Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
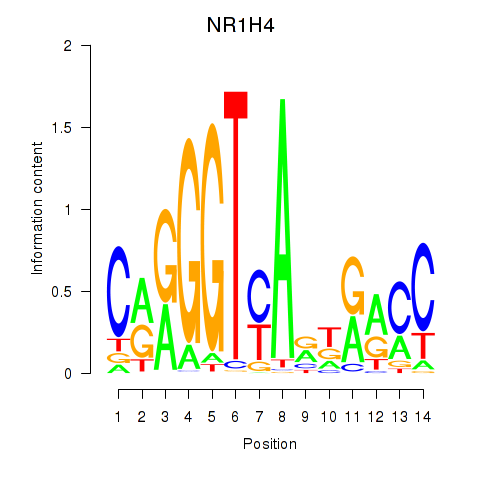
Results for NR1H4
Z-value: 1.11
Transcription factors associated with NR1H4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR1H4
|
ENSG00000012504.9 | nuclear receptor subfamily 1 group H member 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR1H4 | hg19_v2_chr12_+_100897130_100897231 | 0.43 | 5.4e-11 | Click! |
Activity profile of NR1H4 motif
Sorted Z-values of NR1H4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_21492113 | 33.33 |
ENST00000554094.1
|
NDRG2
|
NDRG family member 2 |
| chr14_-_21492251 | 32.32 |
ENST00000554398.1
|
NDRG2
|
NDRG family member 2 |
| chr9_+_74526384 | 29.48 |
ENST00000334731.2
ENST00000377031.3 |
C9orf85
|
chromosome 9 open reading frame 85 |
| chr5_-_139944196 | 22.58 |
ENST00000357560.4
|
APBB3
|
amyloid beta (A4) precursor protein-binding, family B, member 3 |
| chr5_-_139943830 | 21.97 |
ENST00000412920.3
ENST00000511201.2 ENST00000356738.2 ENST00000354402.5 ENST00000358580.5 ENST00000508496.2 |
APBB3
|
amyloid beta (A4) precursor protein-binding, family B, member 3 |
| chr14_-_21945057 | 20.46 |
ENST00000397762.1
|
RAB2B
|
RAB2B, member RAS oncogene family |
| chr22_-_20104700 | 18.15 |
ENST00000439169.2
ENST00000445045.1 ENST00000404751.3 ENST00000252136.7 ENST00000403707.3 |
TRMT2A
|
tRNA methyltransferase 2 homolog A (S. cerevisiae) |
| chr20_+_30640004 | 17.86 |
ENST00000520553.1
ENST00000518730.1 ENST00000375852.2 |
HCK
|
hemopoietic cell kinase |
| chr20_+_30639991 | 15.89 |
ENST00000534862.1
ENST00000538448.1 ENST00000375862.2 |
HCK
|
hemopoietic cell kinase |
| chr12_-_57914275 | 15.57 |
ENST00000547303.1
ENST00000552740.1 ENST00000547526.1 ENST00000551116.1 ENST00000346473.3 |
DDIT3
|
DNA-damage-inducible transcript 3 |
| chr12_+_57914742 | 15.51 |
ENST00000551351.1
|
MBD6
|
methyl-CpG binding domain protein 6 |
| chrX_+_103031421 | 15.27 |
ENST00000433491.1
ENST00000418604.1 ENST00000443502.1 |
PLP1
|
proteolipid protein 1 |
| chr8_+_9046503 | 14.79 |
ENST00000512942.2
|
RP11-10A14.5
|
RP11-10A14.5 |
| chr11_-_64511575 | 14.69 |
ENST00000431822.1
ENST00000377486.3 ENST00000394432.3 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr3_-_57113314 | 13.97 |
ENST00000338458.4
ENST00000468727.1 |
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3 |
| chrX_+_103031758 | 13.94 |
ENST00000303958.2
ENST00000361621.2 |
PLP1
|
proteolipid protein 1 |
| chr3_-_38071122 | 13.86 |
ENST00000334661.4
|
PLCD1
|
phospholipase C, delta 1 |
| chr16_+_23847339 | 13.72 |
ENST00000303531.7
|
PRKCB
|
protein kinase C, beta |
| chr3_+_14989186 | 13.72 |
ENST00000435454.1
ENST00000323373.6 |
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr9_-_137809718 | 13.72 |
ENST00000371806.3
|
FCN1
|
ficolin (collagen/fibrinogen domain containing) 1 |
| chr12_+_58013693 | 13.15 |
ENST00000320442.4
ENST00000379218.2 |
SLC26A10
|
solute carrier family 26, member 10 |
| chr19_+_1071203 | 12.88 |
ENST00000543365.1
|
HMHA1
|
histocompatibility (minor) HA-1 |
| chr22_+_20104947 | 12.55 |
ENST00000402752.1
|
RANBP1
|
RAN binding protein 1 |
| chr14_-_25103472 | 12.51 |
ENST00000216341.4
ENST00000382542.1 ENST00000382540.1 |
GZMB
|
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
| chr3_+_122296465 | 12.10 |
ENST00000483793.1
|
PARP15
|
poly (ADP-ribose) polymerase family, member 15 |
| chr16_+_23847267 | 11.99 |
ENST00000321728.7
|
PRKCB
|
protein kinase C, beta |
| chr19_+_50706866 | 11.96 |
ENST00000440075.2
ENST00000376970.2 ENST00000425460.1 ENST00000599920.1 ENST00000601313.1 |
MYH14
|
myosin, heavy chain 14, non-muscle |
| chr11_+_45868957 | 11.90 |
ENST00000443527.2
|
CRY2
|
cryptochrome 2 (photolyase-like) |
| chr19_+_35629702 | 11.73 |
ENST00000351325.4
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr14_-_25103388 | 11.49 |
ENST00000526004.1
ENST00000415355.3 |
GZMB
|
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
| chr12_+_109273806 | 10.91 |
ENST00000228476.3
ENST00000547768.1 |
DAO
|
D-amino-acid oxidase |
| chr10_-_75634260 | 10.23 |
ENST00000372765.1
ENST00000351293.3 |
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr3_-_49158218 | 9.82 |
ENST00000417901.1
ENST00000306026.5 ENST00000434032.2 |
USP19
|
ubiquitin specific peptidase 19 |
| chr11_-_71639670 | 9.73 |
ENST00000533047.1
ENST00000529844.1 |
RP11-849H4.2
|
Putative short transient receptor potential channel 2-like protein |
| chr15_+_83776137 | 9.73 |
ENST00000322019.9
|
TM6SF1
|
transmembrane 6 superfamily member 1 |
| chr5_+_140588269 | 9.31 |
ENST00000541609.1
ENST00000239450.2 |
PCDHB12
|
protocadherin beta 12 |
| chr10_-_75634326 | 8.97 |
ENST00000322635.3
ENST00000444854.2 ENST00000423381.1 ENST00000322680.3 ENST00000394762.2 |
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr3_-_49158312 | 8.67 |
ENST00000398892.3
ENST00000453664.1 ENST00000398888.2 |
USP19
|
ubiquitin specific peptidase 19 |
| chr12_+_100661156 | 8.59 |
ENST00000360820.2
|
SCYL2
|
SCY1-like 2 (S. cerevisiae) |
| chr1_+_28206150 | 8.47 |
ENST00000456990.1
|
THEMIS2
|
thymocyte selection associated family member 2 |
| chr17_+_43318434 | 8.31 |
ENST00000587489.1
|
FMNL1
|
formin-like 1 |
| chr17_+_73975292 | 8.29 |
ENST00000397640.1
ENST00000416485.1 ENST00000588202.1 ENST00000590676.1 ENST00000586891.1 |
TEN1
|
TEN1 CST complex subunit |
| chr1_+_14026722 | 8.28 |
ENST00000376048.5
|
PRDM2
|
PR domain containing 2, with ZNF domain |
| chr6_+_127587755 | 8.19 |
ENST00000368314.1
ENST00000476956.1 ENST00000609447.1 ENST00000356799.2 ENST00000477776.1 ENST00000609944.1 |
RNF146
|
ring finger protein 146 |
| chr1_+_36789335 | 8.18 |
ENST00000373137.2
|
RP11-268J15.5
|
RP11-268J15.5 |
| chr22_+_32149927 | 8.14 |
ENST00000437411.1
ENST00000535622.1 ENST00000536766.1 ENST00000400242.3 ENST00000266091.3 ENST00000400249.2 ENST00000400246.1 ENST00000382105.2 |
DEPDC5
|
DEP domain containing 5 |
| chr16_+_69166418 | 8.09 |
ENST00000314423.7
ENST00000562237.1 ENST00000567460.1 ENST00000566227.1 ENST00000352319.4 ENST00000563094.1 |
CIRH1A
|
cirrhosis, autosomal recessive 1A (cirhin) |
| chr3_+_52489503 | 8.03 |
ENST00000345716.4
|
NISCH
|
nischarin |
| chr3_-_50383096 | 7.93 |
ENST00000442887.1
ENST00000360165.3 |
ZMYND10
|
zinc finger, MYND-type containing 10 |
| chr6_+_30594619 | 7.87 |
ENST00000318999.7
ENST00000376485.4 ENST00000376478.2 ENST00000319027.5 ENST00000376483.4 ENST00000329992.8 ENST00000330083.5 |
ATAT1
|
alpha tubulin acetyltransferase 1 |
| chr8_-_101321584 | 7.80 |
ENST00000523167.1
|
RNF19A
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr1_+_25598989 | 7.76 |
ENST00000454452.2
|
RHD
|
Rh blood group, D antigen |
| chr12_-_8088871 | 7.65 |
ENST00000075120.7
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr1_+_36690011 | 7.57 |
ENST00000354618.5
ENST00000469141.2 ENST00000478853.1 |
THRAP3
|
thyroid hormone receptor associated protein 3 |
| chr22_+_22786288 | 7.56 |
ENST00000390301.2
|
IGLV1-36
|
immunoglobulin lambda variable 1-36 |
| chr1_-_109584716 | 7.35 |
ENST00000531337.1
ENST00000529074.1 ENST00000369965.4 |
WDR47
|
WD repeat domain 47 |
| chr19_+_10197463 | 7.30 |
ENST00000590378.1
ENST00000397881.3 |
C19orf66
|
chromosome 19 open reading frame 66 |
| chr1_-_109584768 | 7.26 |
ENST00000357672.3
|
WDR47
|
WD repeat domain 47 |
| chr5_+_149569520 | 7.26 |
ENST00000230671.2
ENST00000524041.1 |
SLC6A7
|
solute carrier family 6 (neurotransmitter transporter), member 7 |
| chr12_-_56727487 | 7.05 |
ENST00000548043.1
ENST00000425394.2 |
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr6_+_127588020 | 7.05 |
ENST00000309649.3
ENST00000610162.1 ENST00000610153.1 ENST00000608991.1 ENST00000480444.1 |
RNF146
|
ring finger protein 146 |
| chr3_-_120461378 | 7.04 |
ENST00000273375.3
|
RABL3
|
RAB, member of RAS oncogene family-like 3 |
| chr3_-_28390581 | 6.89 |
ENST00000479665.1
|
AZI2
|
5-azacytidine induced 2 |
| chr1_-_109584608 | 6.74 |
ENST00000400794.3
ENST00000528747.1 ENST00000369962.3 ENST00000361054.3 |
WDR47
|
WD repeat domain 47 |
| chr11_-_58345569 | 6.72 |
ENST00000528954.1
ENST00000528489.1 |
LPXN
|
leupaxin |
| chr19_-_50311896 | 6.51 |
ENST00000529634.2
|
FUZ
|
fuzzy planar cell polarity protein |
| chr20_+_62369623 | 6.42 |
ENST00000467211.1
|
RP4-583P15.14
|
RP4-583P15.14 |
| chr17_-_73975444 | 6.21 |
ENST00000293217.5
ENST00000537812.1 |
ACOX1
|
acyl-CoA oxidase 1, palmitoyl |
| chr12_-_56727676 | 6.13 |
ENST00000547572.1
ENST00000257931.5 ENST00000440411.3 |
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr19_-_633576 | 6.08 |
ENST00000588649.2
|
POLRMT
|
polymerase (RNA) mitochondrial (DNA directed) |
| chr6_-_49681235 | 5.82 |
ENST00000339139.4
|
CRISP2
|
cysteine-rich secretory protein 2 |
| chr3_+_111260954 | 5.79 |
ENST00000283285.5
|
CD96
|
CD96 molecule |
| chr14_+_101297740 | 5.75 |
ENST00000555928.1
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr17_-_73257667 | 5.75 |
ENST00000538886.1
ENST00000580799.1 ENST00000351904.7 ENST00000537686.1 |
GGA3
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 3 |
| chr11_-_7041466 | 5.68 |
ENST00000536068.1
ENST00000278314.4 |
ZNF214
|
zinc finger protein 214 |
| chr3_+_111260856 | 5.61 |
ENST00000352690.4
|
CD96
|
CD96 molecule |
| chr1_+_44412577 | 5.57 |
ENST00000372343.3
|
IPO13
|
importin 13 |
| chr10_-_95360983 | 5.31 |
ENST00000371464.3
|
RBP4
|
retinol binding protein 4, plasma |
| chr5_+_79331164 | 5.28 |
ENST00000350881.2
|
THBS4
|
thrombospondin 4 |
| chr11_-_71639480 | 5.27 |
ENST00000529513.1
|
RP11-849H4.2
|
Putative short transient receptor potential channel 2-like protein |
| chr17_-_1083078 | 5.05 |
ENST00000574266.1
ENST00000302538.5 |
ABR
|
active BCR-related |
| chr7_+_94139105 | 5.04 |
ENST00000297273.4
|
CASD1
|
CAS1 domain containing 1 |
| chr16_+_70258261 | 5.03 |
ENST00000594734.1
|
FKSG63
|
FKSG63 |
| chr12_+_6898638 | 4.97 |
ENST00000011653.4
|
CD4
|
CD4 molecule |
| chr1_-_25256368 | 4.94 |
ENST00000308873.6
|
RUNX3
|
runt-related transcription factor 3 |
| chr1_+_202317815 | 4.89 |
ENST00000608999.1
ENST00000336894.4 ENST00000480184.1 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr22_+_23247030 | 4.86 |
ENST00000390324.2
|
IGLJ3
|
immunoglobulin lambda joining 3 |
| chr7_+_49813255 | 4.80 |
ENST00000340652.4
|
VWC2
|
von Willebrand factor C domain containing 2 |
| chr4_+_123073464 | 4.79 |
ENST00000264501.4
|
KIAA1109
|
KIAA1109 |
| chr2_+_219433281 | 4.75 |
ENST00000273064.6
ENST00000509807.2 ENST00000542068.1 |
RQCD1
|
RCD1 required for cell differentiation1 homolog (S. pombe) |
| chr22_+_31090793 | 4.57 |
ENST00000332585.6
ENST00000382310.3 ENST00000446658.2 |
OSBP2
|
oxysterol binding protein 2 |
| chr5_+_133450365 | 4.52 |
ENST00000342854.5
ENST00000321603.6 ENST00000321584.4 ENST00000378564.1 ENST00000395029.1 |
TCF7
|
transcription factor 7 (T-cell specific, HMG-box) |
| chr17_-_43568062 | 4.43 |
ENST00000421073.2
ENST00000584420.1 ENST00000589780.1 ENST00000430334.3 |
PLEKHM1
|
pleckstrin homology domain containing, family M (with RUN domain) member 1 |
| chr19_+_10196981 | 4.38 |
ENST00000591813.1
|
C19orf66
|
chromosome 19 open reading frame 66 |
| chr11_+_7534999 | 4.29 |
ENST00000528947.1
ENST00000299492.4 |
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr17_-_26879567 | 4.08 |
ENST00000581945.1
ENST00000444148.1 ENST00000301032.4 ENST00000335765.4 |
UNC119
|
unc-119 homolog (C. elegans) |
| chrX_+_152224766 | 4.03 |
ENST00000370265.4
ENST00000447306.1 |
PNMA3
|
paraneoplastic Ma antigen 3 |
| chr17_-_74023291 | 4.03 |
ENST00000586740.1
|
EVPL
|
envoplakin |
| chr18_-_67624160 | 3.80 |
ENST00000581982.1
ENST00000280200.4 |
CD226
|
CD226 molecule |
| chr2_-_178937478 | 3.56 |
ENST00000286063.6
|
PDE11A
|
phosphodiesterase 11A |
| chrX_-_49965009 | 3.49 |
ENST00000437370.1
ENST00000376064.3 ENST00000448865.1 |
AKAP4
|
A kinase (PRKA) anchor protein 4 |
| chr19_-_36304201 | 3.46 |
ENST00000301175.3
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr3_+_121903181 | 3.46 |
ENST00000498619.1
|
CASR
|
calcium-sensing receptor |
| chr6_-_33771757 | 3.44 |
ENST00000507738.1
ENST00000266003.5 ENST00000430124.2 |
MLN
|
motilin |
| chr5_-_16509101 | 3.44 |
ENST00000399793.2
|
FAM134B
|
family with sequence similarity 134, member B |
| chr7_+_94536898 | 3.43 |
ENST00000433360.1
ENST00000340694.4 ENST00000424654.1 |
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A |
| chr17_-_2614927 | 3.40 |
ENST00000435359.1
|
CLUH
|
clustered mitochondria (cluA/CLU1) homolog |
| chr3_+_54157480 | 3.27 |
ENST00000490478.1
|
CACNA2D3
|
calcium channel, voltage-dependent, alpha 2/delta subunit 3 |
| chr1_-_1334685 | 3.26 |
ENST00000400809.3
ENST00000408918.4 |
CCNL2
|
cyclin L2 |
| chr11_-_118305921 | 3.26 |
ENST00000532619.1
|
RP11-770J1.4
|
RP11-770J1.4 |
| chr1_-_27240455 | 3.24 |
ENST00000254227.3
|
NR0B2
|
nuclear receptor subfamily 0, group B, member 2 |
| chr7_-_752074 | 3.13 |
ENST00000360274.4
|
PRKAR1B
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr3_+_186560476 | 3.04 |
ENST00000320741.2
ENST00000444204.2 |
ADIPOQ
|
adiponectin, C1Q and collagen domain containing |
| chr17_+_59529743 | 3.02 |
ENST00000589003.1
ENST00000393853.4 |
TBX4
|
T-box 4 |
| chr3_+_186560462 | 3.00 |
ENST00000412955.2
|
ADIPOQ
|
adiponectin, C1Q and collagen domain containing |
| chr17_+_7533439 | 2.99 |
ENST00000441599.2
ENST00000380450.4 ENST00000416273.3 ENST00000575903.1 ENST00000576830.1 ENST00000571153.1 ENST00000575618.1 ENST00000576152.1 |
SHBG
|
sex hormone-binding globulin |
| chr22_-_29457832 | 2.93 |
ENST00000216071.4
|
C22orf31
|
chromosome 22 open reading frame 31 |
| chr3_+_50606901 | 2.90 |
ENST00000455834.1
|
HEMK1
|
HemK methyltransferase family member 1 |
| chr17_+_64298944 | 2.79 |
ENST00000413366.3
|
PRKCA
|
protein kinase C, alpha |
| chr3_-_194119083 | 2.66 |
ENST00000401815.1
|
GP5
|
glycoprotein V (platelet) |
| chr2_-_182545603 | 2.65 |
ENST00000295108.3
|
NEUROD1
|
neuronal differentiation 1 |
| chrX_-_106449656 | 2.47 |
ENST00000372466.4
ENST00000421752.1 ENST00000372461.3 |
NUP62CL
|
nucleoporin 62kDa C-terminal like |
| chr19_-_44860820 | 2.43 |
ENST00000354340.4
ENST00000337401.4 ENST00000587909.1 |
ZNF112
|
zinc finger protein 112 |
| chr20_-_3644046 | 2.39 |
ENST00000290417.2
ENST00000319242.3 |
GFRA4
|
GDNF family receptor alpha 4 |
| chr12_-_58135903 | 2.37 |
ENST00000257897.3
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr11_-_62380199 | 2.29 |
ENST00000419857.1
ENST00000394773.2 |
EML3
|
echinoderm microtubule associated protein like 3 |
| chr14_+_24025194 | 2.21 |
ENST00000404535.3
ENST00000288014.6 |
THTPA
|
thiamine triphosphatase |
| chr14_+_24025462 | 2.20 |
ENST00000556015.1
ENST00000554970.1 ENST00000554789.1 |
THTPA
|
thiamine triphosphatase |
| chr2_+_73441350 | 2.18 |
ENST00000389501.4
|
SMYD5
|
SMYD family member 5 |
| chr2_+_192542850 | 2.13 |
ENST00000410026.2
|
NABP1
|
nucleic acid binding protein 1 |
| chr17_+_41177220 | 2.13 |
ENST00000587250.2
ENST00000544533.1 |
RND2
|
Rho family GTPase 2 |
| chr12_+_52306113 | 2.13 |
ENST00000547400.1
ENST00000550683.1 ENST00000419526.2 |
ACVRL1
|
activin A receptor type II-like 1 |
| chr11_+_67250490 | 2.01 |
ENST00000528641.2
ENST00000279146.3 |
AIP
|
aryl hydrocarbon receptor interacting protein |
| chr3_+_183873240 | 2.00 |
ENST00000431765.1
|
DVL3
|
dishevelled segment polarity protein 3 |
| chr10_-_119806085 | 1.99 |
ENST00000355624.3
|
RAB11FIP2
|
RAB11 family interacting protein 2 (class I) |
| chr16_+_66995121 | 1.99 |
ENST00000303334.4
|
CES3
|
carboxylesterase 3 |
| chr3_-_128294929 | 1.97 |
ENST00000356020.2
|
C3orf27
|
chromosome 3 open reading frame 27 |
| chr11_-_6341724 | 1.85 |
ENST00000530979.1
|
PRKCDBP
|
protein kinase C, delta binding protein |
| chr1_-_25747283 | 1.82 |
ENST00000346452.4
ENST00000340849.4 ENST00000349438.4 ENST00000294413.7 ENST00000413854.1 ENST00000455194.1 ENST00000243186.6 ENST00000425135.1 |
RHCE
|
Rh blood group, CcEe antigens |
| chr1_+_12123414 | 1.81 |
ENST00000263932.2
|
TNFRSF8
|
tumor necrosis factor receptor superfamily, member 8 |
| chr16_+_89238149 | 1.67 |
ENST00000289746.2
|
CDH15
|
cadherin 15, type 1, M-cadherin (myotubule) |
| chr19_+_10563567 | 1.57 |
ENST00000344979.3
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific |
| chr19_-_11689752 | 1.55 |
ENST00000592659.1
ENST00000592828.1 ENST00000218758.5 ENST00000412435.2 |
ACP5
|
acid phosphatase 5, tartrate resistant |
| chr5_+_32710736 | 1.51 |
ENST00000415685.2
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr5_-_33892204 | 1.45 |
ENST00000504830.1
|
ADAMTS12
|
ADAM metallopeptidase with thrombospondin type 1 motif, 12 |
| chr5_-_33892046 | 1.45 |
ENST00000352040.3
|
ADAMTS12
|
ADAM metallopeptidase with thrombospondin type 1 motif, 12 |
| chr5_-_177423243 | 1.41 |
ENST00000308304.2
|
PROP1
|
PROP paired-like homeobox 1 |
| chr1_+_25598872 | 1.28 |
ENST00000328664.4
|
RHD
|
Rh blood group, D antigen |
| chr16_+_66995144 | 1.28 |
ENST00000394037.1
|
CES3
|
carboxylesterase 3 |
| chr16_-_1429627 | 1.25 |
ENST00000248104.7
|
UNKL
|
unkempt family zinc finger-like |
| chr9_-_138391692 | 1.24 |
ENST00000429260.2
|
C9orf116
|
chromosome 9 open reading frame 116 |
| chr1_-_160040038 | 1.14 |
ENST00000368089.3
|
KCNJ10
|
potassium inwardly-rectifying channel, subfamily J, member 10 |
| chr11_-_65624415 | 1.13 |
ENST00000524553.1
ENST00000527344.1 |
CFL1
|
cofilin 1 (non-muscle) |
| chr19_-_42636617 | 1.12 |
ENST00000529067.1
ENST00000529952.1 ENST00000533720.1 ENST00000389341.5 ENST00000342301.4 |
POU2F2
|
POU class 2 homeobox 2 |
| chr1_-_149908217 | 1.10 |
ENST00000369140.3
|
MTMR11
|
myotubularin related protein 11 |
| chr2_+_207024306 | 1.08 |
ENST00000236957.5
ENST00000392221.1 ENST00000392222.2 ENST00000445505.1 |
EEF1B2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr2_+_33172012 | 1.05 |
ENST00000404816.2
|
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr17_-_9940058 | 1.05 |
ENST00000585266.1
|
GAS7
|
growth arrest-specific 7 |
| chr1_+_25599018 | 1.04 |
ENST00000417538.2
ENST00000357542.4 ENST00000568195.1 ENST00000342055.5 ENST00000423810.2 |
RHD
|
Rh blood group, D antigen |
| chr5_-_83680603 | 1.03 |
ENST00000296591.5
|
EDIL3
|
EGF-like repeats and discoidin I-like domains 3 |
| chr11_-_4414880 | 1.02 |
ENST00000254436.7
ENST00000543625.1 |
TRIM21
|
tripartite motif containing 21 |
| chr22_-_50700140 | 1.00 |
ENST00000215659.8
|
MAPK12
|
mitogen-activated protein kinase 12 |
| chr18_-_3880051 | 0.98 |
ENST00000584874.1
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr2_-_64371546 | 0.77 |
ENST00000358912.4
|
PELI1
|
pellino E3 ubiquitin protein ligase 1 |
| chrX_+_47420516 | 0.73 |
ENST00000377045.4
ENST00000290277.6 ENST00000377039.2 |
ARAF
|
v-raf murine sarcoma 3611 viral oncogene homolog |
| chr17_+_34136459 | 0.68 |
ENST00000588240.1
ENST00000590273.1 ENST00000588441.1 ENST00000587272.1 ENST00000592237.1 ENST00000311979.3 |
TAF15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa |
| chr1_+_26560676 | 0.65 |
ENST00000451429.2
ENST00000252992.4 |
CEP85
|
centrosomal protein 85kDa |
| chr11_-_72070206 | 0.59 |
ENST00000544382.1
|
CLPB
|
ClpB caseinolytic peptidase B homolog (E. coli) |
| chr12_-_71003568 | 0.58 |
ENST00000547715.1
ENST00000451516.2 ENST00000538708.1 ENST00000550857.1 ENST00000261266.5 |
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr19_+_1026566 | 0.57 |
ENST00000348419.3
ENST00000565096.2 ENST00000562958.2 ENST00000562075.2 ENST00000607102.1 |
CNN2
|
calponin 2 |
| chr10_-_49701686 | 0.53 |
ENST00000417247.2
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr10_+_47083454 | 0.52 |
ENST00000374312.1
|
NPY4R
|
neuropeptide Y receptor Y4 |
| chrX_+_148793714 | 0.45 |
ENST00000355220.5
|
MAGEA11
|
melanoma antigen family A, 11 |
| chrX_+_7137475 | 0.33 |
ENST00000217961.4
|
STS
|
steroid sulfatase (microsomal), isozyme S |
| chr3_-_126327398 | 0.31 |
ENST00000383572.2
|
TXNRD3NB
|
thioredoxin reductase 3 neighbor |
| chr10_+_64564469 | 0.24 |
ENST00000373783.1
|
ADO
|
2-aminoethanethiol (cysteamine) dioxygenase |
| chr5_+_139927213 | 0.23 |
ENST00000310331.2
|
EIF4EBP3
|
eukaryotic translation initiation factor 4E binding protein 3 |
| chr7_-_22233442 | 0.21 |
ENST00000401957.2
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chrX_+_66764375 | 0.17 |
ENST00000374690.3
|
AR
|
androgen receptor |
| chr3_+_190105909 | 0.13 |
ENST00000456423.1
|
CLDN16
|
claudin 16 |
| chr2_+_108905095 | 0.11 |
ENST00000251481.6
ENST00000326853.5 |
SULT1C2
|
sulfotransferase family, cytosolic, 1C, member 2 |
| chr19_+_11546153 | 0.09 |
ENST00000591946.1
ENST00000252455.2 ENST00000412601.1 |
PRKCSH
|
protein kinase C substrate 80K-H |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR1H4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.2 | 33.7 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 9.5 | 28.5 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 5.1 | 15.2 | GO:0002727 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) |
| 4.6 | 13.7 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 4.1 | 65.7 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 4.0 | 11.9 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 3.9 | 15.6 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 3.6 | 10.9 | GO:0006524 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) |
| 3.4 | 24.0 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 2.5 | 7.6 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 2.2 | 13.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 2.0 | 12.1 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 2.0 | 6.0 | GO:2000532 | renal albumin absorption(GO:0097018) regulation of renal albumin absorption(GO:2000532) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 2.0 | 11.7 | GO:0010734 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 1.7 | 29.2 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 1.7 | 5.1 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 1.6 | 6.5 | GO:0060825 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 1.5 | 6.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 1.5 | 7.3 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 1.2 | 8.6 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 1.1 | 4.4 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 1.1 | 5.3 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 1.1 | 5.3 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 1.0 | 4.1 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 1.0 | 8.1 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 1.0 | 19.2 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 1.0 | 5.0 | GO:0071350 | interleukin-15-mediated signaling pathway(GO:0035723) cellular response to interleukin-15(GO:0071350) |
| 1.0 | 2.9 | GO:0044728 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) DNA methylation or demethylation(GO:0044728) |
| 0.9 | 18.5 | GO:1900037 | negative regulation of skeletal muscle tissue development(GO:0048642) regulation of cellular response to hypoxia(GO:1900037) |
| 0.9 | 4.5 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.9 | 2.6 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.9 | 3.4 | GO:1904048 | regulation of spontaneous neurotransmitter secretion(GO:1904048) |
| 0.8 | 6.7 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.8 | 7.9 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.7 | 7.9 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.6 | 1.8 | GO:0032639 | TRAIL production(GO:0032639) regulation of TRAIL production(GO:0032679) positive regulation of TRAIL production(GO:0032759) |
| 0.6 | 4.8 | GO:0030856 | regulation of epithelial cell differentiation(GO:0030856) |
| 0.6 | 2.9 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.6 | 3.5 | GO:0010133 | proline catabolic process(GO:0006562) proline catabolic process to glutamate(GO:0010133) |
| 0.4 | 8.0 | GO:0048243 | norepinephrine secretion(GO:0048243) |
| 0.4 | 8.3 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.4 | 8.3 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.4 | 6.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.4 | 12.0 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.4 | 11.9 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.4 | 3.4 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.4 | 18.1 | GO:0001510 | RNA methylation(GO:0001510) |
| 0.3 | 7.6 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.3 | 13.2 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.3 | 2.0 | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.3 | 8.3 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.3 | 3.3 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.3 | 1.6 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.3 | 1.4 | GO:0060126 | hypophysis morphogenesis(GO:0048850) diencephalon morphogenesis(GO:0048852) somatotropin secreting cell differentiation(GO:0060126) |
| 0.3 | 1.0 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.3 | 6.9 | GO:0032607 | interferon-alpha production(GO:0032607) |
| 0.3 | 5.8 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.2 | 2.1 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.2 | 15.2 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.2 | 1.1 | GO:2000769 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) regulation of barbed-end actin filament capping(GO:2000812) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.2 | 2.4 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.2 | 2.1 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.2 | 3.6 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.2 | 3.5 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.2 | 3.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.2 | 0.8 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.2 | 4.9 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.2 | 1.1 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.2 | 20.5 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.2 | 3.5 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.2 | 14.7 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.2 | 0.2 | GO:0060599 | lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) |
| 0.2 | 13.9 | GO:0043647 | inositol phosphate metabolic process(GO:0043647) |
| 0.1 | 2.7 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.1 | 4.8 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.1 | 1.5 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.1 | 1.1 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.1 | 1.6 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 1.0 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 14.0 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.1 | 1.0 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.1 | 3.3 | GO:0034383 | low-density lipoprotein particle clearance(GO:0034383) |
| 0.1 | 8.7 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 2.1 | GO:0009301 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.1 | 2.0 | GO:0003091 | renal water homeostasis(GO:0003091) |
| 0.0 | 3.2 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 9.0 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 3.7 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 7.8 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 3.3 | GO:0061337 | cardiac conduction(GO:0061337) |
| 0.0 | 0.2 | GO:0071692 | sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) |
| 0.0 | 7.0 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 4.9 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.0 | 3.6 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.2 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.0 | 3.0 | GO:0035113 | embryonic limb morphogenesis(GO:0030326) embryonic appendage morphogenesis(GO:0035113) |
| 0.0 | 1.1 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 2.4 | GO:0042177 | negative regulation of protein catabolic process(GO:0042177) |
| 0.0 | 7.8 | GO:0007264 | small GTPase mediated signal transduction(GO:0007264) |
| 0.0 | 0.3 | GO:0006706 | steroid catabolic process(GO:0006706) |
| 0.0 | 0.2 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 15.6 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 4.4 | 13.2 | GO:0031251 | PAN complex(GO:0031251) |
| 2.4 | 12.0 | GO:0097513 | myosin II filament(GO:0097513) |
| 2.3 | 13.7 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 1.4 | 8.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.8 | 7.9 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.7 | 15.5 | GO:0010369 | chromocenter(GO:0010369) |
| 0.7 | 3.4 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.6 | 11.7 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.6 | 8.3 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.5 | 2.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.5 | 2.0 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.5 | 8.3 | GO:0032059 | bleb(GO:0032059) |
| 0.4 | 6.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.4 | 24.0 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.4 | 19.2 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.3 | 35.4 | GO:0005901 | caveola(GO:0005901) |
| 0.3 | 7.9 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.3 | 17.1 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.3 | 59.0 | GO:0030426 | growth cone(GO:0030426) |
| 0.3 | 7.6 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.3 | 8.6 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.2 | 4.6 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.2 | 5.0 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.2 | 4.1 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.2 | 4.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.2 | 1.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.2 | 5.3 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 6.7 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 2.5 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 16.3 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 5.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 14.2 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.1 | 4.5 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 5.8 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 6.1 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 3.6 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 2.1 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 7.6 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.1 | 2.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 18.0 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 10.0 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 0.6 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 22.8 | GO:0005874 | microtubule(GO:0005874) |
| 0.1 | 3.3 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 5.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 3.8 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 15.6 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 6.4 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 92.5 | GO:0005886 | plasma membrane(GO:0005886) |
| 0.0 | 2.6 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.5 | 28.5 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 2.5 | 7.6 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 2.4 | 29.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 2.2 | 13.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 1.8 | 18.5 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 1.6 | 7.9 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 1.5 | 7.3 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 1.4 | 19.8 | GO:0033691 | sialic acid binding(GO:0033691) |
| 1.4 | 19.2 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 1.2 | 32.5 | GO:0071949 | FAD binding(GO:0071949) |
| 1.1 | 22.2 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 1.0 | 15.6 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 1.0 | 5.0 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.8 | 14.7 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.7 | 13.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.6 | 3.6 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.5 | 33.7 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.5 | 2.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.5 | 44.6 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.5 | 12.1 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.5 | 7.6 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.5 | 5.0 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.4 | 5.8 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.4 | 3.2 | GO:0005497 | androgen binding(GO:0005497) |
| 0.4 | 18.1 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.4 | 8.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.4 | 11.9 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.4 | 5.3 | GO:0016918 | retinal binding(GO:0016918) |
| 0.3 | 2.4 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.3 | 1.5 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.2 | 3.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.2 | 7.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.2 | 11.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.2 | 19.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.2 | 1.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.2 | 1.8 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.2 | 0.5 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.2 | 8.3 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.2 | 8.3 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.2 | 3.1 | GO:0008603 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.2 | 2.0 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 5.6 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 2.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 2.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 1.4 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 3.5 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 3.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 1.6 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.1 | 6.1 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.1 | 4.6 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 9.8 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 3.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 3.5 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.1 | 16.0 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 2.9 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.1 | 1.1 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 25.0 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 18.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 1.0 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 1.6 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 2.6 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 4.0 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 0.2 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 2.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 2.2 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 5.3 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 3.4 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.6 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.7 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 3.3 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 1.9 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 2.0 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 2.1 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 4.4 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 8.4 | GO:0008047 | enzyme activator activity(GO:0008047) |
| 0.0 | 0.3 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 1.3 | GO:0005525 | GTP binding(GO:0005525) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 62.3 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.8 | 24.0 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.6 | 81.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.5 | 19.2 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.5 | 11.9 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.4 | 10.7 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.3 | 14.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.3 | 9.7 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.2 | 19.0 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.2 | 1.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 3.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 3.5 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 3.2 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 1.8 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 2.1 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 1.0 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 3.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 8.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 2.4 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 4.5 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 2.0 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.7 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 1.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 4.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 15.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 1.0 | 28.5 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 1.0 | 5.0 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.9 | 33.7 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.4 | 7.6 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.3 | 14.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.3 | 19.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.3 | 7.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.3 | 6.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.3 | 12.0 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.3 | 14.3 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.2 | 11.9 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.2 | 2.7 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.2 | 3.5 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.2 | 15.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 4.7 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 13.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 3.1 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.1 | 2.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 3.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 6.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 5.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 0.8 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 1.1 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 1.0 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 1.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 2.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.1 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 2.0 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 5.3 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 6.1 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.0 | 0.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 3.4 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 5.0 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 2.2 | REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | Genes involved in Transmission across Chemical Synapses |