|
chr1_-_26233423
Show fit
|
11.03 |
ENST00000357865.2
|
STMN1
|
stathmin 1
|
|
chr1_-_41131326
Show fit
|
7.92 |
ENST00000372684.3
|
RIMS3
|
regulating synaptic membrane exocytosis 3
|
|
chr17_-_79139817
Show fit
|
7.68 |
ENST00000326724.4
|
AATK
|
apoptosis-associated tyrosine kinase
|
|
chr1_-_11866034
Show fit
|
6.87 |
ENST00000376590.3
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H)
|
|
chr19_-_23578220
Show fit
|
6.71 |
ENST00000595533.1
ENST00000397082.2
ENST00000599743.1
ENST00000300619.7
|
ZNF91
|
zinc finger protein 91
|
|
chr13_+_113622757
Show fit
|
6.70 |
ENST00000375604.2
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr13_+_113622810
Show fit
|
6.17 |
ENST00000397030.1
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr5_-_111312622
Show fit
|
5.93 |
ENST00000395634.3
|
NREP
|
neuronal regeneration related protein
|
|
chr14_+_24540046
Show fit
|
5.89 |
ENST00000397016.2
ENST00000537691.1
ENST00000560356.1
ENST00000558450.1
|
CPNE6
|
copine VI (neuronal)
|
|
chr1_-_11865982
Show fit
|
5.85 |
ENST00000418034.1
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H)
|
|
chr9_+_126773880
Show fit
|
5.78 |
ENST00000373615.4
|
LHX2
|
LIM homeobox 2
|
|
chr14_-_23770683
Show fit
|
5.59 |
ENST00000561437.1
ENST00000559942.1
ENST00000560913.1
ENST00000559314.1
ENST00000558058.1
|
PPP1R3E
|
protein phosphatase 1, regulatory subunit 3E
|
|
chr2_+_63816087
Show fit
|
5.51 |
ENST00000409908.1
ENST00000442225.1
ENST00000409476.1
ENST00000436321.1
|
MDH1
|
malate dehydrogenase 1, NAD (soluble)
|
|
chr3_-_52002403
Show fit
|
5.25 |
ENST00000490063.1
ENST00000468324.1
ENST00000497653.1
ENST00000484633.1
|
PCBP4
|
poly(rC) binding protein 4
|
|
chr14_+_24540731
Show fit
|
5.18 |
ENST00000558859.1
ENST00000559197.1
ENST00000560828.1
ENST00000216775.2
ENST00000560884.1
|
CPNE6
|
copine VI (neuronal)
|
|
chr19_+_22235310
Show fit
|
5.01 |
ENST00000600162.1
|
ZNF257
|
zinc finger protein 257
|
|
chr10_+_11047259
Show fit
|
4.95 |
ENST00000379261.4
ENST00000416382.2
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr2_+_63816295
Show fit
|
4.91 |
ENST00000539945.1
ENST00000544381.1
|
MDH1
|
malate dehydrogenase 1, NAD (soluble)
|
|
chr7_+_94139105
Show fit
|
4.64 |
ENST00000297273.4
|
CASD1
|
CAS1 domain containing 1
|
|
chr19_-_23433144
Show fit
|
4.62 |
ENST00000418100.1
ENST00000597537.1
ENST00000597037.1
|
ZNF724P
|
zinc finger protein 724, pseudogene
|
|
chr22_-_37880543
Show fit
|
4.53 |
ENST00000442496.1
|
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr1_-_26232522
Show fit
|
4.47 |
ENST00000399728.1
|
STMN1
|
stathmin 1
|
|
chr8_+_105235572
Show fit
|
4.44 |
ENST00000523362.1
|
RIMS2
|
regulating synaptic membrane exocytosis 2
|
|
chr3_+_58223228
Show fit
|
4.36 |
ENST00000478253.1
ENST00000295962.4
|
ABHD6
|
abhydrolase domain containing 6
|
|
chr7_+_139025875
Show fit
|
4.34 |
ENST00000297534.6
|
C7orf55
|
chromosome 7 open reading frame 55
|
|
chrX_+_54947229
Show fit
|
4.30 |
ENST00000442098.1
ENST00000430420.1
ENST00000453081.1
ENST00000173898.7
ENST00000319167.8
ENST00000375022.4
ENST00000399736.1
ENST00000440072.1
ENST00000420798.2
ENST00000431115.1
ENST00000440759.1
ENST00000375041.2
|
TRO
|
trophinin
|
|
chr19_+_21324827
Show fit
|
4.04 |
ENST00000600692.1
ENST00000599296.1
ENST00000594425.1
ENST00000311048.7
|
ZNF431
|
zinc finger protein 431
|
|
chr19_+_21264980
Show fit
|
4.03 |
ENST00000596053.1
ENST00000597086.1
ENST00000596143.1
ENST00000596367.1
ENST00000601416.1
|
ZNF714
|
zinc finger protein 714
|
|
chr2_+_113342163
Show fit
|
3.90 |
ENST00000409719.1
|
CHCHD5
|
coiled-coil-helix-coiled-coil-helix domain containing 5
|
|
chr9_+_103790991
Show fit
|
3.89 |
ENST00000374874.3
|
LPPR1
|
Lipid phosphate phosphatase-related protein type 1
|
|
chr19_-_38806560
Show fit
|
3.75 |
ENST00000591755.1
ENST00000337679.8
ENST00000339413.6
|
YIF1B
|
Yip1 interacting factor homolog B (S. cerevisiae)
|
|
chr16_+_71392616
Show fit
|
3.75 |
ENST00000349553.5
ENST00000302628.4
ENST00000562305.1
|
CALB2
|
calbindin 2
|
|
chr17_-_3599492
Show fit
|
3.73 |
ENST00000435558.1
ENST00000345901.3
|
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5
|
|
chr15_-_81282133
Show fit
|
3.73 |
ENST00000261758.4
|
MESDC2
|
mesoderm development candidate 2
|
|
chr11_+_46366918
Show fit
|
3.72 |
ENST00000528615.1
ENST00000395574.3
|
DGKZ
|
diacylglycerol kinase, zeta
|
|
chr11_+_125365110
Show fit
|
3.40 |
ENST00000527818.1
|
AP000708.1
|
AP000708.1
|
|
chr17_-_38074842
Show fit
|
3.38 |
ENST00000309481.7
|
GSDMB
|
gasdermin B
|
|
chr1_-_26232951
Show fit
|
3.34 |
ENST00000426559.2
ENST00000455785.2
|
STMN1
|
stathmin 1
|
|
chr17_-_38074859
Show fit
|
3.12 |
ENST00000520542.1
ENST00000418519.1
ENST00000394179.1
|
GSDMB
|
gasdermin B
|
|
chr16_-_18468926
Show fit
|
3.11 |
ENST00000545114.1
|
RP11-1212A22.4
|
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1
|
|
chr17_-_3599327
Show fit
|
3.00 |
ENST00000551178.1
ENST00000552276.1
ENST00000547178.1
|
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5
|
|
chr15_+_65903680
Show fit
|
3.00 |
ENST00000537259.1
|
SLC24A1
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 1
|
|
chr19_-_12833164
Show fit
|
2.93 |
ENST00000356861.5
|
TNPO2
|
transportin 2
|
|
chr14_-_50999190
Show fit
|
2.87 |
ENST00000557390.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5
|
|
chr2_-_73053126
Show fit
|
2.75 |
ENST00000272427.6
ENST00000410104.1
|
EXOC6B
|
exocyst complex component 6B
|
|
chr19_+_21203426
Show fit
|
2.66 |
ENST00000261560.5
ENST00000599548.1
ENST00000594110.1
|
ZNF430
|
zinc finger protein 430
|
|
chr1_-_11865351
Show fit
|
2.53 |
ENST00000413656.1
ENST00000376585.1
ENST00000423400.1
ENST00000431243.1
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H)
|
|
chr2_-_103353277
Show fit
|
2.45 |
ENST00000258436.5
|
MFSD9
|
major facilitator superfamily domain containing 9
|
|
chr9_-_35112376
Show fit
|
2.33 |
ENST00000488109.2
|
FAM214B
|
family with sequence similarity 214, member B
|
|
chr7_-_128694927
Show fit
|
2.33 |
ENST00000471166.1
ENST00000265388.5
|
TNPO3
|
transportin 3
|
|
chr19_-_19932501
Show fit
|
2.30 |
ENST00000540806.2
ENST00000590766.1
ENST00000587452.1
ENST00000545006.1
ENST00000590319.1
ENST00000587461.1
ENST00000450683.2
ENST00000443905.2
ENST00000590274.1
|
ZNF506
CTC-559E9.4
|
zinc finger protein 506
CTC-559E9.4
|
|
chrX_-_49089771
Show fit
|
2.16 |
ENST00000376251.1
ENST00000323022.5
ENST00000376265.2
|
CACNA1F
|
calcium channel, voltage-dependent, L type, alpha 1F subunit
|
|
chr19_+_21265028
Show fit
|
2.10 |
ENST00000291770.7
|
ZNF714
|
zinc finger protein 714
|
|
chr1_+_2398876
Show fit
|
1.96 |
ENST00000449969.1
|
PLCH2
|
phospholipase C, eta 2
|
|
chr16_-_70712229
Show fit
|
1.94 |
ENST00000562883.2
|
MTSS1L
|
metastasis suppressor 1-like
|
|
chr7_-_142630820
Show fit
|
1.94 |
ENST00000442623.1
ENST00000265310.1
|
TRPV5
|
transient receptor potential cation channel, subfamily V, member 5
|
|
chr5_-_43313574
Show fit
|
1.83 |
ENST00000325110.6
ENST00000433297.2
|
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble)
|
|
chr17_-_56358287
Show fit
|
1.78 |
ENST00000225275.3
ENST00000340482.3
|
MPO
|
myeloperoxidase
|
|
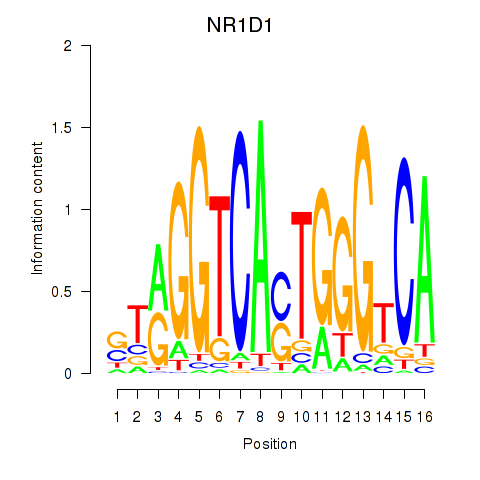
chr17_-_38256973
Show fit
|
1.76 |
ENST00000246672.3
|
NR1D1
|
nuclear receptor subfamily 1, group D, member 1
|
|
chrX_+_24167828
Show fit
|
1.59 |
ENST00000379188.3
ENST00000419690.1
ENST00000379177.1
ENST00000304543.5
|
ZFX
|
zinc finger protein, X-linked
|
|
chr19_+_22235279
Show fit
|
1.53 |
ENST00000594363.1
ENST00000597927.1
ENST00000594947.1
|
ZNF257
|
zinc finger protein 257
|
|
chr2_-_89442621
Show fit
|
1.49 |
ENST00000492167.1
|
IGKV3-20
|
immunoglobulin kappa variable 3-20
|
|
chr3_-_48723268
Show fit
|
1.46 |
ENST00000439518.1
ENST00000416649.2
ENST00000341520.4
ENST00000294129.2
|
NCKIPSD
|
NCK interacting protein with SH3 domain
|
|
chr19_-_22605136
Show fit
|
1.43 |
ENST00000357774.5
ENST00000601553.1
ENST00000593657.1
|
ZNF98
|
zinc finger protein 98
|
|
chr7_-_100493744
Show fit
|
1.39 |
ENST00000428317.1
ENST00000441605.1
|
ACHE
|
acetylcholinesterase (Yt blood group)
|
|
chr1_+_197382957
Show fit
|
1.36 |
ENST00000367397.1
|
CRB1
|
crumbs homolog 1 (Drosophila)
|
|
chr1_-_168513229
Show fit
|
1.32 |
ENST00000367819.2
|
XCL2
|
chemokine (C motif) ligand 2
|
|
chr3_-_133648656
Show fit
|
1.31 |
ENST00000408895.2
|
C3orf36
|
chromosome 3 open reading frame 36
|
|
chr14_+_74815116
Show fit
|
1.31 |
ENST00000256362.4
|
VRTN
|
vertebrae development associated
|
|
chr15_-_65903574
Show fit
|
1.31 |
ENST00000420799.2
ENST00000313182.2
ENST00000431261.2
ENST00000442903.3
|
VWA9
|
von Willebrand factor A domain containing 9
|
|
chr15_-_65903407
Show fit
|
1.23 |
ENST00000395644.4
ENST00000567744.1
ENST00000568573.1
ENST00000562830.1
ENST00000569491.1
ENST00000561769.1
|
VWA9
|
von Willebrand factor A domain containing 9
|
|
chr1_+_110210644
Show fit
|
1.22 |
ENST00000369831.2
ENST00000442650.1
ENST00000369827.3
ENST00000460717.3
ENST00000241337.4
ENST00000467579.3
ENST00000414179.2
ENST00000369829.2
|
GSTM2
|
glutathione S-transferase mu 2 (muscle)
|
|
chr19_-_58071201
Show fit
|
1.21 |
ENST00000325134.5
ENST00000457177.1
|
ZNF550
|
zinc finger protein 550
|
|
chr2_+_90153696
Show fit
|
1.17 |
ENST00000417279.2
|
IGKV3D-15
|
immunoglobulin kappa variable 3D-15 (gene/pseudogene)
|
|
chrY_+_2803322
Show fit
|
1.12 |
ENST00000383052.1
ENST00000155093.3
ENST00000449237.1
ENST00000443793.1
|
ZFY
|
zinc finger protein, Y-linked
|
|
chr17_+_6899366
Show fit
|
1.10 |
ENST00000251535.6
|
ALOX12
|
arachidonate 12-lipoxygenase
|
|
chr2_+_203776937
Show fit
|
1.03 |
ENST00000402905.3
ENST00000414490.1
ENST00000431787.1
ENST00000444724.1
ENST00000414857.1
ENST00000430899.1
ENST00000445120.1
ENST00000441569.1
ENST00000432024.1
ENST00000443740.1
ENST00000414439.1
ENST00000428585.1
ENST00000545253.1
ENST00000545262.1
ENST00000447539.1
ENST00000456821.2
ENST00000434998.1
ENST00000320443.8
|
CARF
|
calcium responsive transcription factor
|
|
chr6_+_167525277
Show fit
|
0.99 |
ENST00000400926.2
|
CCR6
|
chemokine (C-C motif) receptor 6
|
|
chr7_+_64126535
Show fit
|
0.99 |
ENST00000344930.3
|
ZNF107
|
zinc finger protein 107
|
|
chr7_-_100493482
Show fit
|
0.88 |
ENST00000411582.1
ENST00000419336.2
ENST00000241069.5
ENST00000302913.4
|
ACHE
|
acetylcholinesterase (Yt blood group)
|
|
chr7_-_128695147
Show fit
|
0.87 |
ENST00000482320.1
ENST00000393245.1
ENST00000471234.1
|
TNPO3
|
transportin 3
|
|
chr19_-_12833361
Show fit
|
0.86 |
ENST00000592287.1
|
TNPO2
|
transportin 2
|
|
chr22_+_27017921
Show fit
|
0.85 |
ENST00000354760.3
|
CRYBA4
|
crystallin, beta A4
|
|
chr6_-_27440837
Show fit
|
0.84 |
ENST00000211936.6
|
ZNF184
|
zinc finger protein 184
|
|
chr8_+_11561660
Show fit
|
0.79 |
ENST00000526716.1
ENST00000335135.4
ENST00000528027.1
|
GATA4
|
GATA binding protein 4
|
|
chr17_-_76128488
Show fit
|
0.76 |
ENST00000322914.3
|
TMC6
|
transmembrane channel-like 6
|
|
chr11_+_46740730
Show fit
|
0.73 |
ENST00000311907.5
ENST00000530231.1
ENST00000442468.1
|
F2
|
coagulation factor II (thrombin)
|
|
chr13_+_100741269
Show fit
|
0.65 |
ENST00000376286.4
ENST00000376279.3
ENST00000376285.1
|
PCCA
|
propionyl CoA carboxylase, alpha polypeptide
|
|
chr7_-_44229022
Show fit
|
0.62 |
ENST00000403799.3
|
GCK
|
glucokinase (hexokinase 4)
|
|
chr1_+_44398943
Show fit
|
0.61 |
ENST00000372359.5
ENST00000414809.3
|
ARTN
|
artemin
|
|
chr6_+_31543334
Show fit
|
0.59 |
ENST00000449264.2
|
TNF
|
tumor necrosis factor
|
|
chr19_-_42927251
Show fit
|
0.50 |
ENST00000597001.1
|
LIPE
|
lipase, hormone-sensitive
|
|
chr1_+_168545711
Show fit
|
0.44 |
ENST00000367818.3
|
XCL1
|
chemokine (C motif) ligand 1
|
|
chr19_+_21688366
Show fit
|
0.29 |
ENST00000358491.4
ENST00000597078.1
|
ZNF429
|
zinc finger protein 429
|
|
chrX_+_24167746
Show fit
|
0.28 |
ENST00000428571.1
ENST00000539115.1
|
ZFX
|
zinc finger protein, X-linked
|
|
chrX_+_16964985
Show fit
|
0.26 |
ENST00000303843.7
|
REPS2
|
RALBP1 associated Eps domain containing 2
|
|
chr11_+_7534999
Show fit
|
0.26 |
ENST00000528947.1
ENST00000299492.4
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2)
|
|
chr18_-_44336998
Show fit
|
0.24 |
ENST00000315087.7
|
ST8SIA5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5
|
|
chr14_-_50999307
Show fit
|
0.22 |
ENST00000013125.4
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5
|
|
chr2_+_220306745
Show fit
|
0.18 |
ENST00000431523.1
ENST00000396698.1
ENST00000396695.2
|
SPEG
|
SPEG complex locus
|
|
chr6_+_76458909
Show fit
|
0.13 |
ENST00000369981.3
ENST00000369985.4
|
MYO6
|
myosin VI
|
|
chr16_+_1832902
Show fit
|
0.05 |
ENST00000262302.9
ENST00000563136.1
ENST00000565987.1
ENST00000543305.1
ENST00000568287.1
ENST00000565134.1
|
NUBP2
|
nucleotide binding protein 2
|
|
chr17_-_36347030
Show fit
|
0.04 |
ENST00000518551.1
|
TBC1D3
|
TBC1 domain family, member 3
|