Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
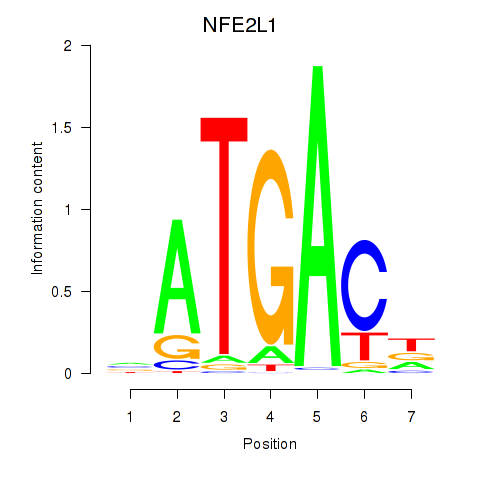
Results for NFE2L1
Z-value: 0.78
Transcription factors associated with NFE2L1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFE2L1
|
ENSG00000082641.11 | nuclear factor, erythroid 2 like 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFE2L1 | hg19_v2_chr17_+_46125707_46125746 | 0.23 | 7.5e-04 | Click! |
Activity profile of NFE2L1 motif
Sorted Z-values of NFE2L1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_35198243 | 12.02 |
ENST00000528455.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr11_+_35211511 | 11.16 |
ENST00000524922.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr11_+_35201826 | 10.42 |
ENST00000531873.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr11_+_35198118 | 10.38 |
ENST00000525211.1
ENST00000526000.1 ENST00000279452.6 ENST00000527889.1 |
CD44
|
CD44 molecule (Indian blood group) |
| chr2_-_106015527 | 10.04 |
ENST00000344213.4
ENST00000358129.4 |
FHL2
|
four and a half LIM domains 2 |
| chr16_-_69760409 | 9.33 |
ENST00000561500.1
ENST00000439109.2 ENST00000564043.1 ENST00000379046.2 ENST00000379047.3 |
NQO1
|
NAD(P)H dehydrogenase, quinone 1 |
| chr11_+_35211429 | 9.17 |
ENST00000525688.1
ENST00000278385.6 ENST00000533222.1 |
CD44
|
CD44 molecule (Indian blood group) |
| chr1_+_26605618 | 8.61 |
ENST00000270792.5
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3 |
| chr18_+_657733 | 8.50 |
ENST00000323250.5
ENST00000323224.7 |
TYMS
|
thymidylate synthetase |
| chr9_-_128246769 | 8.20 |
ENST00000444226.1
|
MAPKAP1
|
mitogen-activated protein kinase associated protein 1 |
| chr7_+_872107 | 8.12 |
ENST00000405266.1
ENST00000401592.1 ENST00000403868.1 ENST00000425407.2 |
SUN1
|
Sad1 and UNC84 domain containing 1 |
| chr20_-_33539655 | 8.09 |
ENST00000451957.2
|
GSS
|
glutathione synthetase |
| chr1_-_24126023 | 7.95 |
ENST00000429356.1
|
GALE
|
UDP-galactose-4-epimerase |
| chr17_+_73230799 | 7.90 |
ENST00000579838.1
|
NUP85
|
nucleoporin 85kDa |
| chr2_-_106015491 | 7.88 |
ENST00000408995.1
ENST00000393353.3 ENST00000322142.8 |
FHL2
|
four and a half LIM domains 2 |
| chr16_+_3068393 | 7.83 |
ENST00000573001.1
|
TNFRSF12A
|
tumor necrosis factor receptor superfamily, member 12A |
| chr19_+_35645618 | 7.77 |
ENST00000392218.2
ENST00000543307.1 ENST00000392219.2 ENST00000541435.2 ENST00000590686.1 ENST00000342879.3 ENST00000588699.1 |
FXYD5
|
FXYD domain containing ion transport regulator 5 |
| chr16_+_31044413 | 7.75 |
ENST00000394998.1
|
STX4
|
syntaxin 4 |
| chr2_+_201994042 | 7.66 |
ENST00000417748.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr3_+_69928256 | 6.92 |
ENST00000394355.2
|
MITF
|
microphthalmia-associated transcription factor |
| chr6_-_43027105 | 6.70 |
ENST00000230413.5
ENST00000487429.1 ENST00000489623.1 ENST00000468957.1 |
MRPL2
|
mitochondrial ribosomal protein L2 |
| chr5_+_68462944 | 6.69 |
ENST00000506572.1
|
CCNB1
|
cyclin B1 |
| chr7_-_35077653 | 6.29 |
ENST00000310974.4
|
DPY19L1
|
dpy-19-like 1 (C. elegans) |
| chr9_-_117853297 | 6.08 |
ENST00000542877.1
ENST00000537320.1 ENST00000341037.4 |
TNC
|
tenascin C |
| chr11_+_85339623 | 6.03 |
ENST00000358867.6
ENST00000534341.1 ENST00000393375.1 ENST00000531274.1 |
TMEM126B
|
transmembrane protein 126B |
| chr6_+_63921399 | 5.95 |
ENST00000356170.3
|
FKBP1C
|
FK506 binding protein 1C |
| chr3_-_81792780 | 5.91 |
ENST00000489715.1
|
GBE1
|
glucan (1,4-alpha-), branching enzyme 1 |
| chr2_+_201994208 | 5.90 |
ENST00000440180.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr22_+_38142235 | 5.81 |
ENST00000407319.2
ENST00000403663.2 ENST00000428075.1 |
TRIOBP
|
TRIO and F-actin binding protein |
| chr18_+_55888767 | 5.78 |
ENST00000431212.2
ENST00000586268.1 ENST00000587190.1 |
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr1_-_222885770 | 5.74 |
ENST00000355727.2
ENST00000340020.6 |
AIDA
|
axin interactor, dorsalization associated |
| chr3_-_149095652 | 5.63 |
ENST00000305366.3
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr19_-_44160768 | 5.53 |
ENST00000593447.1
|
PLAUR
|
plasminogen activator, urokinase receptor |
| chr18_+_3252265 | 5.30 |
ENST00000580887.1
ENST00000536605.1 |
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr11_-_107729887 | 5.26 |
ENST00000525815.1
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr10_+_121410882 | 5.25 |
ENST00000369085.3
|
BAG3
|
BCL2-associated athanogene 3 |
| chr2_+_242289502 | 5.12 |
ENST00000451310.1
|
SEPT2
|
septin 2 |
| chr16_-_58768177 | 4.88 |
ENST00000434819.2
ENST00000245206.5 |
GOT2
|
glutamic-oxaloacetic transaminase 2, mitochondrial |
| chr5_+_159848854 | 4.76 |
ENST00000517480.1
ENST00000520452.1 ENST00000393964.1 |
PTTG1
|
pituitary tumor-transforming 1 |
| chr1_-_115259337 | 4.53 |
ENST00000369535.4
|
NRAS
|
neuroblastoma RAS viral (v-ras) oncogene homolog |
| chr2_-_64881018 | 4.49 |
ENST00000313349.3
|
SERTAD2
|
SERTA domain containing 2 |
| chr17_-_57784755 | 4.46 |
ENST00000537860.1
ENST00000393038.2 ENST00000409433.2 |
PTRH2
|
peptidyl-tRNA hydrolase 2 |
| chrX_+_70503037 | 4.33 |
ENST00000535149.1
|
NONO
|
non-POU domain containing, octamer-binding |
| chr4_+_69962185 | 4.28 |
ENST00000305231.7
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr11_+_101983176 | 4.25 |
ENST00000524575.1
|
YAP1
|
Yes-associated protein 1 |
| chr2_-_220119280 | 4.24 |
ENST00000392088.2
|
TUBA4A
|
tubulin, alpha 4a |
| chr4_-_103749179 | 4.21 |
ENST00000502690.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr17_+_57784997 | 4.17 |
ENST00000537567.1
ENST00000539763.1 ENST00000587945.1 ENST00000536180.1 ENST00000589823.2 ENST00000592106.1 ENST00000591315.1 ENST00000545362.1 |
VMP1
|
vacuole membrane protein 1 |
| chr6_+_63921351 | 4.15 |
ENST00000370659.1
|
FKBP1C
|
FK506 binding protein 1C |
| chr5_+_33440802 | 4.13 |
ENST00000502553.1
ENST00000514259.1 ENST00000265112.3 |
TARS
|
threonyl-tRNA synthetase |
| chr17_+_73663470 | 4.09 |
ENST00000583536.1
|
SAP30BP
|
SAP30 binding protein |
| chr9_-_36276966 | 4.07 |
ENST00000543356.2
ENST00000396594.3 |
GNE
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr15_+_89182178 | 4.06 |
ENST00000559876.1
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr2_+_234545092 | 4.06 |
ENST00000344644.5
|
UGT1A10
|
UDP glucuronosyltransferase 1 family, polypeptide A10 |
| chr12_+_20963632 | 4.05 |
ENST00000540853.1
ENST00000261196.2 |
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr17_+_57784826 | 4.04 |
ENST00000262291.4
|
VMP1
|
vacuole membrane protein 1 |
| chr11_+_12399071 | 4.04 |
ENST00000539723.1
ENST00000550549.1 |
PARVA
|
parvin, alpha |
| chr15_+_80364901 | 4.00 |
ENST00000560228.1
ENST00000559835.1 ENST00000559775.1 ENST00000558688.1 ENST00000560392.1 ENST00000560976.1 ENST00000558272.1 ENST00000558390.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr4_-_103749205 | 3.97 |
ENST00000508249.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr3_-_47934234 | 3.95 |
ENST00000420772.2
|
MAP4
|
microtubule-associated protein 4 |
| chr1_+_45478568 | 3.94 |
ENST00000428106.1
|
UROD
|
uroporphyrinogen decarboxylase |
| chr10_+_81272287 | 3.91 |
ENST00000520547.2
|
EIF5AL1
|
eukaryotic translation initiation factor 5A-like 1 |
| chr10_+_94352956 | 3.78 |
ENST00000260731.3
|
KIF11
|
kinesin family member 11 |
| chr17_+_73663402 | 3.76 |
ENST00000355423.3
|
SAP30BP
|
SAP30 binding protein |
| chr4_+_69962212 | 3.76 |
ENST00000508661.1
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr5_+_33441053 | 3.67 |
ENST00000541634.1
ENST00000455217.2 ENST00000414361.2 |
TARS
|
threonyl-tRNA synthetase |
| chr18_+_3252206 | 3.57 |
ENST00000578562.2
|
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr2_+_234602305 | 3.53 |
ENST00000406651.1
|
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr17_-_40169429 | 3.53 |
ENST00000316603.7
ENST00000588641.1 |
DNAJC7
|
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr16_+_31044812 | 3.50 |
ENST00000313843.3
|
STX4
|
syntaxin 4 |
| chr2_+_219110149 | 3.49 |
ENST00000456575.1
|
ARPC2
|
actin related protein 2/3 complex, subunit 2, 34kDa |
| chr11_-_6640585 | 3.41 |
ENST00000533371.1
ENST00000528657.1 ENST00000436873.2 ENST00000299427.6 |
TPP1
|
tripeptidyl peptidase I |
| chr1_+_32645269 | 3.41 |
ENST00000373610.3
|
TXLNA
|
taxilin alpha |
| chr12_-_112856623 | 3.36 |
ENST00000551291.2
|
RPL6
|
ribosomal protein L6 |
| chr14_+_64680854 | 3.36 |
ENST00000458046.2
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2 |
| chr11_+_59480899 | 3.31 |
ENST00000300150.7
|
STX3
|
syntaxin 3 |
| chr12_+_123237321 | 3.29 |
ENST00000280557.6
ENST00000455982.2 |
DENR
|
density-regulated protein |
| chr15_+_89181974 | 3.27 |
ENST00000306072.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr6_+_160183492 | 3.23 |
ENST00000541436.1
|
ACAT2
|
acetyl-CoA acetyltransferase 2 |
| chr11_+_71938925 | 3.20 |
ENST00000538751.1
|
INPPL1
|
inositol polyphosphate phosphatase-like 1 |
| chr3_+_69812877 | 3.09 |
ENST00000457080.1
ENST00000328528.6 |
MITF
|
microphthalmia-associated transcription factor |
| chr11_+_65337901 | 3.05 |
ENST00000309328.3
ENST00000531405.1 ENST00000527920.1 ENST00000526877.1 ENST00000533115.1 ENST00000526433.1 |
SSSCA1
|
Sjogren syndrome/scleroderma autoantigen 1 |
| chr1_-_94312706 | 3.04 |
ENST00000370244.1
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr1_-_6445809 | 3.04 |
ENST00000377855.2
|
ACOT7
|
acyl-CoA thioesterase 7 |
| chr13_+_48611665 | 3.01 |
ENST00000258662.2
|
NUDT15
|
nudix (nucleoside diphosphate linked moiety X)-type motif 15 |
| chr6_-_43595039 | 2.99 |
ENST00000307114.7
|
GTPBP2
|
GTP binding protein 2 |
| chr11_-_87908600 | 2.98 |
ENST00000531138.1
ENST00000526372.1 ENST00000243662.6 |
RAB38
|
RAB38, member RAS oncogene family |
| chr17_+_76210267 | 2.98 |
ENST00000301633.4
ENST00000350051.3 ENST00000374948.2 ENST00000590449.1 |
BIRC5
|
baculoviral IAP repeat containing 5 |
| chr9_+_37422663 | 2.96 |
ENST00000318158.6
ENST00000607784.1 |
GRHPR
|
glyoxylate reductase/hydroxypyruvate reductase |
| chr12_-_44200146 | 2.95 |
ENST00000395510.2
ENST00000325127.4 |
TWF1
|
twinfilin actin-binding protein 1 |
| chr14_+_102276209 | 2.95 |
ENST00000445439.3
ENST00000334743.5 ENST00000557095.1 |
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr1_-_155211017 | 2.94 |
ENST00000536770.1
ENST00000368373.3 |
GBA
|
glucosidase, beta, acid |
| chr8_+_21823726 | 2.93 |
ENST00000433566.4
|
XPO7
|
exportin 7 |
| chr3_+_138340049 | 2.92 |
ENST00000464668.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr12_-_106641728 | 2.87 |
ENST00000378026.4
|
CKAP4
|
cytoskeleton-associated protein 4 |
| chr14_+_61447927 | 2.87 |
ENST00000451406.1
|
SLC38A6
|
solute carrier family 38, member 6 |
| chr9_+_5890802 | 2.85 |
ENST00000381477.3
ENST00000381476.1 ENST00000381471.1 |
MLANA
|
melan-A |
| chrX_-_99891796 | 2.82 |
ENST00000373020.4
|
TSPAN6
|
tetraspanin 6 |
| chr12_-_65146636 | 2.78 |
ENST00000418919.2
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase |
| chr6_+_32146131 | 2.76 |
ENST00000375094.3
|
RNF5
|
ring finger protein 5, E3 ubiquitin protein ligase |
| chr9_+_70856899 | 2.75 |
ENST00000377342.5
ENST00000478048.1 |
CBWD3
|
COBW domain containing 3 |
| chr2_+_234590556 | 2.73 |
ENST00000373426.3
|
UGT1A7
|
UDP glucuronosyltransferase 1 family, polypeptide A7 |
| chr6_-_3157760 | 2.67 |
ENST00000333628.3
|
TUBB2A
|
tubulin, beta 2A class IIa |
| chr5_+_72143988 | 2.65 |
ENST00000506351.2
|
TNPO1
|
transportin 1 |
| chr8_-_13372395 | 2.61 |
ENST00000276297.4
ENST00000511869.1 |
DLC1
|
deleted in liver cancer 1 |
| chr2_+_234545148 | 2.60 |
ENST00000373445.1
|
UGT1A10
|
UDP glucuronosyltransferase 1 family, polypeptide A10 |
| chr7_+_107220660 | 2.60 |
ENST00000465919.1
ENST00000445771.2 ENST00000479917.1 ENST00000421217.1 ENST00000457837.1 |
BCAP29
|
B-cell receptor-associated protein 29 |
| chrX_+_11129388 | 2.59 |
ENST00000321143.4
ENST00000380763.3 ENST00000380762.4 |
HCCS
|
holocytochrome c synthase |
| chr3_-_49142178 | 2.54 |
ENST00000452739.1
ENST00000414533.1 ENST00000417025.1 |
QARS
|
glutaminyl-tRNA synthetase |
| chr19_+_40477062 | 2.51 |
ENST00000455878.2
|
PSMC4
|
proteasome (prosome, macropain) 26S subunit, ATPase, 4 |
| chr17_-_64216748 | 2.50 |
ENST00000585162.1
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr4_-_103748696 | 2.50 |
ENST00000321805.7
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr5_+_133707252 | 2.48 |
ENST00000506787.1
ENST00000507277.1 |
UBE2B
|
ubiquitin-conjugating enzyme E2B |
| chr5_+_65440032 | 2.47 |
ENST00000334121.6
|
SREK1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chr7_+_107220899 | 2.44 |
ENST00000379117.2
ENST00000473124.1 |
BCAP29
|
B-cell receptor-associated protein 29 |
| chr3_-_47950745 | 2.44 |
ENST00000429422.1
|
MAP4
|
microtubule-associated protein 4 |
| chr6_+_30539153 | 2.41 |
ENST00000326195.8
ENST00000376545.3 ENST00000396515.4 ENST00000441867.1 ENST00000468958.1 |
ABCF1
|
ATP-binding cassette, sub-family F (GCN20), member 1 |
| chr17_-_29233221 | 2.39 |
ENST00000580840.1
|
TEFM
|
transcription elongation factor, mitochondrial |
| chrX_+_70503433 | 2.38 |
ENST00000276079.8
ENST00000373856.3 ENST00000373841.1 ENST00000420903.1 |
NONO
|
non-POU domain containing, octamer-binding |
| chr7_+_116312411 | 2.30 |
ENST00000456159.1
ENST00000397752.3 ENST00000318493.6 |
MET
|
met proto-oncogene |
| chr11_+_7618413 | 2.30 |
ENST00000528883.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr7_-_148580563 | 2.30 |
ENST00000476773.1
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila) |
| chr22_+_38349670 | 2.30 |
ENST00000442738.2
ENST00000460648.1 ENST00000407936.1 ENST00000488684.1 ENST00000492213.1 ENST00000606538.1 ENST00000405557.1 |
POLR2F
|
polymerase (RNA) II (DNA directed) polypeptide F |
| chr1_+_95616933 | 2.29 |
ENST00000604203.1
|
RP11-57H12.6
|
TMEM56-RWDD3 readthrough |
| chr17_+_7155819 | 2.28 |
ENST00000570322.1
ENST00000576496.1 ENST00000574841.2 |
ELP5
|
elongator acetyltransferase complex subunit 5 |
| chr5_-_37371278 | 2.27 |
ENST00000231498.3
|
NUP155
|
nucleoporin 155kDa |
| chr17_+_57274914 | 2.24 |
ENST00000582004.1
ENST00000577660.1 |
PRR11
CTD-2510F5.6
|
proline rich 11 Uncharacterized protein |
| chr22_-_32651326 | 2.23 |
ENST00000266086.4
|
SLC5A4
|
solute carrier family 5 (glucose activated ion channel), member 4 |
| chr5_+_162887556 | 2.21 |
ENST00000393915.4
ENST00000432118.2 ENST00000358715.3 |
HMMR
|
hyaluronan-mediated motility receptor (RHAMM) |
| chr17_-_40169161 | 2.21 |
ENST00000589586.2
ENST00000426588.3 ENST00000589576.1 |
DNAJC7
|
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr22_+_20861858 | 2.20 |
ENST00000414658.1
ENST00000432052.1 ENST00000425759.2 ENST00000292733.7 ENST00000542773.1 ENST00000263205.7 ENST00000406969.1 ENST00000382974.2 |
MED15
|
mediator complex subunit 15 |
| chr2_+_234580525 | 2.20 |
ENST00000609637.1
|
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr2_+_109237717 | 2.16 |
ENST00000409441.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr14_+_64854958 | 2.11 |
ENST00000555709.2
ENST00000554739.1 ENST00000554768.1 ENST00000216605.8 |
MTHFD1
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase |
| chr15_+_89182156 | 2.11 |
ENST00000379224.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr2_-_113594279 | 2.10 |
ENST00000416750.1
ENST00000418817.1 ENST00000263341.2 |
IL1B
|
interleukin 1, beta |
| chr6_-_144416737 | 2.09 |
ENST00000367569.2
|
SF3B5
|
splicing factor 3b, subunit 5, 10kDa |
| chrX_+_47077632 | 2.08 |
ENST00000457458.2
|
CDK16
|
cyclin-dependent kinase 16 |
| chr2_+_234580499 | 2.08 |
ENST00000354728.4
|
UGT1A9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr14_+_103801140 | 2.08 |
ENST00000561325.1
ENST00000392715.2 ENST00000559130.1 ENST00000559532.1 ENST00000558506.1 |
EIF5
|
eukaryotic translation initiation factor 5 |
| chr12_+_16035307 | 2.07 |
ENST00000538352.1
ENST00000025399.6 ENST00000419869.2 |
STRAP
|
serine/threonine kinase receptor associated protein |
| chr7_-_142232071 | 2.07 |
ENST00000390364.3
|
TRBV10-1
|
T cell receptor beta variable 10-1(gene/pseudogene) |
| chr3_-_127541194 | 2.06 |
ENST00000453507.2
|
MGLL
|
monoglyceride lipase |
| chr20_-_1447467 | 2.05 |
ENST00000353088.2
ENST00000350991.4 |
NSFL1C
|
NSFL1 (p97) cofactor (p47) |
| chr2_+_169923577 | 2.05 |
ENST00000432060.2
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr19_-_5719860 | 2.03 |
ENST00000590729.1
|
LONP1
|
lon peptidase 1, mitochondrial |
| chr17_+_33914276 | 2.03 |
ENST00000592545.1
ENST00000538556.1 ENST00000312678.8 ENST00000589344.1 |
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr3_-_49142504 | 1.98 |
ENST00000306125.6
ENST00000420147.2 |
QARS
|
glutaminyl-tRNA synthetase |
| chr11_-_66103867 | 1.98 |
ENST00000424433.2
|
RIN1
|
Ras and Rab interactor 1 |
| chr4_+_169013666 | 1.96 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chr4_+_113558272 | 1.95 |
ENST00000509061.1
ENST00000508577.1 ENST00000513553.1 |
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr14_+_22919081 | 1.94 |
ENST00000390473.1
|
TRDJ1
|
T cell receptor delta joining 1 |
| chr9_-_14180778 | 1.93 |
ENST00000380924.1
ENST00000543693.1 |
NFIB
|
nuclear factor I/B |
| chr2_-_55496174 | 1.91 |
ENST00000417363.1
ENST00000412530.1 ENST00000394600.3 ENST00000366137.2 ENST00000420637.1 |
MTIF2
|
mitochondrial translational initiation factor 2 |
| chr11_-_126138808 | 1.91 |
ENST00000332118.6
ENST00000532259.1 |
SRPR
|
signal recognition particle receptor (docking protein) |
| chr7_-_22539771 | 1.90 |
ENST00000406890.2
ENST00000424363.1 |
STEAP1B
|
STEAP family member 1B |
| chr1_-_214638146 | 1.89 |
ENST00000543945.1
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14 |
| chr7_-_41742697 | 1.86 |
ENST00000242208.4
|
INHBA
|
inhibin, beta A |
| chr2_-_120980939 | 1.86 |
ENST00000426077.2
|
TMEM185B
|
transmembrane protein 185B |
| chr1_+_45241186 | 1.84 |
ENST00000372209.3
|
RPS8
|
ribosomal protein S8 |
| chr9_-_130966497 | 1.84 |
ENST00000393608.1
ENST00000372948.3 |
CIZ1
|
CDKN1A interacting zinc finger protein 1 |
| chr2_+_27651519 | 1.83 |
ENST00000379863.3
|
NRBP1
|
nuclear receptor binding protein 1 |
| chr14_-_75530693 | 1.81 |
ENST00000555135.1
ENST00000357971.3 ENST00000553302.1 ENST00000555694.1 ENST00000238618.3 |
ACYP1
|
acylphosphatase 1, erythrocyte (common) type |
| chr16_-_4395905 | 1.81 |
ENST00000571941.1
|
PAM16
|
presequence translocase-associated motor 16 homolog (S. cerevisiae) |
| chr1_-_111506562 | 1.79 |
ENST00000485275.2
ENST00000369763.4 |
LRIF1
|
ligand dependent nuclear receptor interacting factor 1 |
| chr7_+_107531580 | 1.78 |
ENST00000537148.1
ENST00000440410.1 ENST00000437604.2 |
DLD
|
dihydrolipoamide dehydrogenase |
| chr21_-_33975547 | 1.77 |
ENST00000431599.1
|
C21orf59
|
chromosome 21 open reading frame 59 |
| chr2_+_234526272 | 1.77 |
ENST00000373450.4
|
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr6_-_131321863 | 1.76 |
ENST00000528282.1
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr20_-_1447547 | 1.76 |
ENST00000476071.1
|
NSFL1C
|
NSFL1 (p97) cofactor (p47) |
| chrX_-_41449204 | 1.72 |
ENST00000378179.3
|
CASK
|
calcium/calmodulin-dependent serine protein kinase (MAGUK family) |
| chr12_+_28343353 | 1.71 |
ENST00000539107.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chr1_-_155211065 | 1.68 |
ENST00000427500.3
|
GBA
|
glucosidase, beta, acid |
| chr4_+_74606223 | 1.67 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr12_+_51633061 | 1.64 |
ENST00000551313.1
|
DAZAP2
|
DAZ associated protein 2 |
| chr15_+_91478493 | 1.64 |
ENST00000418476.2
|
UNC45A
|
unc-45 homolog A (C. elegans) |
| chr19_+_9361606 | 1.63 |
ENST00000456448.1
|
OR7E24
|
olfactory receptor, family 7, subfamily E, member 24 |
| chr12_-_102874102 | 1.61 |
ENST00000392905.2
|
IGF1
|
insulin-like growth factor 1 (somatomedin C) |
| chr19_-_39881777 | 1.60 |
ENST00000595564.1
ENST00000221265.3 |
PAF1
|
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae) |
| chr11_-_64052111 | 1.60 |
ENST00000394532.3
ENST00000394531.3 ENST00000309032.3 |
BAD
|
BCL2-associated agonist of cell death |
| chr2_+_228337079 | 1.58 |
ENST00000409315.1
ENST00000373671.3 ENST00000409171.1 |
AGFG1
|
ArfGAP with FG repeats 1 |
| chr4_+_4861385 | 1.58 |
ENST00000382723.4
|
MSX1
|
msh homeobox 1 |
| chr19_+_40476912 | 1.58 |
ENST00000157812.2
|
PSMC4
|
proteasome (prosome, macropain) 26S subunit, ATPase, 4 |
| chr10_+_115674530 | 1.58 |
ENST00000451472.1
|
AL162407.1
|
CDNA FLJ20147 fis, clone COL07954; HCG1781466; Uncharacterized protein |
| chr10_+_5488564 | 1.57 |
ENST00000449083.1
ENST00000380359.3 |
NET1
|
neuroepithelial cell transforming 1 |
| chr9_-_123676827 | 1.54 |
ENST00000546084.1
|
TRAF1
|
TNF receptor-associated factor 1 |
| chr12_-_123752624 | 1.53 |
ENST00000542174.1
ENST00000535796.1 |
CDK2AP1
|
cyclin-dependent kinase 2 associated protein 1 |
| chr5_+_138940742 | 1.53 |
ENST00000398733.3
ENST00000253815.2 ENST00000505007.1 |
UBE2D2
|
ubiquitin-conjugating enzyme E2D 2 |
| chr22_-_43040515 | 1.51 |
ENST00000361740.4
|
CYB5R3
|
cytochrome b5 reductase 3 |
| chr8_+_104831554 | 1.50 |
ENST00000408894.2
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr2_+_46769798 | 1.47 |
ENST00000238738.4
|
RHOQ
|
ras homolog family member Q |
| chr3_-_12705600 | 1.41 |
ENST00000542177.1
ENST00000442415.2 ENST00000251849.4 |
RAF1
|
v-raf-1 murine leukemia viral oncogene homolog 1 |
| chr8_+_9953061 | 1.40 |
ENST00000522907.1
ENST00000528246.1 |
MSRA
|
methionine sulfoxide reductase A |
| chr14_+_22963806 | 1.39 |
ENST00000390493.1
|
TRAJ44
|
T cell receptor alpha joining 44 |
| chr6_+_27114861 | 1.38 |
ENST00000377459.1
|
HIST1H2AH
|
histone cluster 1, H2ah |
| chr3_-_11685345 | 1.38 |
ENST00000430365.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr14_+_22739823 | 1.36 |
ENST00000390464.2
|
TRAV38-1
|
T cell receptor alpha variable 38-1 |
| chr11_-_8680383 | 1.35 |
ENST00000299550.6
|
TRIM66
|
tripartite motif containing 66 |
| chr17_+_39240459 | 1.34 |
ENST00000391417.4
|
KRTAP4-7
|
keratin associated protein 4-7 |
| chr1_+_43148059 | 1.30 |
ENST00000321358.7
ENST00000332220.6 |
YBX1
|
Y box binding protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NFE2L1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.6 | 53.2 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 3.7 | 11.2 | GO:1902568 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 3.6 | 17.9 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 3.1 | 9.4 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 2.6 | 7.8 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 2.2 | 6.7 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) response to DDT(GO:0046680) histone H3-S10 phosphorylation involved in chromosome condensation(GO:2000775) |
| 2.1 | 8.5 | GO:0019860 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) transformation of host cell by virus(GO:0019087) uracil metabolic process(GO:0019860) |
| 2.1 | 6.3 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 2.0 | 8.1 | GO:0070197 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 2.0 | 6.1 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 2.0 | 6.0 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 1.8 | 8.8 | GO:0030047 | actin modification(GO:0030047) |
| 1.6 | 4.9 | GO:0006106 | fumarate metabolic process(GO:0006106) L-kynurenine metabolic process(GO:0097052) |
| 1.5 | 15.4 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 1.5 | 4.5 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 1.4 | 5.5 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 1.4 | 8.2 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 1.4 | 13.6 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 1.2 | 6.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 1.2 | 4.6 | GO:1901805 | beta-glucoside metabolic process(GO:1901804) beta-glucoside catabolic process(GO:1901805) positive regulation of neuronal action potential(GO:1904457) |
| 1.1 | 6.7 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 1.1 | 3.3 | GO:0002188 | translation reinitiation(GO:0002188) |
| 1.1 | 6.4 | GO:0051012 | microtubule sliding(GO:0051012) |
| 1.0 | 7.8 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.9 | 3.5 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.9 | 2.6 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.8 | 8.0 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.8 | 2.3 | GO:1904772 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.8 | 3.0 | GO:1900533 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.8 | 3.0 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.7 | 2.1 | GO:0070487 | monocyte aggregation(GO:0070487) |
| 0.7 | 2.0 | GO:0070407 | oxidation-dependent protein catabolic process(GO:0070407) |
| 0.6 | 1.9 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.6 | 7.9 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.6 | 2.3 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.6 | 4.5 | GO:0021815 | modulation of microtubule cytoskeleton involved in cerebral cortex radial glia guided migration(GO:0021815) |
| 0.5 | 2.7 | GO:1904073 | trophectodermal cell proliferation(GO:0001834) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.5 | 1.6 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.5 | 3.6 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.5 | 2.0 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.5 | 5.0 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.5 | 3.9 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.5 | 4.3 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.5 | 2.3 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.5 | 1.8 | GO:0044533 | induction of programmed cell death(GO:0012502) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) |
| 0.4 | 1.8 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.4 | 5.7 | GO:0048262 | determination of dorsal/ventral asymmetry(GO:0048262) |
| 0.4 | 2.1 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.4 | 5.7 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.4 | 4.5 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.4 | 1.9 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.4 | 3.0 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.4 | 8.1 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.4 | 1.1 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.4 | 1.8 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.4 | 0.4 | GO:2000777 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.3 | 1.0 | GO:0060459 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) left lung development(GO:0060459) left lung morphogenesis(GO:0060460) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) |
| 0.3 | 1.9 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.3 | 4.1 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.3 | 1.5 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.3 | 3.9 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.3 | 1.8 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.3 | 5.4 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.3 | 2.1 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.3 | 2.1 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.3 | 1.4 | GO:0072233 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.3 | 14.1 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.3 | 1.9 | GO:2000795 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.3 | 4.0 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
| 0.3 | 0.8 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.3 | 2.4 | GO:0006390 | mitochondrial DNA replication(GO:0006264) transcription from mitochondrial promoter(GO:0006390) |
| 0.3 | 1.1 | GO:0032972 | diaphragm contraction(GO:0002086) regulation of muscle filament sliding speed(GO:0032972) |
| 0.3 | 5.2 | GO:0010664 | negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.3 | 1.8 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.3 | 15.1 | GO:0035666 | TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 0.3 | 2.3 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.3 | 1.0 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.3 | 1.3 | GO:0098502 | DNA dephosphorylation(GO:0098502) |
| 0.3 | 1.0 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.2 | 2.8 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.2 | 3.0 | GO:0000278 | mitotic cell cycle(GO:0000278) |
| 0.2 | 10.1 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.2 | 1.3 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.2 | 1.4 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.2 | 1.6 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.2 | 1.0 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.2 | 9.3 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.2 | 2.3 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.2 | 2.6 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.2 | 1.3 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.2 | 1.1 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.2 | 3.3 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.2 | 2.7 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.2 | 7.7 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.2 | 0.5 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.2 | 2.8 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.2 | 8.2 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.2 | 3.5 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.2 | 0.8 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.2 | 1.4 | GO:0030091 | protein repair(GO:0030091) |
| 0.2 | 0.9 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.1 | 2.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.4 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.1 | 0.6 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 3.5 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 0.6 | GO:1903672 | positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.1 | 1.8 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 0.6 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.1 | 0.9 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 3.8 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.1 | 1.7 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 | 1.2 | GO:0015846 | polyamine transport(GO:0015846) |
| 0.1 | 0.4 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.1 | 1.8 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 1.6 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.1 | 1.0 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.1 | 0.8 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 1.7 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 6.2 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 2.0 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.1 | 0.8 | GO:0021903 | rostrocaudal neural tube patterning(GO:0021903) positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.1 | 2.6 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.1 | 8.8 | GO:0043535 | regulation of blood vessel endothelial cell migration(GO:0043535) |
| 0.1 | 0.2 | GO:2000110 | negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.1 | 1.9 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.1 | 3.2 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.1 | 3.3 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.1 | 9.2 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.1 | 0.7 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 1.7 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.1 | 10.1 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 0.9 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 2.6 | GO:0017004 | cytochrome complex assembly(GO:0017004) |
| 0.1 | 0.6 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 0.1 | 0.2 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.1 | 0.6 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.1 | 1.6 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.1 | 6.0 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.5 | GO:0009597 | detection of virus(GO:0009597) |
| 0.1 | 1.5 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.1 | 2.2 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 1.0 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 1.0 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.1 | 3.0 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.1 | 0.8 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 0.4 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 | 1.3 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.1 | 0.1 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.1 | 2.9 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 1.6 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.9 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.1 | 0.8 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.1 | 0.5 | GO:0042167 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.1 | 2.3 | GO:2000816 | negative regulation of mitotic sister chromatid separation(GO:2000816) |
| 0.1 | 1.3 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.1 | 1.2 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.1 | 0.4 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 2.3 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.8 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.1 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.9 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 1.1 | GO:0072663 | protein targeting to peroxisome(GO:0006625) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 1.7 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.5 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 2.1 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.0 | 0.7 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 1.2 | GO:0001501 | skeletal system development(GO:0001501) |
| 0.0 | 1.0 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.5 | GO:0055003 | actin filament network formation(GO:0051639) cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.9 | GO:0043586 | tongue development(GO:0043586) |
| 0.0 | 0.4 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.0 | 0.5 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 3.2 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 2.8 | GO:0007588 | excretion(GO:0007588) |
| 0.0 | 0.9 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 1.1 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.6 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 1.0 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.0 | 0.6 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 1.8 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.0 | 5.0 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 1.0 | GO:0060135 | response to progesterone(GO:0032570) maternal process involved in female pregnancy(GO:0060135) |
| 0.0 | 0.4 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.7 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.3 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.0 | 1.4 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 0.5 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 2.3 | GO:0010389 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) |
| 0.0 | 0.9 | GO:0007249 | I-kappaB kinase/NF-kappaB signaling(GO:0007249) |
| 0.0 | 0.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.3 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.2 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.1 | GO:0051058 | negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.0 | 0.8 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 1.9 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.0 | 0.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.5 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.0 | 1.0 | GO:0032204 | regulation of telomere maintenance(GO:0032204) |
| 0.0 | 0.6 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.5 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.7 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 2.1 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.6 | 53.2 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 1.4 | 11.5 | GO:0000322 | storage vacuole(GO:0000322) |
| 1.3 | 6.7 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 1.2 | 3.5 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 1.0 | 3.8 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.9 | 3.6 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.9 | 4.3 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.8 | 6.7 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.8 | 13.6 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.7 | 9.4 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.6 | 1.9 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.6 | 3.0 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.6 | 1.8 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.6 | 2.3 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.6 | 1.7 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.5 | 4.3 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.5 | 2.6 | GO:0033503 | HULC complex(GO:0033503) |
| 0.5 | 5.5 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.5 | 7.9 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.5 | 8.2 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.5 | 1.8 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.4 | 2.7 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.3 | 17.9 | GO:0031430 | M band(GO:0031430) |
| 0.3 | 1.0 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.3 | 4.1 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.3 | 3.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.3 | 4.1 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.3 | 2.3 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.3 | 2.8 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.2 | 2.3 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.2 | 8.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.2 | 4.5 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.2 | 0.6 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.2 | 4.0 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 2.1 | GO:0032797 | SMN complex(GO:0032797) |
| 0.2 | 1.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.2 | 2.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.2 | 6.7 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.2 | 2.3 | GO:0045120 | pronucleus(GO:0045120) |
| 0.2 | 1.3 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.2 | 1.9 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.2 | 2.9 | GO:0042599 | lamellar body(GO:0042599) |
| 0.1 | 2.8 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.1 | 1.0 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 2.5 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 2.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 1.5 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 6.2 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 1.7 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.1 | 0.4 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 3.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 1.2 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.1 | 2.1 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 6.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 6.0 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 5.1 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 7.8 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 8.2 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 1.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 1.5 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 0.2 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 2.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 1.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.5 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) |
| 0.1 | 10.3 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.1 | 2.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.9 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 1.2 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.1 | 10.0 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 3.4 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 6.0 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 2.1 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 1.4 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 3.0 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 1.8 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.1 | 6.4 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 7.7 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.4 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.6 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.8 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 1.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 9.8 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 5.8 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 18.7 | GO:0019866 | mitochondrial inner membrane(GO:0005743) organelle inner membrane(GO:0019866) |
| 0.0 | 4.3 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 3.6 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 1.5 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 1.3 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.2 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.6 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 2.8 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.4 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 1.4 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 1.6 | GO:0005741 | mitochondrial outer membrane(GO:0005741) organelle outer membrane(GO:0031968) |
| 0.0 | 2.7 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.3 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 2.5 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.9 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 11.5 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 9.4 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 2.8 | 11.2 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 2.6 | 7.9 | GO:0003978 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 2.6 | 7.8 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 1.8 | 5.5 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 1.8 | 54.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 1.6 | 4.9 | GO:0070546 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 1.5 | 4.5 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 1.5 | 4.5 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 1.5 | 2.9 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 1.3 | 3.9 | GO:0004853 | uroporphyrinogen decarboxylase activity(GO:0004853) |
| 1.2 | 4.6 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 1.0 | 9.3 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 1.0 | 3.0 | GO:0008413 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) NADH pyrophosphatase activity(GO:0035529) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 1.0 | 6.7 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.8 | 3.2 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.8 | 6.2 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.7 | 3.0 | GO:0035651 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.7 | 27.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.7 | 2.1 | GO:0004488 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.7 | 2.8 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.7 | 2.0 | GO:0070364 | mitochondrial light strand promoter anti-sense binding(GO:0070361) mitochondrial heavy strand promoter anti-sense binding(GO:0070362) mitochondrial heavy strand promoter sense binding(GO:0070364) |
| 0.7 | 6.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.6 | 1.9 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.6 | 8.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.6 | 1.2 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.6 | 8.5 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.6 | 2.4 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.6 | 8.1 | GO:0016594 | glycine binding(GO:0016594) |
| 0.6 | 3.4 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.5 | 2.5 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.5 | 1.5 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.5 | 5.8 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.5 | 1.9 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.5 | 4.1 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.4 | 13.6 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.4 | 5.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.4 | 3.0 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.4 | 2.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.4 | 8.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.4 | 8.2 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.4 | 4.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.4 | 2.6 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.4 | 2.9 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.4 | 1.8 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.4 | 9.9 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.3 | 10.1 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.3 | 15.3 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.3 | 1.3 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.3 | 6.3 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.3 | 3.6 | GO:0050544 | icosanoid binding(GO:0050542) arachidonic acid binding(GO:0050544) fatty acid derivative binding(GO:1901567) |
| 0.3 | 3.2 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.3 | 1.3 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) polynucleotide phosphatase activity(GO:0098518) |
| 0.3 | 3.0 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.3 | 1.4 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.3 | 0.8 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.3 | 2.3 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.3 | 1.0 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.2 | 2.8 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.2 | 0.7 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.2 | 1.8 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.2 | 16.1 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.2 | 1.3 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.2 | 2.2 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.2 | 1.8 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.2 | 6.9 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.2 | 7.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.2 | 2.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.2 | 5.7 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.2 | 2.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.2 | 4.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 1.2 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 3.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 1.5 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 1.9 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 2.3 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 7.4 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 3.8 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.9 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 0.5 | GO:0019862 | IgA binding(GO:0019862) |
| 0.1 | 0.2 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.1 | 0.9 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 2.5 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 0.8 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.1 | 2.7 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 1.5 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 1.5 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 0.9 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 1.6 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 2.0 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 1.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 3.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 0.3 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.1 | 1.4 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 0.9 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 1.0 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 0.3 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.1 | 3.3 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 2.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 5.9 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 0.4 | GO:0070404 | NADH binding(GO:0070404) |
| 0.1 | 1.3 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 1.8 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.1 | 2.8 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 2.1 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.5 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.1 | 1.0 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.1 | 0.2 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.1 | 1.4 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.1 | 2.1 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 0.4 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 0.6 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.1 | 2.6 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 1.7 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 3.8 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.2 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 2.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 10.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 8.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.5 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 1.0 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 2.9 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 1.6 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.5 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 1.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 6.3 | GO:0016758 | transferase activity, transferring hexosyl groups(GO:0016758) |
| 0.0 | 2.1 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 8.4 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 1.9 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.6 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.0 | 2.4 | GO:0008135 | translation factor activity, RNA binding(GO:0008135) |
| 0.0 | 3.1 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.2 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.9 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.6 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 1.0 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 2.8 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 0.4 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 4.9 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.4 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 7.3 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 2.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.6 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 1.0 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 1.3 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.6 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 1.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.6 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.3 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 54.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.6 | 20.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.5 | 13.6 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.3 | 9.8 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.3 | 5.9 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.3 | 6.7 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.3 | 9.9 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.2 | 9.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.2 | 5.0 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.2 | 11.8 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.2 | 6.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 12.6 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 10.0 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 9.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 7.9 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 4.0 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 1.9 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 6.4 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 6.4 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.1 | 1.6 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 8.5 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 1.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 0.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 1.5 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.1 | 2.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 2.4 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 1.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.2 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 1.0 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 1.0 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.6 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.9 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.6 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 1.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.0 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.9 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 2.2 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.5 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.2 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.3 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 54.9 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.8 | 20.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.5 | 5.9 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.5 | 7.0 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.5 | 13.6 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.4 | 11.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.4 | 8.5 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.3 | 12.8 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.3 | 12.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.3 | 7.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.3 | 9.9 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.2 | 6.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 9.8 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.2 | 15.5 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.2 | 4.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.2 | 3.9 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 4.9 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.2 | 6.0 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 5.5 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.2 | 4.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.2 | 2.7 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.2 | 2.6 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.2 | 3.6 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 2.0 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 5.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 1.9 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 4.2 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 1.0 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 1.1 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.1 | 1.7 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.1 | 9.4 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 1.8 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.1 | 13.0 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 1.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 1.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 2.0 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 2.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 2.2 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF MITOTIC PROTEINS | Genes involved in APC/C:Cdc20 mediated degradation of mitotic proteins |
| 0.1 | 1.8 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 5.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 2.6 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 2.1 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 2.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 2.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 4.8 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 0.8 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 2.1 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.8 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.9 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.4 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 1.0 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 5.1 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 1.2 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.4 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 2.1 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 0.8 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 1.0 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 1.6 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 1.9 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.0 | 0.2 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 2.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.6 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.3 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |