 8.8 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.3 | 47.3 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.3 | 11.8 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.2 | 18.6 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.2 | 3.8 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.2 | 9.4 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.2 | 6.2 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.2 | 22.7 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.2 | 3.5 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.2 | 9.9 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.2 | 17.0 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.2 | 8.1 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.2 | 5.5 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.2 | 10.9 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.2 | 4.3 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.2 | 8.5 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.2 | 7.4 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 2.8 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 1.9 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 9.2 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 3.5 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 2.8 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 6.9 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 3.2 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 12.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 0.9 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 1.1 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.8 | 51.9 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 5.0 | 130.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 2.7 | 75.0 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 2.6 | 106.5 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 2.5 | 204.4 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 1.7 | 65.3 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 1.7 | 28.9 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 1.5 | 42.5 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 1.5 | 60.4 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 1.4 | 48.7 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 1.1 | 25.9 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 1.1 | 24.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 1.0 | 31.1 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.9 | 45.9 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.9 | 30.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.9 | 126.1 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.8 | 27.9 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.7 | 15.7 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.7 | 11.4 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.6 | 20.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.6 | 17.0 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.6 | 49.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.5 | 21.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.5 | 17.2 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.5 | 26.3 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.5 | 24.7 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.5 | 13.5 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.4 | 6.2 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.4 | 77.5 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.4 | 9.6 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.4 | 10.4 | REACTOME S PHASE | Genes involved in S Phase |
| 0.4 | 4.2 | REACTOME TELOMERE MAINTENANCE | Genes involved in Telomere Maintenance |
| 0.4 | 8.3 | REACTOME TRANSCRIPTION COUPLED NER TC NER | Genes involved in Transcription-coupled NER (TC-NER) |
| 0.4 | 30.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.4 | 8.5 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.3 | 7.8 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.3 | 6.0 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.3 | 2.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.2 | 17.4 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.2 | 2.5 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.2 | 3.2 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.2 | 4.0 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.2 | 7.2 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.2 | 5.1 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 1.9 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.2 | 4.9 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.2 | 5.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 2.8 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.1 | 3.0 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.1 | 3.9 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.1 | 21.6 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.1 | 1.9 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 2.7 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 2.1 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 4.9 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 4.2 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 1.7 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 5.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 4.8 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.7 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.7 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.9 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.6 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.6 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 6.9 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 1.0 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.2 | REACTOME NEF MEDIATES DOWN MODULATION OF CELL SURFACE RECEPTORS BY RECRUITING THEM TO CLATHRIN ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.0 | 0.6 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 1.7 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |