Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
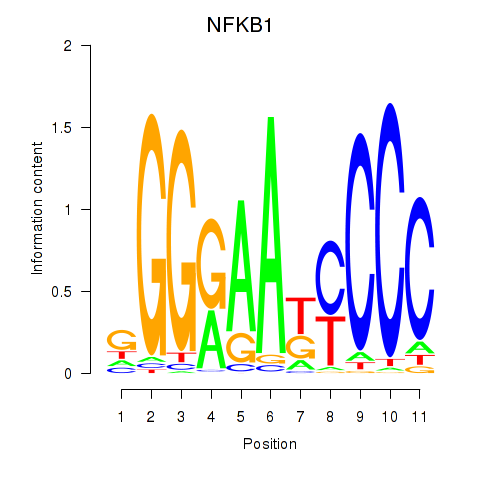
Results for NFKB1
Z-value: 1.55
Transcription factors associated with NFKB1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFKB1
|
ENSG00000109320.7 | nuclear factor kappa B subunit 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFKB1 | hg19_v2_chr4_+_103423055_103423112, hg19_v2_chr4_+_103422471_103422495 | 0.31 | 2.3e-06 | Click! |
Activity profile of NFKB1 motif
Sorted Z-values of NFKB1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_149792295 | 62.13 |
ENST00000518797.1
ENST00000524315.1 ENST00000009530.7 ENST00000377795.3 |
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain |
| chr19_+_1067492 | 34.80 |
ENST00000586866.1
|
HMHA1
|
histocompatibility (minor) HA-1 |
| chr6_+_32605195 | 34.68 |
ENST00000374949.2
|
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1 |
| chr19_+_1067144 | 34.53 |
ENST00000313093.2
|
HMHA1
|
histocompatibility (minor) HA-1 |
| chr19_+_1067271 | 33.34 |
ENST00000536472.1
ENST00000590214.1 |
HMHA1
|
histocompatibility (minor) HA-1 |
| chr6_+_138188551 | 32.31 |
ENST00000237289.4
ENST00000433680.1 |
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr1_+_6845384 | 30.19 |
ENST00000303635.7
|
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr22_-_37640277 | 28.82 |
ENST00000401529.3
ENST00000249071.6 |
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr16_+_50776021 | 28.35 |
ENST00000566679.2
ENST00000564634.1 ENST00000398568.2 |
CYLD
|
cylindromatosis (turban tumor syndrome) |
| chr19_+_42381173 | 26.51 |
ENST00000221972.3
|
CD79A
|
CD79a molecule, immunoglobulin-associated alpha |
| chr17_+_40440481 | 25.74 |
ENST00000590726.2
ENST00000452307.2 ENST00000444283.1 ENST00000588868.1 |
STAT5A
|
signal transducer and activator of transcription 5A |
| chr6_-_31550192 | 25.40 |
ENST00000429299.2
ENST00000446745.2 |
LTB
|
lymphotoxin beta (TNF superfamily, member 3) |
| chr22_-_37640456 | 25.09 |
ENST00000405484.1
ENST00000441619.1 ENST00000406508.1 |
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr6_+_32605134 | 24.19 |
ENST00000343139.5
ENST00000395363.1 ENST00000496318.1 |
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1 |
| chr16_+_50775971 | 23.38 |
ENST00000311559.9
ENST00000564326.1 ENST00000566206.1 |
CYLD
|
cylindromatosis (turban tumor syndrome) |
| chr16_+_50775948 | 22.22 |
ENST00000569681.1
ENST00000569418.1 ENST00000540145.1 |
CYLD
|
cylindromatosis (turban tumor syndrome) |
| chr19_+_42381337 | 22.12 |
ENST00000597454.1
ENST00000444740.2 |
CD79A
|
CD79a molecule, immunoglobulin-associated alpha |
| chr15_+_85923797 | 22.06 |
ENST00000559362.1
|
AKAP13
|
A kinase (PRKA) anchor protein 13 |
| chr15_+_85923856 | 19.48 |
ENST00000560302.1
ENST00000394518.2 ENST00000361243.2 ENST00000560256.1 |
AKAP13
|
A kinase (PRKA) anchor protein 13 |
| chr11_-_58345569 | 19.42 |
ENST00000528954.1
ENST00000528489.1 |
LPXN
|
leupaxin |
| chr6_+_29691198 | 19.27 |
ENST00000440587.2
ENST00000434407.2 |
HLA-F
|
major histocompatibility complex, class I, F |
| chr14_-_35873856 | 19.13 |
ENST00000553342.1
ENST00000216797.5 ENST00000557140.1 |
NFKBIA
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
| chr6_+_14117872 | 18.31 |
ENST00000379153.3
|
CD83
|
CD83 molecule |
| chr6_+_29691056 | 18.17 |
ENST00000414333.1
ENST00000334668.4 ENST00000259951.7 |
HLA-F
|
major histocompatibility complex, class I, F |
| chr2_-_177502659 | 18.05 |
ENST00000295549.4
|
AC017048.3
|
long intergenic non-protein coding RNA 1116 |
| chr3_+_101546827 | 17.87 |
ENST00000461724.1
ENST00000483180.1 ENST00000394054.2 |
NFKBIZ
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| chr14_+_73704201 | 17.81 |
ENST00000340738.5
ENST00000427855.1 ENST00000381166.3 |
PAPLN
|
papilin, proteoglycan-like sulfated glycoprotein |
| chr1_-_202130702 | 17.53 |
ENST00000309017.3
ENST00000477554.1 ENST00000492451.1 |
PTPN7
|
protein tyrosine phosphatase, non-receptor type 7 |
| chr11_+_128562372 | 16.59 |
ENST00000344954.6
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr2_+_61108771 | 16.26 |
ENST00000394479.3
|
REL
|
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr19_+_50922187 | 16.18 |
ENST00000595883.1
ENST00000597855.1 ENST00000596074.1 ENST00000439922.2 ENST00000594685.1 ENST00000270632.7 |
SPIB
|
Spi-B transcription factor (Spi-1/PU.1 related) |
| chr12_-_9913489 | 15.75 |
ENST00000228434.3
ENST00000536709.1 |
CD69
|
CD69 molecule |
| chr4_-_134070250 | 15.68 |
ENST00000505289.1
ENST00000509715.1 |
RP11-9G1.3
|
RP11-9G1.3 |
| chr2_+_61108650 | 15.49 |
ENST00000295025.8
|
REL
|
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr1_-_236030216 | 14.95 |
ENST00000389794.3
ENST00000389793.2 |
LYST
|
lysosomal trafficking regulator |
| chr6_+_32821924 | 14.90 |
ENST00000374859.2
ENST00000453265.2 |
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr1_-_202129105 | 14.61 |
ENST00000367279.4
|
PTPN7
|
protein tyrosine phosphatase, non-receptor type 7 |
| chr19_-_39390440 | 14.50 |
ENST00000249396.7
ENST00000414941.1 ENST00000392081.2 |
SIRT2
|
sirtuin 2 |
| chr17_-_61777090 | 14.35 |
ENST00000578061.1
|
LIMD2
|
LIM domain containing 2 |
| chr19_-_39390350 | 14.05 |
ENST00000447739.1
ENST00000358931.5 ENST00000407552.1 |
SIRT2
|
sirtuin 2 |
| chr2_+_97202480 | 13.30 |
ENST00000357485.3
|
ARID5A
|
AT rich interactive domain 5A (MRF1-like) |
| chr19_+_45504688 | 13.04 |
ENST00000221452.8
ENST00000540120.1 ENST00000505236.1 |
RELB
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr14_+_75988851 | 13.04 |
ENST00000555504.1
|
BATF
|
basic leucine zipper transcription factor, ATF-like |
| chr16_+_28834531 | 12.67 |
ENST00000570200.1
|
ATXN2L
|
ataxin 2-like |
| chr4_+_103422471 | 12.65 |
ENST00000226574.4
ENST00000394820.4 |
NFKB1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
| chr11_+_113930291 | 12.55 |
ENST00000335953.4
|
ZBTB16
|
zinc finger and BTB domain containing 16 |
| chr22_-_39151947 | 12.39 |
ENST00000216064.4
|
SUN2
|
Sad1 and UNC84 domain containing 2 |
| chr22_-_39151995 | 12.11 |
ENST00000405018.1
ENST00000438058.1 |
SUN2
|
Sad1 and UNC84 domain containing 2 |
| chr1_-_202129704 | 11.83 |
ENST00000476061.1
ENST00000544762.1 ENST00000467283.1 ENST00000464870.1 ENST00000435759.2 ENST00000486116.1 ENST00000543735.1 ENST00000308986.5 ENST00000477625.1 |
PTPN7
|
protein tyrosine phosphatase, non-receptor type 7 |
| chr19_+_2476116 | 11.62 |
ENST00000215631.4
ENST00000587345.1 |
GADD45B
|
growth arrest and DNA-damage-inducible, beta |
| chr5_-_127418755 | 11.44 |
ENST00000501702.2
ENST00000501173.2 ENST00000514573.1 ENST00000499346.2 ENST00000606251.1 |
CTC-228N24.3
|
CTC-228N24.3 |
| chr6_+_106534192 | 11.34 |
ENST00000369091.2
ENST00000369096.4 |
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr1_+_183441500 | 11.26 |
ENST00000456731.2
|
SMG7
|
SMG7 nonsense mediated mRNA decay factor |
| chr19_+_42363917 | 11.15 |
ENST00000598742.1
|
RPS19
|
ribosomal protein S19 |
| chr10_+_104154229 | 11.10 |
ENST00000428099.1
ENST00000369966.3 |
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr1_-_161168834 | 11.03 |
ENST00000367995.3
ENST00000367996.5 |
ADAMTS4
|
ADAM metallopeptidase with thrombospondin type 1 motif, 4 |
| chr6_-_32821599 | 10.98 |
ENST00000354258.4
|
TAP1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr1_+_156123359 | 10.69 |
ENST00000368284.1
ENST00000368286.2 ENST00000438830.1 |
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr14_-_64970494 | 10.69 |
ENST00000608382.1
|
ZBTB25
|
zinc finger and BTB domain containing 25 |
| chr12_+_52445191 | 10.62 |
ENST00000243050.1
ENST00000394825.1 ENST00000550763.1 ENST00000394824.2 ENST00000548232.1 ENST00000562373.1 |
NR4A1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr19_+_41222998 | 10.58 |
ENST00000263370.2
|
ITPKC
|
inositol-trisphosphate 3-kinase C |
| chr11_+_102188272 | 10.49 |
ENST00000532808.1
|
BIRC3
|
baculoviral IAP repeat containing 3 |
| chr10_+_30722866 | 10.38 |
ENST00000263056.1
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr1_+_156123318 | 10.33 |
ENST00000368285.3
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr2_+_176957619 | 10.22 |
ENST00000392539.3
|
HOXD13
|
homeobox D13 |
| chr3_-_50605077 | 9.74 |
ENST00000426034.1
ENST00000441239.1 |
C3orf18
|
chromosome 3 open reading frame 18 |
| chr12_+_100661156 | 9.73 |
ENST00000360820.2
|
SCYL2
|
SCY1-like 2 (S. cerevisiae) |
| chr6_+_29910301 | 9.55 |
ENST00000376809.5
ENST00000376802.2 |
HLA-A
|
major histocompatibility complex, class I, A |
| chrX_-_73072534 | 9.49 |
ENST00000429829.1
|
XIST
|
X inactive specific transcript (non-protein coding) |
| chr14_+_61789382 | 9.45 |
ENST00000555082.1
|
PRKCH
|
protein kinase C, eta |
| chr4_-_5891918 | 9.33 |
ENST00000512574.1
|
CRMP1
|
collapsin response mediator protein 1 |
| chr11_+_102188224 | 9.32 |
ENST00000263464.3
|
BIRC3
|
baculoviral IAP repeat containing 3 |
| chr16_+_3096638 | 9.24 |
ENST00000336577.4
|
MMP25
|
matrix metallopeptidase 25 |
| chr1_+_111770278 | 9.22 |
ENST00000369748.4
|
CHI3L2
|
chitinase 3-like 2 |
| chr19_-_54784353 | 9.21 |
ENST00000391746.1
|
LILRB2
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 2 |
| chr3_+_53195136 | 9.13 |
ENST00000394729.2
ENST00000330452.3 |
PRKCD
|
protein kinase C, delta |
| chr14_+_75536280 | 9.12 |
ENST00000238686.8
|
ZC2HC1C
|
zinc finger, C2HC-type containing 1C |
| chr19_+_42364460 | 9.06 |
ENST00000593863.1
|
RPS19
|
ribosomal protein S19 |
| chr1_+_37940153 | 9.00 |
ENST00000373087.6
|
ZC3H12A
|
zinc finger CCCH-type containing 12A |
| chr19_+_496454 | 8.97 |
ENST00000346144.4
ENST00000215637.3 ENST00000382683.4 |
MADCAM1
|
mucosal vascular addressin cell adhesion molecule 1 |
| chr11_-_64570706 | 8.96 |
ENST00000294066.2
ENST00000377350.3 |
MAP4K2
|
mitogen-activated protein kinase kinase kinase kinase 2 |
| chr9_+_82187630 | 8.92 |
ENST00000265284.6
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr3_+_10857885 | 8.82 |
ENST00000254488.2
ENST00000454147.1 |
SLC6A11
|
solute carrier family 6 (neurotransmitter transporter), member 11 |
| chr6_-_150039249 | 8.80 |
ENST00000543571.1
|
LATS1
|
large tumor suppressor kinase 1 |
| chr2_-_157189180 | 8.80 |
ENST00000539077.1
ENST00000424077.1 ENST00000426264.1 ENST00000339562.4 ENST00000421709.1 |
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr17_-_7197881 | 8.76 |
ENST00000007699.5
|
YBX2
|
Y box binding protein 2 |
| chr1_+_100818156 | 8.76 |
ENST00000336454.3
|
CDC14A
|
cell division cycle 14A |
| chr6_+_292051 | 8.71 |
ENST00000344450.5
|
DUSP22
|
dual specificity phosphatase 22 |
| chr9_+_82188077 | 8.71 |
ENST00000425506.1
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr14_+_75536335 | 8.32 |
ENST00000554763.1
ENST00000439583.2 ENST00000526130.1 ENST00000525046.1 |
ZC2HC1C
|
zinc finger, C2HC-type containing 1C |
| chr19_+_51728316 | 8.29 |
ENST00000436584.2
ENST00000421133.2 ENST00000391796.3 ENST00000262262.4 |
CD33
|
CD33 molecule |
| chr3_-_50605150 | 8.26 |
ENST00000357203.3
|
C3orf18
|
chromosome 3 open reading frame 18 |
| chr9_+_82187487 | 8.16 |
ENST00000435650.1
ENST00000414465.1 ENST00000376537.4 ENST00000376534.4 |
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr9_-_123691047 | 8.09 |
ENST00000373887.3
|
TRAF1
|
TNF receptor-associated factor 1 |
| chr4_-_74964904 | 8.05 |
ENST00000508487.2
|
CXCL2
|
chemokine (C-X-C motif) ligand 2 |
| chr17_-_1419878 | 7.95 |
ENST00000449479.1
ENST00000477910.1 ENST00000542125.1 ENST00000575172.1 |
INPP5K
|
inositol polyphosphate-5-phosphatase K |
| chr1_+_100818009 | 7.95 |
ENST00000370125.2
ENST00000361544.6 ENST00000370124.3 |
CDC14A
|
cell division cycle 14A |
| chr2_+_97203082 | 7.92 |
ENST00000454558.2
|
ARID5A
|
AT rich interactive domain 5A (MRF1-like) |
| chr11_-_72853091 | 7.90 |
ENST00000311172.7
ENST00000409314.1 |
FCHSD2
|
FCH and double SH3 domains 2 |
| chrX_-_37706661 | 7.84 |
ENST00000432389.2
ENST00000378581.3 |
DYNLT3
|
dynein, light chain, Tctex-type 3 |
| chr1_+_27561104 | 7.83 |
ENST00000361771.3
|
WDTC1
|
WD and tetratricopeptide repeats 1 |
| chr12_-_54982300 | 7.82 |
ENST00000547431.1
|
PPP1R1A
|
protein phosphatase 1, regulatory (inhibitor) subunit 1A |
| chr4_+_185395947 | 7.81 |
ENST00000605834.1
|
RP11-326I11.3
|
RP11-326I11.3 |
| chr12_+_11802753 | 7.80 |
ENST00000396373.4
|
ETV6
|
ets variant 6 |
| chr4_-_76944621 | 7.70 |
ENST00000306602.1
|
CXCL10
|
chemokine (C-X-C motif) ligand 10 |
| chr6_-_29527702 | 7.67 |
ENST00000377050.4
|
UBD
|
ubiquitin D |
| chr19_+_42364313 | 7.66 |
ENST00000601492.1
ENST00000600467.1 ENST00000221975.2 |
RPS19
|
ribosomal protein S19 |
| chr6_+_135502466 | 7.65 |
ENST00000367814.4
|
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr17_+_4634705 | 7.51 |
ENST00000575284.1
ENST00000573708.1 ENST00000293777.5 |
MED11
|
mediator complex subunit 11 |
| chr17_-_1419914 | 7.48 |
ENST00000397335.3
ENST00000574561.1 |
INPP5K
|
inositol polyphosphate-5-phosphatase K |
| chr17_-_7123021 | 7.45 |
ENST00000399510.2
|
DLG4
|
discs, large homolog 4 (Drosophila) |
| chr4_-_120550146 | 7.41 |
ENST00000354960.3
|
PDE5A
|
phosphodiesterase 5A, cGMP-specific |
| chr11_-_133826852 | 7.38 |
ENST00000533871.2
ENST00000321016.8 |
IGSF9B
|
immunoglobulin superfamily, member 9B |
| chr1_-_161279749 | 7.30 |
ENST00000533357.1
ENST00000360451.6 ENST00000336559.4 |
MPZ
|
myelin protein zero |
| chr17_-_1420182 | 7.29 |
ENST00000421807.2
|
INPP5K
|
inositol polyphosphate-5-phosphatase K |
| chr19_-_2042065 | 7.29 |
ENST00000591588.1
ENST00000591142.1 |
MKNK2
|
MAP kinase interacting serine/threonine kinase 2 |
| chr19_+_40697514 | 7.10 |
ENST00000253055.3
|
MAP3K10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr1_+_211432775 | 6.98 |
ENST00000419091.2
|
RCOR3
|
REST corepressor 3 |
| chr16_+_28835437 | 6.92 |
ENST00000568266.1
|
ATXN2L
|
ataxin 2-like |
| chr4_-_76928641 | 6.91 |
ENST00000264888.5
|
CXCL9
|
chemokine (C-X-C motif) ligand 9 |
| chr11_+_118754475 | 6.90 |
ENST00000292174.4
|
CXCR5
|
chemokine (C-X-C motif) receptor 5 |
| chr19_-_17958832 | 6.87 |
ENST00000458235.1
|
JAK3
|
Janus kinase 3 |
| chr5_-_95297534 | 6.87 |
ENST00000513343.1
ENST00000431061.2 |
ELL2
|
elongation factor, RNA polymerase II, 2 |
| chr17_-_1420006 | 6.86 |
ENST00000320345.6
ENST00000406424.4 |
INPP5K
|
inositol polyphosphate-5-phosphatase K |
| chr5_-_95297678 | 6.79 |
ENST00000237853.4
|
ELL2
|
elongation factor, RNA polymerase II, 2 |
| chr17_-_40333150 | 6.75 |
ENST00000264661.3
|
KCNH4
|
potassium voltage-gated channel, subfamily H (eag-related), member 4 |
| chrX_+_147582228 | 6.68 |
ENST00000342251.3
|
AFF2
|
AF4/FMR2 family, member 2 |
| chr6_-_44233361 | 6.68 |
ENST00000275015.5
|
NFKBIE
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon |
| chr6_-_33160231 | 6.67 |
ENST00000395194.1
ENST00000457788.1 ENST00000341947.2 ENST00000357486.1 ENST00000374714.1 ENST00000374713.1 ENST00000395197.1 ENST00000374712.1 ENST00000361917.1 ENST00000374708.4 |
COL11A2
|
collagen, type XI, alpha 2 |
| chr11_+_94706973 | 6.61 |
ENST00000536741.1
|
KDM4D
|
lysine (K)-specific demethylase 4D |
| chr16_+_28834303 | 6.58 |
ENST00000340394.8
ENST00000325215.6 ENST00000395547.2 ENST00000336783.4 ENST00000382686.4 ENST00000564304.1 |
ATXN2L
|
ataxin 2-like |
| chr3_-_194072019 | 6.57 |
ENST00000429275.1
ENST00000323830.3 |
CPN2
|
carboxypeptidase N, polypeptide 2 |
| chr6_+_144471643 | 6.56 |
ENST00000367568.4
|
STX11
|
syntaxin 11 |
| chr2_-_206950781 | 6.47 |
ENST00000403263.1
|
INO80D
|
INO80 complex subunit D |
| chr7_+_143013198 | 6.45 |
ENST00000343257.2
|
CLCN1
|
chloride channel, voltage-sensitive 1 |
| chr15_-_77712477 | 6.39 |
ENST00000560626.2
|
PEAK1
|
pseudopodium-enriched atypical kinase 1 |
| chr11_+_1860682 | 6.38 |
ENST00000381906.1
|
TNNI2
|
troponin I type 2 (skeletal, fast) |
| chr17_-_40333099 | 6.26 |
ENST00000607371.1
|
KCNH4
|
potassium voltage-gated channel, subfamily H (eag-related), member 4 |
| chr8_+_73449625 | 6.22 |
ENST00000523207.1
|
KCNB2
|
potassium voltage-gated channel, Shab-related subfamily, member 2 |
| chr19_-_13617247 | 6.22 |
ENST00000573710.2
|
CACNA1A
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
| chr1_+_211432593 | 6.17 |
ENST00000367006.4
|
RCOR3
|
REST corepressor 3 |
| chr1_+_27561007 | 6.15 |
ENST00000319394.3
|
WDTC1
|
WD and tetratricopeptide repeats 1 |
| chr2_-_100721923 | 6.13 |
ENST00000356421.2
|
AFF3
|
AF4/FMR2 family, member 3 |
| chr20_+_44746885 | 6.03 |
ENST00000372285.3
|
CD40
|
CD40 molecule, TNF receptor superfamily member 5 |
| chr1_+_111770232 | 5.93 |
ENST00000369744.2
|
CHI3L2
|
chitinase 3-like 2 |
| chr9_-_136344197 | 5.89 |
ENST00000414172.1
ENST00000371897.4 |
SLC2A6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr11_-_47399942 | 5.76 |
ENST00000227163.4
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr11_-_47400078 | 5.66 |
ENST00000378538.3
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr1_-_57889687 | 5.59 |
ENST00000371236.2
ENST00000371230.1 |
DAB1
|
Dab, reelin signal transducer, homolog 1 (Drosophila) |
| chr11_-_3862206 | 5.53 |
ENST00000351018.4
|
RHOG
|
ras homolog family member G |
| chr12_-_54982420 | 5.48 |
ENST00000257905.8
|
PPP1R1A
|
protein phosphatase 1, regulatory (inhibitor) subunit 1A |
| chr12_-_57504069 | 5.47 |
ENST00000543873.2
ENST00000554663.1 ENST00000557635.1 |
STAT6
|
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr1_+_54359854 | 5.43 |
ENST00000361921.3
ENST00000322679.6 ENST00000532493.1 ENST00000525202.1 ENST00000524406.1 ENST00000388876.3 |
DIO1
|
deiodinase, iodothyronine, type I |
| chr6_+_31949801 | 5.43 |
ENST00000428956.2
ENST00000498271.1 |
C4A
|
complement component 4A (Rodgers blood group) |
| chr9_-_115480303 | 5.34 |
ENST00000374234.1
ENST00000374238.1 ENST00000374236.1 ENST00000374242.4 |
INIP
|
INTS3 and NABP interacting protein |
| chr5_-_135231516 | 5.34 |
ENST00000274520.1
|
IL9
|
interleukin 9 |
| chr6_-_33168391 | 5.33 |
ENST00000374685.4
ENST00000413614.2 ENST00000374680.3 |
RXRB
|
retinoid X receptor, beta |
| chr4_-_76957214 | 5.27 |
ENST00000306621.3
|
CXCL11
|
chemokine (C-X-C motif) ligand 11 |
| chr15_-_27018884 | 5.26 |
ENST00000299267.4
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr2_-_152955213 | 5.18 |
ENST00000427385.1
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr11_+_63997750 | 5.05 |
ENST00000321685.3
|
DNAJC4
|
DnaJ (Hsp40) homolog, subfamily C, member 4 |
| chr20_+_44746939 | 5.04 |
ENST00000372276.3
|
CD40
|
CD40 molecule, TNF receptor superfamily member 5 |
| chr6_-_109762344 | 4.98 |
ENST00000521072.2
ENST00000424445.2 ENST00000440797.2 |
PPIL6
|
peptidylprolyl isomerase (cyclophilin)-like 6 |
| chr5_-_150521192 | 4.96 |
ENST00000523714.1
ENST00000521749.1 |
ANXA6
|
annexin A6 |
| chr3_+_137483579 | 4.96 |
ENST00000306087.1
|
SOX14
|
SRY (sex determining region Y)-box 14 |
| chr10_-_51371321 | 4.93 |
ENST00000602930.1
|
AGAP8
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 8 |
| chr2_+_208394658 | 4.89 |
ENST00000421139.1
|
CREB1
|
cAMP responsive element binding protein 1 |
| chr4_-_120549163 | 4.82 |
ENST00000394439.1
ENST00000420633.1 |
PDE5A
|
phosphodiesterase 5A, cGMP-specific |
| chr1_-_16302565 | 4.79 |
ENST00000537142.1
ENST00000448462.2 |
ZBTB17
|
zinc finger and BTB domain containing 17 |
| chr2_-_191885686 | 4.78 |
ENST00000432058.1
|
STAT1
|
signal transducer and activator of transcription 1, 91kDa |
| chr11_+_118307179 | 4.76 |
ENST00000534358.1
ENST00000531904.2 ENST00000389506.5 ENST00000354520.4 |
KMT2A
|
lysine (K)-specific methyltransferase 2A |
| chr4_-_76598326 | 4.74 |
ENST00000503660.1
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr15_+_89346699 | 4.72 |
ENST00000558207.1
|
ACAN
|
aggrecan |
| chr12_-_54813229 | 4.71 |
ENST00000293379.4
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| chr19_-_40324255 | 4.66 |
ENST00000593685.1
ENST00000600611.1 |
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr19_+_39390320 | 4.65 |
ENST00000576510.1
|
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr19_-_2050852 | 4.58 |
ENST00000541165.1
ENST00000591601.1 |
MKNK2
|
MAP kinase interacting serine/threonine kinase 2 |
| chr5_-_96143602 | 4.55 |
ENST00000443439.2
ENST00000503921.1 ENST00000508227.1 ENST00000507154.1 |
ERAP1
|
endoplasmic reticulum aminopeptidase 1 |
| chr5_+_139781445 | 4.53 |
ENST00000532219.1
ENST00000394722.3 |
ANKHD1-EIF4EBP3
ANKHD1
|
ANKHD1-EIF4EBP3 readthrough ankyrin repeat and KH domain containing 1 |
| chr12_+_8185288 | 4.52 |
ENST00000162391.3
|
FOXJ2
|
forkhead box J2 |
| chr3_+_9773409 | 4.50 |
ENST00000433861.2
ENST00000424362.1 ENST00000383829.2 ENST00000302054.3 ENST00000420291.1 |
BRPF1
|
bromodomain and PHD finger containing, 1 |
| chr14_+_76618242 | 4.44 |
ENST00000557542.1
ENST00000557263.1 ENST00000557207.1 ENST00000312858.5 ENST00000261530.7 |
GPATCH2L
|
G patch domain containing 2-like |
| chr2_-_68694390 | 4.38 |
ENST00000377957.3
|
FBXO48
|
F-box protein 48 |
| chr1_-_16302608 | 4.37 |
ENST00000375743.4
ENST00000375733.2 |
ZBTB17
|
zinc finger and BTB domain containing 17 |
| chr4_-_185395672 | 4.33 |
ENST00000393593.3
|
IRF2
|
interferon regulatory factor 2 |
| chr1_+_154301264 | 4.31 |
ENST00000341822.2
|
ATP8B2
|
ATPase, aminophospholipid transporter, class I, type 8B, member 2 |
| chr14_+_32798462 | 4.28 |
ENST00000280979.4
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr17_-_1389228 | 4.27 |
ENST00000438665.2
|
MYO1C
|
myosin IC |
| chr2_-_152955537 | 4.27 |
ENST00000201943.5
ENST00000539935.1 |
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr10_+_12391481 | 4.24 |
ENST00000378847.3
|
CAMK1D
|
calcium/calmodulin-dependent protein kinase ID |
| chr19_-_45826125 | 4.21 |
ENST00000221476.3
|
CKM
|
creatine kinase, muscle |
| chr3_-_49131013 | 4.14 |
ENST00000424300.1
|
QRICH1
|
glutamine-rich 1 |
| chr2_-_100721178 | 4.14 |
ENST00000409236.2
|
AFF3
|
AF4/FMR2 family, member 3 |
| chr14_+_64970662 | 4.06 |
ENST00000556965.1
ENST00000554015.1 |
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr15_-_71055878 | 4.05 |
ENST00000322954.6
|
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr17_+_63133587 | 4.05 |
ENST00000449996.3
ENST00000262406.9 |
RGS9
|
regulator of G-protein signaling 9 |
| chr1_-_146644036 | 4.03 |
ENST00000425272.2
|
PRKAB2
|
protein kinase, AMP-activated, beta 2 non-catalytic subunit |
| chr6_+_149068464 | 4.03 |
ENST00000367463.4
|
UST
|
uronyl-2-sulfotransferase |
| chr11_+_94706804 | 4.02 |
ENST00000335080.5
|
KDM4D
|
lysine (K)-specific demethylase 4D |
| chr19_-_17958771 | 4.02 |
ENST00000534444.1
|
JAK3
|
Janus kinase 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NFKB1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 24.7 | 74.0 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 13.8 | 41.5 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 12.4 | 62.1 | GO:0002905 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 11.0 | 33.1 | GO:0090291 | negative regulation of osteoclast proliferation(GO:0090291) |
| 9.5 | 28.5 | GO:2000777 | positive regulation of oocyte maturation(GO:1900195) negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 7.4 | 29.6 | GO:2001153 | regulation of renal water transport(GO:2001151) positive regulation of renal water transport(GO:2001153) |
| 7.0 | 27.9 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 6.4 | 25.7 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 5.4 | 59.3 | GO:0060263 | regulation of respiratory burst(GO:0060263) |
| 5.0 | 15.0 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 4.8 | 47.5 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 4.3 | 21.6 | GO:2000630 | positive regulation of miRNA metabolic process(GO:2000630) |
| 4.2 | 25.4 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 4.1 | 24.5 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 4.1 | 12.2 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 3.8 | 19.1 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 3.8 | 15.1 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 3.6 | 10.9 | GO:0045210 | negative regulation of dendritic cell cytokine production(GO:0002731) FasL biosynthetic process(GO:0045210) |
| 3.5 | 10.6 | GO:1904617 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 3.4 | 23.9 | GO:0051136 | regulation of NK T cell differentiation(GO:0051136) |
| 3.2 | 12.7 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 2.9 | 8.8 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 2.7 | 11.0 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 2.6 | 44.8 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 2.5 | 7.6 | GO:1904897 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 2.4 | 7.3 | GO:0021589 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 2.4 | 19.4 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 2.3 | 9.2 | GO:0002774 | Fc receptor mediated inhibitory signaling pathway(GO:0002774) |
| 2.3 | 9.1 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 2.3 | 2.3 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 2.2 | 11.1 | GO:0033590 | response to cobalamin(GO:0033590) |
| 2.2 | 19.8 | GO:0070424 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) |
| 2.2 | 8.8 | GO:0009386 | translational attenuation(GO:0009386) |
| 2.1 | 10.6 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 2.0 | 18.3 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 2.0 | 26.2 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 2.0 | 19.9 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 1.9 | 13.0 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 1.8 | 7.3 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 1.8 | 8.8 | GO:0001826 | inner cell mass cell differentiation(GO:0001826) |
| 1.7 | 5.2 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 1.7 | 10.2 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 1.7 | 6.7 | GO:0060023 | soft palate development(GO:0060023) |
| 1.7 | 9.9 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 1.6 | 9.4 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 1.5 | 7.7 | GO:0070842 | aggresome assembly(GO:0070842) |
| 1.5 | 10.6 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 1.5 | 7.4 | GO:0097688 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 1.4 | 9.7 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 1.4 | 15.2 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 1.4 | 5.5 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 1.3 | 6.7 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 1.3 | 7.8 | GO:0007296 | vitellogenesis(GO:0007296) |
| 1.3 | 3.9 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) regulation of chromatin silencing at telomere(GO:0031938) |
| 1.3 | 3.8 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 1.2 | 7.4 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 1.2 | 2.5 | GO:0006405 | RNA export from nucleus(GO:0006405) |
| 1.2 | 18.1 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 1.2 | 4.8 | GO:0072308 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of nephron tubule epithelial cell differentiation(GO:0072183) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) negative regulation of epithelial cell differentiation involved in kidney development(GO:2000697) |
| 1.2 | 4.8 | GO:1901674 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 1.1 | 6.7 | GO:0072553 | terminal button organization(GO:0072553) |
| 1.1 | 3.3 | GO:0070507 | regulation of microtubule-based process(GO:0032886) regulation of microtubule cytoskeleton organization(GO:0070507) |
| 1.1 | 3.3 | GO:0031394 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 1.0 | 9.2 | GO:0060022 | hard palate development(GO:0060022) |
| 1.0 | 4.1 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 1.0 | 7.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 1.0 | 2.0 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) |
| 1.0 | 4.0 | GO:0051142 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 1.0 | 21.9 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 1.0 | 3.8 | GO:0014859 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.9 | 5.4 | GO:1904550 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.9 | 16.6 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.9 | 13.1 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.9 | 3.4 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.8 | 2.3 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) leukocyte migration involved in immune response(GO:0002522) |
| 0.8 | 6.3 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.8 | 1.6 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.7 | 2.2 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.7 | 7.7 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.7 | 13.9 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.7 | 14.5 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.7 | 8.7 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.7 | 6.7 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.7 | 72.2 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.6 | 3.8 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.6 | 1.9 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.6 | 11.9 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.6 | 25.8 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.5 | 2.7 | GO:0030070 | insulin processing(GO:0030070) |
| 0.5 | 30.2 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.5 | 45.1 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.5 | 4.2 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.5 | 4.7 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.5 | 8.4 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.5 | 8.8 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.4 | 4.0 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.4 | 2.2 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.4 | 5.4 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.4 | 17.1 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.4 | 1.2 | GO:0036378 | calcitriol biosynthetic process from calciol(GO:0036378) |
| 0.4 | 2.0 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.4 | 1.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.4 | 1.5 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.4 | 7.8 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.4 | 11.1 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.4 | 1.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.3 | 9.0 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.3 | 6.6 | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.3 | 21.2 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.3 | 6.1 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.3 | 2.4 | GO:0060371 | regulation of atrial cardiac muscle cell membrane depolarization(GO:0060371) |
| 0.3 | 1.5 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.3 | 2.6 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.3 | 30.8 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.3 | 7.3 | GO:0098743 | cell aggregation(GO:0098743) |
| 0.3 | 0.8 | GO:0044501 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.3 | 2.7 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.3 | 0.5 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.3 | 1.6 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.3 | 0.3 | GO:0003099 | positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.2 | 5.4 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.2 | 10.0 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.2 | 0.2 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.2 | 5.3 | GO:0032754 | positive regulation of interleukin-5 production(GO:0032754) |
| 0.2 | 1.8 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.2 | 5.0 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.2 | 9.4 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.2 | 9.9 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.2 | 9.8 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.2 | 5.5 | GO:0090004 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.2 | 5.9 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.2 | 1.0 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.2 | 7.9 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.2 | 1.5 | GO:0042428 | serotonin metabolic process(GO:0042428) |
| 0.2 | 4.3 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.2 | 5.1 | GO:0035966 | response to unfolded protein(GO:0006986) response to topologically incorrect protein(GO:0035966) |
| 0.2 | 6.2 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.2 | 4.7 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.2 | 13.3 | GO:0005977 | glycogen metabolic process(GO:0005977) |
| 0.2 | 3.1 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.1 | 1.6 | GO:0003084 | positive regulation of systemic arterial blood pressure(GO:0003084) |
| 0.1 | 5.0 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.1 | 0.5 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.1 | 7.4 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.1 | 2.5 | GO:0050855 | regulation of B cell receptor signaling pathway(GO:0050855) |
| 0.1 | 2.7 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 10.6 | GO:0043647 | inositol phosphate metabolic process(GO:0043647) |
| 0.1 | 18.4 | GO:0035303 | regulation of dephosphorylation(GO:0035303) |
| 0.1 | 0.8 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 | 0.6 | GO:0009629 | response to gravity(GO:0009629) |
| 0.1 | 5.0 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 2.9 | GO:0043403 | skeletal muscle tissue regeneration(GO:0043403) |
| 0.1 | 5.3 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 1.2 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.1 | 0.4 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.1 | 1.5 | GO:0006941 | striated muscle contraction(GO:0006941) |
| 0.1 | 0.9 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.1 | 1.5 | GO:0090003 | regulation of establishment of protein localization to plasma membrane(GO:0090003) |
| 0.1 | 2.5 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.1 | 3.3 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.1 | 1.7 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.1 | 4.5 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.1 | 1.7 | GO:0006833 | water transport(GO:0006833) |
| 0.1 | 6.2 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.1 | 4.7 | GO:0060337 | type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.1 | 0.2 | GO:1902741 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) interferon-beta secretion(GO:0035546) regulation of interferon-beta secretion(GO:0035547) positive regulation of interferon-beta secretion(GO:0035549) type I interferon secretion(GO:0072641) interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 0.1 | 6.6 | GO:0030449 | regulation of complement activation(GO:0030449) regulation of protein activation cascade(GO:2000257) |
| 0.0 | 4.0 | GO:0042795 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 2.8 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 2.6 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 1.9 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 6.5 | GO:0006310 | DNA recombination(GO:0006310) |
| 0.0 | 1.7 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 5.3 | GO:0010212 | response to ionizing radiation(GO:0010212) |
| 0.0 | 1.7 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.3 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 2.2 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 2.8 | GO:0071222 | cellular response to lipopolysaccharide(GO:0071222) |
| 0.0 | 1.3 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 3.9 | GO:2000116 | regulation of cysteine-type endopeptidase activity(GO:2000116) |
| 0.0 | 4.4 | GO:0031098 | stress-activated protein kinase signaling cascade(GO:0031098) |
| 0.0 | 0.3 | GO:0043524 | negative regulation of neuron apoptotic process(GO:0043524) |
| 0.0 | 0.3 | GO:0097190 | apoptotic signaling pathway(GO:0097190) |
| 0.0 | 1.8 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 1.2 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.9 | 62.1 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 7.4 | 89.3 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 6.9 | 48.6 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 4.3 | 47.5 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 3.2 | 28.5 | GO:0072687 | meiotic spindle(GO:0072687) |
| 2.9 | 58.9 | GO:0042611 | MHC protein complex(GO:0042611) |
| 2.6 | 18.1 | GO:0060091 | kinocilium(GO:0060091) |
| 1.9 | 24.5 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 1.9 | 14.9 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 1.9 | 7.4 | GO:0045160 | myosin I complex(GO:0045160) |
| 1.8 | 11.0 | GO:0042825 | TAP complex(GO:0042825) |
| 1.7 | 74.0 | GO:0097542 | ciliary tip(GO:0097542) |
| 1.5 | 9.0 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 1.5 | 17.8 | GO:0015630 | microtubule cytoskeleton(GO:0015630) |
| 1.4 | 11.1 | GO:0043196 | varicosity(GO:0043196) |
| 1.3 | 5.3 | GO:0070876 | SOSS complex(GO:0070876) |
| 1.3 | 4.0 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 1.2 | 4.7 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 1.1 | 7.8 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 1.0 | 9.3 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.8 | 5.4 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.8 | 56.6 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.7 | 1.5 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.6 | 3.1 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.6 | 7.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.6 | 1.7 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.6 | 6.7 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.5 | 2.7 | GO:1990393 | 3M complex(GO:1990393) |
| 0.5 | 3.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.5 | 2.5 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.5 | 2.9 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.5 | 1.9 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.5 | 4.5 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.4 | 22.0 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.4 | 18.4 | GO:0002102 | podosome(GO:0002102) |
| 0.4 | 9.9 | GO:0005861 | troponin complex(GO:0005861) |
| 0.4 | 2.0 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.4 | 10.6 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.4 | 27.9 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.4 | 26.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.4 | 41.1 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.3 | 33.9 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.3 | 6.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.3 | 8.8 | GO:0000922 | spindle pole(GO:0000922) |
| 0.3 | 2.6 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.3 | 31.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.2 | 3.8 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.2 | 7.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.2 | 6.2 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.2 | 47.9 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.2 | 5.3 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.2 | 1.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 56.7 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.2 | 11.0 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.2 | 1.6 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.2 | 9.4 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.2 | 3.9 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.2 | 6.4 | GO:0005925 | cell-substrate adherens junction(GO:0005924) focal adhesion(GO:0005925) cell-substrate junction(GO:0030055) |
| 0.2 | 7.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.2 | 1.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 6.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 13.1 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 7.9 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 6.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 4.8 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 0.4 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.1 | 6.6 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 11.5 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 9.2 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.1 | 16.8 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.1 | 5.1 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 1.1 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 43.5 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 28.2 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.1 | 0.8 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 2.7 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.1 | 9.2 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.1 | 2.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 1.3 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 5.5 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 10.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 23.8 | GO:0016604 | nuclear body(GO:0016604) |
| 0.1 | 10.6 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 3.3 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.1 | 1.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 5.1 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 5.5 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 2.7 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 5.0 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 9.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 24.8 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.6 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 6.6 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 2.3 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 12.1 | GO:0000323 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 4.6 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.5 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 12.4 | GO:0005615 | extracellular space(GO:0005615) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.4 | 62.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 10.6 | 106.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 9.5 | 28.5 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 8.1 | 48.4 | GO:0046979 | TAP2 binding(GO:0046979) |
| 4.9 | 29.6 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 4.5 | 58.9 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 4.0 | 19.9 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 3.7 | 44.9 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 3.7 | 18.6 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 3.0 | 9.0 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 2.5 | 7.4 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 2.4 | 2.4 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 2.2 | 13.0 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 1.8 | 7.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 1.8 | 10.6 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 1.7 | 13.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 1.6 | 4.8 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 1.6 | 12.5 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 1.6 | 6.3 | GO:0005151 | interleukin-1, Type II receptor binding(GO:0005151) |
| 1.5 | 75.7 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 1.5 | 8.8 | GO:0015185 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 1.4 | 15.2 | GO:0008061 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 1.3 | 5.3 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 1.2 | 21.0 | GO:0038191 | neuropilin binding(GO:0038191) |
| 1.2 | 9.2 | GO:0032396 | inhibitory MHC class I receptor activity(GO:0032396) |
| 1.1 | 6.7 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 1.0 | 30.0 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 1.0 | 19.6 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.9 | 18.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.9 | 5.4 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.9 | 5.4 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.9 | 4.5 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.8 | 3.3 | GO:0050473 | prostaglandin-endoperoxide synthase activity(GO:0004666) arachidonate 15-lipoxygenase activity(GO:0050473) |
| 0.8 | 27.9 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.8 | 3.1 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.8 | 3.9 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.8 | 10.5 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.7 | 35.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.7 | 9.9 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.7 | 7.6 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.6 | 9.0 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.6 | 1.9 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) oncostatin-M receptor activity(GO:0004924) interleukin-6 binding(GO:0019981) |
| 0.6 | 23.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.6 | 6.9 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.6 | 11.9 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.6 | 6.7 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.6 | 26.8 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.6 | 3.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.6 | 9.5 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.6 | 10.6 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.6 | 5.5 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.5 | 1.6 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.5 | 7.4 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.5 | 3.2 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.5 | 4.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.5 | 1.5 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.5 | 10.6 | GO:0031005 | filamin binding(GO:0031005) |
| 0.5 | 14.9 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.5 | 2.9 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.4 | 6.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.4 | 13.1 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.4 | 1.7 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.4 | 12.3 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.4 | 3.8 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.4 | 12.2 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.4 | 5.9 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.4 | 5.3 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.4 | 17.9 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.4 | 1.2 | GO:0070643 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.4 | 25.8 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.4 | 2.3 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.4 | 11.3 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.4 | 1.8 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.3 | 30.8 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.3 | 7.0 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.3 | 11.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.3 | 38.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.3 | 19.7 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.3 | 1.9 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.3 | 2.3 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.3 | 6.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.3 | 0.6 | GO:0035877 | death effector domain binding(GO:0035877) cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.3 | 6.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.3 | 3.8 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.2 | 60.6 | GO:0019887 | protein kinase regulator activity(GO:0019887) |
| 0.2 | 8.8 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.2 | 0.9 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.2 | 2.0 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.2 | 18.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.2 | 7.3 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.2 | 6.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 5.4 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.2 | 6.0 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.2 | 2.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.2 | 2.2 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.2 | 4.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.2 | 8.9 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.2 | 12.0 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.2 | 3.8 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.2 | 7.5 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.1 | 48.5 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.1 | 6.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 2.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 0.5 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.1 | 1.5 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 5.5 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 26.2 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 7.4 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.1 | 7.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 5.0 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 22.2 | GO:0003712 | transcription cofactor activity(GO:0003712) |
| 0.1 | 0.6 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 1.6 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 2.5 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 0.4 | GO:0070905 | serine binding(GO:0070905) |
| 0.1 | 2.3 | GO:0008200 | ion channel inhibitor activity(GO:0008200) |
| 0.1 | 47.5 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.1 | 2.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 4.0 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.1 | 0.5 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 0.2 | GO:0019976 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.1 | 1.1 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 0.3 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 14.0 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 1.7 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.6 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 6.1 | GO:0019900 | kinase binding(GO:0019900) |
| 0.0 | 3.1 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.3 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.8 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 2.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 138.3 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 2.2 | 48.0 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 1.3 | 42.6 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 1.2 | 50.7 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 1.1 | 33.3 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 1.1 | 52.0 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 1.0 | 25.6 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.7 | 20.0 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.7 | 48.6 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.7 | 19.9 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.7 | 23.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.5 | 17.3 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.5 | 34.5 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.5 | 8.1 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.5 | 2.9 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.5 | 24.5 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.5 | 17.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.4 | 29.1 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.4 | 18.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.3 | 9.0 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.3 | 1.9 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.3 | 4.7 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.3 | 7.4 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.2 | 6.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 7.8 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.2 | 9.4 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.2 | 9.2 | PID MYC PATHWAY | C-MYC pathway |
| 0.2 | 10.6 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.2 | 8.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 2.7 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 23.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 5.6 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 3.3 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 4.0 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 26.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 6.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 16.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 1.9 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 1.8 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 2.5 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 5.5 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 4.2 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 3.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 3.8 | PID P73PATHWAY | p73 transcription factor network |
| 0.1 | 4.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 1.7 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 2.4 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.3 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.2 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 47.5 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 2.6 | 128.3 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 2.6 | 58.9 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 1.8 | 37.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 1.7 | 19.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 1.3 | 48.6 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 1.2 | 59.4 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 1.1 | 30.9 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 1.0 | 18.9 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.9 | 21.0 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.7 | 2.9 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.7 | 16.0 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.7 | 35.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.7 | 17.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.6 | 18.1 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.6 | 10.4 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.6 | 65.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.5 | 12.8 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.5 | 23.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.5 | 8.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.5 | 10.6 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.5 | 18.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.4 | 6.7 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.4 | 8.8 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.4 | 44.3 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.4 | 25.8 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.4 | 10.7 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.4 | 27.9 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.3 | 12.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.3 | 5.4 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.3 | 5.9 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.3 | 20.3 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.3 | 16.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.3 | 7.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.2 | 9.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 6.6 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.2 | 5.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.2 | 17.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.2 | 1.2 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.2 | 11.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.2 | 0.5 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.2 | 2.5 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.2 | 5.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 1.5 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.2 | 15.3 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.2 | 9.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.2 | 2.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 6.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 3.1 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 0.6 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.1 | 11.3 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.1 | 9.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 2.0 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.1 | 4.7 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.1 | 2.7 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 4.5 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 2.7 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.1 | 1.7 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 2.0 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 5.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 2.8 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 2.9 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 4.1 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 5.0 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 1.7 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 2.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 1.9 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 1.7 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.2 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |