Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
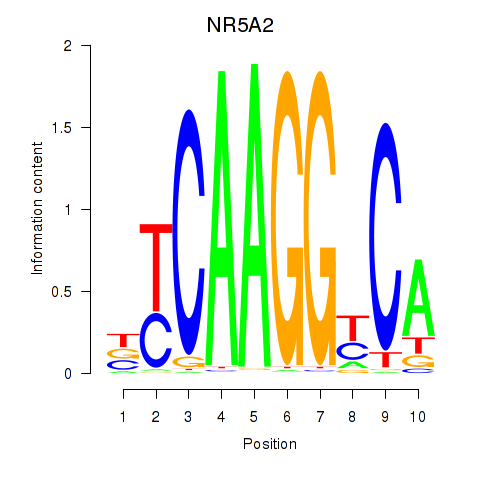
Results for NR5A2
Z-value: 1.53
Transcription factors associated with NR5A2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR5A2
|
ENSG00000116833.9 | nuclear receptor subfamily 5 group A member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR5A2 | hg19_v2_chr1_+_199996702_199996732 | 0.32 | 2.0e-06 | Click! |
Activity profile of NR5A2 motif
Sorted Z-values of NR5A2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_21491477 | 65.98 |
ENST00000298684.5
ENST00000557169.1 ENST00000553563.1 |
NDRG2
|
NDRG family member 2 |
| chr14_-_21492251 | 58.22 |
ENST00000554398.1
|
NDRG2
|
NDRG family member 2 |
| chr14_-_21492113 | 57.00 |
ENST00000554094.1
|
NDRG2
|
NDRG family member 2 |
| chr16_+_56623433 | 36.02 |
ENST00000570176.1
|
MT3
|
metallothionein 3 |
| chrX_+_103031421 | 35.60 |
ENST00000433491.1
ENST00000418604.1 ENST00000443502.1 |
PLP1
|
proteolipid protein 1 |
| chr8_-_110704014 | 35.09 |
ENST00000529190.1
ENST00000422135.1 ENST00000419099.1 |
SYBU
|
syntabulin (syntaxin-interacting) |
| chrX_+_103031758 | 34.81 |
ENST00000303958.2
ENST00000361621.2 |
PLP1
|
proteolipid protein 1 |
| chr2_+_220491973 | 28.49 |
ENST00000358055.3
|
SLC4A3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr16_+_226658 | 28.47 |
ENST00000320868.5
ENST00000397797.1 |
HBA1
|
hemoglobin, alpha 1 |
| chr14_-_21490958 | 26.64 |
ENST00000554104.1
|
NDRG2
|
NDRG family member 2 |
| chr8_+_21911054 | 25.20 |
ENST00000519850.1
ENST00000381470.3 |
DMTN
|
dematin actin binding protein |
| chr5_-_176056974 | 24.53 |
ENST00000510387.1
ENST00000506696.1 |
SNCB
|
synuclein, beta |
| chr11_-_790060 | 23.46 |
ENST00000330106.4
|
CEND1
|
cell cycle exit and neuronal differentiation 1 |
| chr2_+_220492116 | 21.74 |
ENST00000373760.2
|
SLC4A3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr3_+_133465228 | 21.06 |
ENST00000482271.1
ENST00000264998.3 |
TF
|
transferrin |
| chrX_-_49056635 | 20.96 |
ENST00000472598.1
ENST00000538567.1 ENST00000479808.1 ENST00000263233.4 |
SYP
|
synaptophysin |
| chr14_-_21491305 | 20.82 |
ENST00000554531.1
|
NDRG2
|
NDRG family member 2 |
| chr19_-_36643329 | 19.62 |
ENST00000589154.1
|
COX7A1
|
cytochrome c oxidase subunit VIIa polypeptide 1 (muscle) |
| chr3_-_58613323 | 19.31 |
ENST00000474531.1
ENST00000465970.1 |
FAM107A
|
family with sequence similarity 107, member A |
| chr14_-_60337684 | 18.26 |
ENST00000267484.5
|
RTN1
|
reticulon 1 |
| chr17_-_8066250 | 17.15 |
ENST00000488857.1
ENST00000481878.1 ENST00000316509.6 ENST00000498285.1 |
VAMP2
RP11-599B13.6
|
vesicle-associated membrane protein 2 (synaptobrevin 2) Uncharacterized protein |
| chr11_-_111781454 | 16.86 |
ENST00000533280.1
|
CRYAB
|
crystallin, alpha B |
| chr22_-_39239987 | 16.86 |
ENST00000333039.2
|
NPTXR
|
neuronal pentraxin receptor |
| chr6_-_52860171 | 15.57 |
ENST00000370963.4
|
GSTA4
|
glutathione S-transferase alpha 4 |
| chr1_-_149889382 | 15.41 |
ENST00000369145.1
ENST00000369146.3 |
SV2A
|
synaptic vesicle glycoprotein 2A |
| chr7_+_29519486 | 14.94 |
ENST00000409041.4
|
CHN2
|
chimerin 2 |
| chr8_-_110703819 | 14.79 |
ENST00000532779.1
ENST00000534578.1 |
SYBU
|
syntabulin (syntaxin-interacting) |
| chr11_-_111781554 | 14.75 |
ENST00000526167.1
ENST00000528961.1 |
CRYAB
|
crystallin, alpha B |
| chr22_-_37213045 | 14.54 |
ENST00000406910.2
ENST00000417718.2 |
PVALB
|
parvalbumin |
| chr20_-_62130474 | 14.53 |
ENST00000217182.3
|
EEF1A2
|
eukaryotic translation elongation factor 1 alpha 2 |
| chr7_+_29519662 | 14.35 |
ENST00000424025.2
ENST00000439711.2 ENST00000421775.2 |
CHN2
|
chimerin 2 |
| chr2_+_220492373 | 14.26 |
ENST00000317151.3
|
SLC4A3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr22_+_19710468 | 14.22 |
ENST00000366425.3
|
GP1BB
|
glycoprotein Ib (platelet), beta polypeptide |
| chr19_+_19322758 | 14.19 |
ENST00000252575.6
|
NCAN
|
neurocan |
| chr15_+_43985084 | 13.99 |
ENST00000434505.1
ENST00000411750.1 |
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr17_-_56606705 | 13.98 |
ENST00000317268.3
|
SEPT4
|
septin 4 |
| chr5_+_71403061 | 13.97 |
ENST00000512974.1
ENST00000296755.7 |
MAP1B
|
microtubule-associated protein 1B |
| chr17_-_56606664 | 13.88 |
ENST00000580844.1
|
SEPT4
|
septin 4 |
| chr22_-_29711704 | 13.73 |
ENST00000216101.6
|
RASL10A
|
RAS-like, family 10, member A |
| chr15_+_43886057 | 13.69 |
ENST00000441322.1
ENST00000413657.2 ENST00000453733.1 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr17_-_56606639 | 13.69 |
ENST00000579371.1
|
SEPT4
|
septin 4 |
| chr12_+_130554803 | 13.68 |
ENST00000535487.1
|
RP11-474D1.2
|
RP11-474D1.2 |
| chr15_+_43985725 | 13.68 |
ENST00000413453.2
|
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr10_-_75634219 | 13.56 |
ENST00000305762.7
|
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr6_-_34360413 | 13.51 |
ENST00000607016.1
|
NUDT3
|
nudix (nucleoside diphosphate linked moiety X)-type motif 3 |
| chr6_+_111580508 | 13.11 |
ENST00000368847.4
|
KIAA1919
|
KIAA1919 |
| chr22_-_29711645 | 13.10 |
ENST00000401450.3
|
RASL10A
|
RAS-like, family 10, member A |
| chr12_-_50290839 | 12.90 |
ENST00000552863.1
|
FAIM2
|
Fas apoptotic inhibitory molecule 2 |
| chr20_+_37434329 | 12.88 |
ENST00000299824.1
ENST00000373331.2 |
PPP1R16B
|
protein phosphatase 1, regulatory subunit 16B |
| chr15_+_43885252 | 12.60 |
ENST00000453782.1
ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr7_-_105029812 | 12.60 |
ENST00000482897.1
|
SRPK2
|
SRSF protein kinase 2 |
| chr2_+_220492287 | 12.51 |
ENST00000273063.6
ENST00000373762.3 |
SLC4A3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr3_-_10547333 | 12.46 |
ENST00000383800.4
|
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2 |
| chr12_-_56882136 | 12.45 |
ENST00000311966.4
|
GLS2
|
glutaminase 2 (liver, mitochondrial) |
| chr4_-_681114 | 12.37 |
ENST00000503156.1
|
MFSD7
|
major facilitator superfamily domain containing 7 |
| chr1_-_2323140 | 12.26 |
ENST00000378531.3
ENST00000378529.3 |
MORN1
|
MORN repeat containing 1 |
| chr19_+_5623186 | 12.15 |
ENST00000538656.1
|
SAFB
|
scaffold attachment factor B |
| chr10_-_75634260 | 12.07 |
ENST00000372765.1
ENST00000351293.3 |
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr20_-_48532019 | 11.95 |
ENST00000289431.5
|
SPATA2
|
spermatogenesis associated 2 |
| chr22_-_37213554 | 11.85 |
ENST00000443735.1
|
PVALB
|
parvalbumin |
| chr22_+_32149927 | 11.79 |
ENST00000437411.1
ENST00000535622.1 ENST00000536766.1 ENST00000400242.3 ENST00000266091.3 ENST00000400249.2 ENST00000400246.1 ENST00000382105.2 |
DEPDC5
|
DEP domain containing 5 |
| chr7_-_87104963 | 11.76 |
ENST00000359206.3
ENST00000358400.3 ENST00000265723.4 |
ABCB4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr1_+_169077172 | 11.50 |
ENST00000499679.3
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr12_+_110718921 | 11.40 |
ENST00000308664.6
|
ATP2A2
|
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 |
| chr3_-_10547192 | 11.22 |
ENST00000360273.2
ENST00000343816.4 |
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2 |
| chr10_-_75634326 | 11.21 |
ENST00000322635.3
ENST00000444854.2 ENST00000423381.1 ENST00000322680.3 ENST00000394762.2 |
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr7_+_45613958 | 11.21 |
ENST00000297323.7
|
ADCY1
|
adenylate cyclase 1 (brain) |
| chr8_+_136469684 | 11.12 |
ENST00000355849.5
|
KHDRBS3
|
KH domain containing, RNA binding, signal transduction associated 3 |
| chr1_+_202995611 | 11.11 |
ENST00000367240.2
|
PPFIA4
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 4 |
| chr19_+_41284121 | 11.10 |
ENST00000594800.1
ENST00000357052.2 ENST00000602173.1 |
RAB4B
|
RAB4B, member RAS oncogene family |
| chr17_-_27503770 | 11.10 |
ENST00000533112.1
|
MYO18A
|
myosin XVIIIA |
| chr8_+_86376081 | 10.99 |
ENST00000285379.5
|
CA2
|
carbonic anhydrase II |
| chr1_+_84609944 | 10.97 |
ENST00000370685.3
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr3_+_10068095 | 10.87 |
ENST00000287647.3
ENST00000383807.1 ENST00000383806.1 ENST00000419585.1 |
FANCD2
|
Fanconi anemia, complementation group D2 |
| chr17_-_8066843 | 10.78 |
ENST00000404970.3
|
VAMP2
|
vesicle-associated membrane protein 2 (synaptobrevin 2) |
| chr9_+_131873227 | 10.73 |
ENST00000358994.4
ENST00000455292.1 |
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr22_+_40390930 | 10.37 |
ENST00000333407.6
|
FAM83F
|
family with sequence similarity 83, member F |
| chr13_+_113622757 | 10.36 |
ENST00000375604.2
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like |
| chr7_+_99156314 | 10.23 |
ENST00000425063.1
ENST00000493277.1 |
ZNF655
|
zinc finger protein 655 |
| chr8_-_67341208 | 10.13 |
ENST00000499642.1
|
RP11-346I3.4
|
RP11-346I3.4 |
| chr10_-_50970322 | 10.01 |
ENST00000374103.4
|
OGDHL
|
oxoglutarate dehydrogenase-like |
| chr15_+_43809797 | 9.92 |
ENST00000399453.1
ENST00000300231.5 |
MAP1A
|
microtubule-associated protein 1A |
| chr1_-_241520525 | 9.81 |
ENST00000366565.1
|
RGS7
|
regulator of G-protein signaling 7 |
| chr17_-_73844722 | 9.60 |
ENST00000586257.1
|
WBP2
|
WW domain binding protein 2 |
| chr1_+_19923454 | 9.54 |
ENST00000602662.1
ENST00000602293.1 ENST00000322753.6 |
MINOS1-NBL1
MINOS1
|
MINOS1-NBL1 readthrough mitochondrial inner membrane organizing system 1 |
| chr12_-_49393092 | 9.46 |
ENST00000421952.2
|
DDN
|
dendrin |
| chr19_+_589893 | 9.40 |
ENST00000251287.2
|
HCN2
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 2 |
| chr2_-_38604398 | 9.31 |
ENST00000443098.1
ENST00000449130.1 ENST00000378954.4 ENST00000539122.1 ENST00000419554.2 ENST00000451483.1 ENST00000406122.1 |
ATL2
|
atlastin GTPase 2 |
| chr17_-_76778339 | 9.29 |
ENST00000591455.1
ENST00000446868.3 ENST00000361101.4 ENST00000589296.1 |
CYTH1
|
cytohesin 1 |
| chr11_-_64512273 | 9.22 |
ENST00000377497.3
ENST00000377487.1 ENST00000430645.1 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr5_+_141303373 | 9.13 |
ENST00000432126.2
ENST00000194118.4 |
KIAA0141
|
KIAA0141 |
| chr1_+_1260147 | 9.05 |
ENST00000343938.4
|
GLTPD1
|
glycolipid transfer protein domain containing 1 |
| chr7_+_99156434 | 9.02 |
ENST00000424881.1
ENST00000440391.1 ENST00000394163.2 |
ZNF655
|
zinc finger protein 655 |
| chr13_+_113622810 | 8.93 |
ENST00000397030.1
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like |
| chr4_-_5894777 | 8.89 |
ENST00000324989.7
|
CRMP1
|
collapsin response mediator protein 1 |
| chr11_-_9025541 | 8.84 |
ENST00000525100.1
ENST00000309166.3 ENST00000531090.1 |
NRIP3
|
nuclear receptor interacting protein 3 |
| chr1_-_200992827 | 8.75 |
ENST00000332129.2
ENST00000422435.2 |
KIF21B
|
kinesin family member 21B |
| chr10_-_50970382 | 8.73 |
ENST00000419399.1
ENST00000432695.1 |
OGDHL
|
oxoglutarate dehydrogenase-like |
| chr3_-_10749696 | 8.72 |
ENST00000397077.1
|
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2 |
| chr7_+_156742399 | 8.64 |
ENST00000275820.3
|
NOM1
|
nucleolar protein with MIF4G domain 1 |
| chr3_-_50605077 | 8.55 |
ENST00000426034.1
ENST00000441239.1 |
C3orf18
|
chromosome 3 open reading frame 18 |
| chr22_-_44708731 | 8.36 |
ENST00000381176.4
|
KIAA1644
|
KIAA1644 |
| chr4_+_7045042 | 8.23 |
ENST00000310074.7
ENST00000512388.1 |
TADA2B
|
transcriptional adaptor 2B |
| chr12_-_12491608 | 8.20 |
ENST00000545735.1
|
MANSC1
|
MANSC domain containing 1 |
| chr7_+_29237354 | 8.13 |
ENST00000546235.1
|
CHN2
|
chimerin 2 |
| chr1_+_27719148 | 8.03 |
ENST00000374024.3
|
GPR3
|
G protein-coupled receptor 3 |
| chr12_+_6644443 | 8.02 |
ENST00000396858.1
|
GAPDH
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr16_-_2264779 | 7.93 |
ENST00000333503.7
|
PGP
|
phosphoglycolate phosphatase |
| chr5_+_66254698 | 7.90 |
ENST00000405643.1
ENST00000407621.1 ENST00000432426.1 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr12_+_57857475 | 7.83 |
ENST00000528467.1
|
GLI1
|
GLI family zinc finger 1 |
| chr22_+_20104947 | 7.79 |
ENST00000402752.1
|
RANBP1
|
RAN binding protein 1 |
| chr6_+_134758827 | 7.66 |
ENST00000431422.1
|
LINC01010
|
long intergenic non-protein coding RNA 1010 |
| chr3_+_4535155 | 7.58 |
ENST00000544951.1
|
ITPR1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr1_-_241520385 | 7.57 |
ENST00000366564.1
|
RGS7
|
regulator of G-protein signaling 7 |
| chr16_+_67694849 | 7.51 |
ENST00000602551.1
ENST00000458121.2 ENST00000219255.3 |
PARD6A
|
par-6 family cell polarity regulator alpha |
| chr22_+_46546494 | 7.51 |
ENST00000396000.2
ENST00000262735.5 ENST00000420804.1 |
PPARA
|
peroxisome proliferator-activated receptor alpha |
| chr12_+_120875887 | 7.43 |
ENST00000229379.2
|
COX6A1
|
cytochrome c oxidase subunit VIa polypeptide 1 |
| chr16_+_19729586 | 7.38 |
ENST00000564186.1
ENST00000541926.1 ENST00000433597.2 |
IQCK
|
IQ motif containing K |
| chr11_-_118047376 | 7.35 |
ENST00000278947.5
|
SCN2B
|
sodium channel, voltage-gated, type II, beta subunit |
| chr6_-_27782548 | 7.25 |
ENST00000333151.3
|
HIST1H2AJ
|
histone cluster 1, H2aj |
| chr12_-_117537240 | 7.25 |
ENST00000392545.4
ENST00000541210.1 ENST00000335209.7 |
TESC
|
tescalcin |
| chr14_+_75761099 | 7.21 |
ENST00000561000.1
ENST00000558575.1 |
RP11-293M10.5
|
RP11-293M10.5 |
| chr15_-_78526855 | 7.21 |
ENST00000541759.1
ENST00000558130.1 |
ACSBG1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr12_+_48513570 | 7.18 |
ENST00000551804.1
|
PFKM
|
phosphofructokinase, muscle |
| chr20_-_45280066 | 7.17 |
ENST00000279027.4
|
SLC13A3
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 3 |
| chr6_-_27806117 | 7.16 |
ENST00000330180.2
|
HIST1H2AK
|
histone cluster 1, H2ak |
| chr10_-_101190202 | 7.13 |
ENST00000543866.1
ENST00000370508.5 |
GOT1
|
glutamic-oxaloacetic transaminase 1, soluble |
| chr14_-_31889782 | 7.05 |
ENST00000543095.2
|
HEATR5A
|
HEAT repeat containing 5A |
| chr11_-_3862059 | 7.05 |
ENST00000396978.1
|
RHOG
|
ras homolog family member G |
| chr6_-_33285505 | 7.01 |
ENST00000431845.2
|
ZBTB22
|
zinc finger and BTB domain containing 22 |
| chr5_+_80529104 | 6.93 |
ENST00000254035.4
ENST00000511719.1 ENST00000437669.1 ENST00000424301.2 ENST00000505060.1 |
CKMT2
|
creatine kinase, mitochondrial 2 (sarcomeric) |
| chr5_-_218251 | 6.93 |
ENST00000296824.3
|
CCDC127
|
coiled-coil domain containing 127 |
| chr20_-_45280091 | 6.88 |
ENST00000396360.1
ENST00000435032.1 ENST00000413164.2 ENST00000372121.1 ENST00000339636.3 |
SLC13A3
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 3 |
| chr3_+_4535025 | 6.83 |
ENST00000302640.8
ENST00000354582.6 ENST00000423119.2 ENST00000357086.4 ENST00000456211.2 |
ITPR1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr17_+_48585958 | 6.82 |
ENST00000436259.2
|
MYCBPAP
|
MYCBP associated protein |
| chr2_-_100939195 | 6.79 |
ENST00000393437.3
|
LONRF2
|
LON peptidase N-terminal domain and ring finger 2 |
| chr16_+_2588012 | 6.77 |
ENST00000354836.5
ENST00000389224.3 |
PDPK1
|
3-phosphoinositide dependent protein kinase-1 |
| chr3_-_49466686 | 6.73 |
ENST00000273598.3
ENST00000436744.2 |
NICN1
|
nicolin 1 |
| chr10_-_104179682 | 6.70 |
ENST00000406432.1
|
PSD
|
pleckstrin and Sec7 domain containing |
| chr2_-_97304009 | 6.69 |
ENST00000431828.1
ENST00000435669.1 ENST00000440133.1 ENST00000448075.1 |
KANSL3
|
KAT8 regulatory NSL complex subunit 3 |
| chr17_+_30814707 | 6.67 |
ENST00000584792.1
|
CDK5R1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
| chr19_+_58694396 | 6.65 |
ENST00000326804.4
ENST00000345813.3 ENST00000424679.2 |
ZNF274
|
zinc finger protein 274 |
| chr6_+_88757507 | 6.60 |
ENST00000237201.1
|
SPACA1
|
sperm acrosome associated 1 |
| chr16_+_15031300 | 6.57 |
ENST00000328085.6
|
NPIPA1
|
nuclear pore complex interacting protein family, member A1 |
| chr19_+_42381173 | 6.54 |
ENST00000221972.3
|
CD79A
|
CD79a molecule, immunoglobulin-associated alpha |
| chr1_-_149858227 | 6.53 |
ENST00000369155.2
|
HIST2H2BE
|
histone cluster 2, H2be |
| chr1_+_207226574 | 6.53 |
ENST00000367080.3
ENST00000367079.2 |
PFKFB2
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
| chr12_-_9268707 | 6.45 |
ENST00000318602.7
|
A2M
|
alpha-2-macroglobulin |
| chr20_+_32077880 | 6.43 |
ENST00000342704.6
ENST00000375279.2 |
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2 |
| chr18_-_51750948 | 6.41 |
ENST00000583046.1
ENST00000398398.2 |
MBD2
|
methyl-CpG binding domain protein 2 |
| chr2_-_97304105 | 6.41 |
ENST00000599854.1
ENST00000441706.2 |
KANSL3
|
KAT8 regulatory NSL complex subunit 3 |
| chr4_-_170947522 | 6.41 |
ENST00000361618.3
|
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr19_+_48773337 | 6.39 |
ENST00000595607.1
|
ZNF114
|
zinc finger protein 114 |
| chr3_-_183735731 | 6.36 |
ENST00000334444.6
|
ABCC5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr22_+_30792980 | 6.32 |
ENST00000403484.1
ENST00000405717.3 ENST00000402592.3 |
SEC14L2
|
SEC14-like 2 (S. cerevisiae) |
| chr1_-_160492994 | 6.25 |
ENST00000368055.1
ENST00000368057.3 ENST00000368059.3 |
SLAMF6
|
SLAM family member 6 |
| chr1_+_15764931 | 6.24 |
ENST00000375949.4
ENST00000375943.2 |
CTRC
|
chymotrypsin C (caldecrin) |
| chr3_-_122512619 | 6.23 |
ENST00000383659.1
ENST00000306103.2 |
HSPBAP1
|
HSPB (heat shock 27kDa) associated protein 1 |
| chr7_-_158622210 | 6.22 |
ENST00000251527.5
|
ESYT2
|
extended synaptotagmin-like protein 2 |
| chr5_-_134914673 | 6.22 |
ENST00000512158.1
|
CXCL14
|
chemokine (C-X-C motif) ligand 14 |
| chr12_+_48513009 | 6.18 |
ENST00000359794.5
ENST00000551339.1 ENST00000395233.2 ENST00000548345.1 |
PFKM
|
phosphofructokinase, muscle |
| chr2_-_27718052 | 6.13 |
ENST00000264703.3
|
FNDC4
|
fibronectin type III domain containing 4 |
| chr22_+_38035459 | 6.12 |
ENST00000357436.4
|
SH3BP1
|
SH3-domain binding protein 1 |
| chr1_-_203155868 | 6.12 |
ENST00000255409.3
|
CHI3L1
|
chitinase 3-like 1 (cartilage glycoprotein-39) |
| chr5_+_140753444 | 6.11 |
ENST00000517434.1
|
PCDHGA6
|
protocadherin gamma subfamily A, 6 |
| chr10_-_124768300 | 6.11 |
ENST00000368886.5
|
IKZF5
|
IKAROS family zinc finger 5 (Pegasus) |
| chr11_+_63742050 | 6.08 |
ENST00000314133.3
ENST00000535431.1 |
COX8A
AP000721.4
|
cytochrome c oxidase subunit VIIIA (ubiquitous) Uncharacterized protein |
| chr16_+_28875126 | 6.05 |
ENST00000359285.5
ENST00000538342.1 |
SH2B1
|
SH2B adaptor protein 1 |
| chrX_+_30671476 | 6.01 |
ENST00000378946.3
ENST00000378943.3 ENST00000378945.3 ENST00000427190.1 ENST00000378941.3 |
GK
|
glycerol kinase |
| chr11_+_1411503 | 5.98 |
ENST00000526678.1
|
BRSK2
|
BR serine/threonine kinase 2 |
| chr20_+_54933971 | 5.95 |
ENST00000371384.3
ENST00000437418.1 |
FAM210B
|
family with sequence similarity 210, member B |
| chr20_+_1875942 | 5.95 |
ENST00000358771.4
|
SIRPA
|
signal-regulatory protein alpha |
| chr11_-_64511789 | 5.94 |
ENST00000419843.1
ENST00000394430.1 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr20_+_53092123 | 5.81 |
ENST00000262593.5
|
DOK5
|
docking protein 5 |
| chr1_+_203096831 | 5.76 |
ENST00000337894.4
|
ADORA1
|
adenosine A1 receptor |
| chr9_+_103235365 | 5.76 |
ENST00000374879.4
|
TMEFF1
|
transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chr6_-_137113604 | 5.74 |
ENST00000359015.4
|
MAP3K5
|
mitogen-activated protein kinase kinase kinase 5 |
| chr3_-_49158312 | 5.72 |
ENST00000398892.3
ENST00000453664.1 ENST00000398888.2 |
USP19
|
ubiquitin specific peptidase 19 |
| chr5_-_88179017 | 5.68 |
ENST00000514028.1
ENST00000514015.1 ENST00000503075.1 ENST00000437473.2 |
MEF2C
|
myocyte enhancer factor 2C |
| chrX_-_43741594 | 5.67 |
ENST00000536181.1
ENST00000378069.4 |
MAOB
|
monoamine oxidase B |
| chr5_+_65018017 | 5.55 |
ENST00000380985.5
ENST00000502464.1 |
NLN
|
neurolysin (metallopeptidase M3 family) |
| chr16_+_3451184 | 5.53 |
ENST00000575752.1
ENST00000571936.1 ENST00000344823.5 |
ZNF174
|
zinc finger protein 174 |
| chr22_+_30792846 | 5.52 |
ENST00000312932.9
ENST00000428195.1 |
SEC14L2
|
SEC14-like 2 (S. cerevisiae) |
| chr1_+_29138654 | 5.51 |
ENST00000234961.2
|
OPRD1
|
opioid receptor, delta 1 |
| chr14_+_91580732 | 5.49 |
ENST00000519019.1
ENST00000523816.1 ENST00000517518.1 |
C14orf159
|
chromosome 14 open reading frame 159 |
| chrX_+_48681768 | 5.48 |
ENST00000430858.1
|
HDAC6
|
histone deacetylase 6 |
| chr16_+_8814563 | 5.46 |
ENST00000425191.2
ENST00000569156.1 |
ABAT
|
4-aminobutyrate aminotransferase |
| chr1_+_44584522 | 5.43 |
ENST00000372299.3
|
KLF17
|
Kruppel-like factor 17 |
| chr20_-_18447667 | 5.33 |
ENST00000262547.5
ENST00000329494.5 ENST00000357236.4 |
DZANK1
|
double zinc ribbon and ankyrin repeat domains 1 |
| chr1_+_29213678 | 5.33 |
ENST00000347529.3
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
| chr21_+_34697258 | 5.31 |
ENST00000442071.1
ENST00000442357.2 |
IFNAR1
|
interferon (alpha, beta and omega) receptor 1 |
| chr6_+_27100811 | 5.29 |
ENST00000359193.2
|
HIST1H2AG
|
histone cluster 1, H2ag |
| chrX_+_22056165 | 5.27 |
ENST00000535894.1
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked |
| chr9_-_127269661 | 5.23 |
ENST00000373588.4
|
NR5A1
|
nuclear receptor subfamily 5, group A, member 1 |
| chr22_+_23412479 | 5.19 |
ENST00000248996.4
|
GNAZ
|
guanine nucleotide binding protein (G protein), alpha z polypeptide |
| chr17_+_66539369 | 5.18 |
ENST00000600820.1
|
AC079210.1
|
Uncharacterized protein; cDNA FLJ45097 fis, clone BRAWH3031054 |
| chr5_+_34929677 | 5.11 |
ENST00000342382.4
ENST00000382021.2 ENST00000303525.7 |
DNAJC21
|
DnaJ (Hsp40) homolog, subfamily C, member 21 |
| chr5_-_88178964 | 5.11 |
ENST00000513252.1
ENST00000508569.1 ENST00000510942.1 ENST00000506554.1 |
MEF2C
|
myocyte enhancer factor 2C |
| chr11_+_10472223 | 5.00 |
ENST00000396554.3
ENST00000524866.1 |
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr1_-_20834586 | 4.99 |
ENST00000264198.3
|
MUL1
|
mitochondrial E3 ubiquitin protein ligase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR5A2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.3 | 228.6 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 12.0 | 36.0 | GO:2000296 | cadmium ion homeostasis(GO:0055073) negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 8.3 | 41.6 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 4.7 | 23.5 | GO:0021699 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 4.5 | 13.4 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 4.3 | 12.9 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 4.2 | 25.2 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 4.1 | 69.5 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 4.0 | 27.9 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 3.9 | 11.8 | GO:0097327 | response to antineoplastic agent(GO:0097327) glycoside transport(GO:1901656) cellular response to bile acid(GO:1903413) |
| 3.9 | 11.7 | GO:1903233 | regulation of calcium ion-dependent exocytosis of neurotransmitter(GO:1903233) |
| 3.8 | 15.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 3.7 | 11.1 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 3.7 | 11.0 | GO:0042938 | dipeptide transport(GO:0042938) |
| 3.5 | 14.1 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 3.5 | 21.1 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 3.1 | 34.6 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 3.0 | 3.0 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 3.0 | 8.9 | GO:1904530 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 2.9 | 14.4 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 2.9 | 11.5 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) positive regulation of potassium ion import(GO:1903288) |
| 2.8 | 8.4 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 2.7 | 13.5 | GO:1901908 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 2.7 | 10.8 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 2.5 | 12.6 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 2.5 | 12.5 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 2.4 | 9.5 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 2.3 | 7.0 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 2.3 | 20.9 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 2.3 | 9.1 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 2.2 | 6.7 | GO:0021718 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 2.2 | 11.0 | GO:0097338 | response to clozapine(GO:0097338) |
| 2.0 | 8.1 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 2.0 | 29.7 | GO:0015671 | oxygen transport(GO:0015671) |
| 2.0 | 7.8 | GO:0007418 | ventral midline development(GO:0007418) smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 1.9 | 36.8 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 1.9 | 5.8 | GO:0032242 | regulation of nucleoside transport(GO:0032242) negative regulation of neurotrophin production(GO:0032900) |
| 1.9 | 21.0 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 1.9 | 11.3 | GO:1903301 | fructose 2,6-bisphosphate metabolic process(GO:0006003) positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 1.9 | 7.5 | GO:0072366 | regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 1.9 | 9.3 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 1.9 | 5.6 | GO:1902809 | skeletal muscle fiber differentiation(GO:0098528) regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 1.8 | 5.5 | GO:0090034 | regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 1.8 | 10.9 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 1.8 | 31.6 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 1.6 | 9.4 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 1.5 | 15.0 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 1.5 | 6.0 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 1.5 | 3.0 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 1.5 | 78.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 1.5 | 11.8 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 1.5 | 7.4 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 1.5 | 4.4 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 1.5 | 4.4 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 1.4 | 4.3 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 1.4 | 4.3 | GO:0010902 | positive regulation of very-low-density lipoprotein particle remodeling(GO:0010902) |
| 1.4 | 9.6 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 1.4 | 6.8 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 1.3 | 6.6 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 1.3 | 3.9 | GO:0021571 | rhombomere 5 development(GO:0021571) |
| 1.3 | 5.2 | GO:0007538 | primary sex determination(GO:0007538) |
| 1.2 | 11.2 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 1.2 | 3.7 | GO:0018874 | benzoate metabolic process(GO:0018874) butyrate metabolic process(GO:0019605) |
| 1.2 | 7.3 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 1.2 | 3.6 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 1.1 | 4.5 | GO:0072255 | metanephric glomerular mesangial cell development(GO:0072255) reversible differentiation(GO:0090677) cell dedifferentiation involved in phenotypic switching(GO:0090678) positive regulation of phenotypic switching(GO:1900241) regulation of vascular smooth muscle cell dedifferentiation(GO:1905174) positive regulation of vascular smooth muscle cell dedifferentiation(GO:1905176) vascular smooth muscle cell dedifferentiation(GO:1990936) |
| 1.1 | 5.7 | GO:0045915 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 1.1 | 4.5 | GO:2000314 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 1.1 | 3.3 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 1.1 | 3.2 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 1.1 | 5.3 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 1.0 | 5.2 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 1.0 | 4.0 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 1.0 | 19.3 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.9 | 2.8 | GO:0048075 | positive regulation of eye pigmentation(GO:0048075) |
| 0.9 | 12.9 | GO:0021702 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.9 | 7.3 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.9 | 20.7 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.9 | 2.7 | GO:0045643 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.9 | 6.2 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.9 | 2.6 | GO:0018262 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.9 | 13.1 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.9 | 2.6 | GO:0007497 | posterior midgut development(GO:0007497) |
| 0.9 | 6.0 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.9 | 2.6 | GO:2001016 | positive regulation of skeletal muscle cell differentiation(GO:2001016) |
| 0.8 | 17.6 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.8 | 7.5 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.8 | 5.0 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.8 | 18.2 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.8 | 4.7 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.8 | 2.3 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 0.8 | 2.3 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.8 | 14.5 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.7 | 3.0 | GO:1904048 | regulation of spontaneous neurotransmitter secretion(GO:1904048) |
| 0.7 | 3.6 | GO:0046968 | peptide antigen transport(GO:0046968) |
| 0.7 | 3.6 | GO:0051142 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.7 | 5.0 | GO:1904925 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.7 | 4.9 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.7 | 4.1 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.7 | 2.7 | GO:1990535 | transformation of host cell by virus(GO:0019087) neuron projection maintenance(GO:1990535) |
| 0.7 | 11.1 | GO:0061339 | establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.6 | 3.8 | GO:1901523 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 0.6 | 6.4 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.6 | 3.2 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.6 | 4.4 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.6 | 6.2 | GO:0001787 | natural killer cell proliferation(GO:0001787) |
| 0.6 | 1.2 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.6 | 4.3 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.6 | 1.9 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.6 | 9.3 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.6 | 1.8 | GO:0009726 | detection of endogenous stimulus(GO:0009726) |
| 0.6 | 6.1 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.6 | 3.6 | GO:0042636 | negative regulation of hair cycle(GO:0042636) |
| 0.6 | 7.6 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.6 | 9.4 | GO:0006705 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.6 | 2.3 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.6 | 1.7 | GO:0072014 | proximal tubule development(GO:0072014) negative regulation of androgen receptor activity(GO:2000824) |
| 0.6 | 1.7 | GO:1900073 | regulation of neuromuscular synaptic transmission(GO:1900073) positive regulation of neuromuscular synaptic transmission(GO:1900075) |
| 0.6 | 1.1 | GO:0044036 | cell wall macromolecule metabolic process(GO:0044036) cell wall organization or biogenesis(GO:0071554) |
| 0.6 | 14.3 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.6 | 2.3 | GO:0061364 | apoptotic process involved in luteolysis(GO:0061364) |
| 0.6 | 10.2 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.6 | 2.3 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.6 | 4.4 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.6 | 3.9 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.6 | 6.7 | GO:1900112 | regulation of histone H3-K9 trimethylation(GO:1900112) |
| 0.5 | 2.7 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.5 | 7.5 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.5 | 2.1 | GO:0042335 | cuticle development(GO:0042335) |
| 0.5 | 2.1 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.5 | 5.3 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.5 | 2.1 | GO:2000969 | positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.5 | 1.6 | GO:0036378 | calcitriol biosynthetic process from calciol(GO:0036378) |
| 0.5 | 3.1 | GO:0071910 | determination of pancreatic left/right asymmetry(GO:0035469) determination of liver left/right asymmetry(GO:0071910) |
| 0.5 | 3.6 | GO:0006921 | cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.5 | 8.6 | GO:0048820 | hair follicle maturation(GO:0048820) |
| 0.5 | 2.0 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.5 | 20.2 | GO:0042417 | dopamine metabolic process(GO:0042417) |
| 0.5 | 7.0 | GO:0010666 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.5 | 1.0 | GO:1902612 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 0.5 | 4.4 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.5 | 2.0 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.5 | 5.8 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.5 | 3.4 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.5 | 3.8 | GO:0040034 | regulation of development, heterochronic(GO:0040034) regulation of timing of cell differentiation(GO:0048505) |
| 0.5 | 18.1 | GO:0006536 | glutamate metabolic process(GO:0006536) |
| 0.5 | 1.9 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.5 | 14.7 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.5 | 4.6 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.4 | 0.4 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.4 | 2.7 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.4 | 4.8 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.4 | 1.7 | GO:0061304 | extracellular matrix-cell signaling(GO:0035426) retinal blood vessel morphogenesis(GO:0061304) |
| 0.4 | 5.0 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.4 | 1.2 | GO:0035565 | regulation of pronephros size(GO:0035565) renal glucose absorption(GO:0035623) |
| 0.4 | 6.0 | GO:1904152 | regulation of retrograde protein transport, ER to cytosol(GO:1904152) |
| 0.4 | 3.2 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.4 | 1.6 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.4 | 0.7 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.4 | 1.4 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.4 | 3.6 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.4 | 5.7 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.4 | 9.1 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.3 | 3.8 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.3 | 13.8 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.3 | 3.5 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.3 | 3.7 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.3 | 6.4 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.3 | 4.0 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.3 | 3.7 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.3 | 3.6 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.3 | 4.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.3 | 1.6 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.3 | 2.5 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.3 | 4.3 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.3 | 9.9 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.3 | 4.1 | GO:0043306 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.3 | 0.8 | GO:1904469 | positive regulation of tumor necrosis factor secretion(GO:1904469) |
| 0.3 | 2.2 | GO:0035912 | dorsal aorta morphogenesis(GO:0035912) |
| 0.3 | 3.2 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.3 | 2.1 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.3 | 2.1 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.3 | 11.1 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.3 | 3.3 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.2 | 4.0 | GO:0071028 | nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.2 | 2.2 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.2 | 3.3 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.2 | 15.4 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.2 | 1.8 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.2 | 4.5 | GO:0001976 | neurological system process involved in regulation of systemic arterial blood pressure(GO:0001976) |
| 0.2 | 4.4 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.2 | 1.3 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.2 | 1.1 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) |
| 0.2 | 1.5 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.2 | 3.2 | GO:0043536 | positive regulation of blood vessel endothelial cell migration(GO:0043536) |
| 0.2 | 0.6 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.2 | 5.3 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.2 | 10.3 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.2 | 4.8 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.2 | 4.6 | GO:0018195 | peptidyl-arginine modification(GO:0018195) |
| 0.2 | 0.9 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.2 | 1.1 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.2 | 7.7 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.2 | 2.7 | GO:0009083 | branched-chain amino acid metabolic process(GO:0009081) branched-chain amino acid catabolic process(GO:0009083) |
| 0.2 | 4.1 | GO:2000811 | negative regulation of anoikis(GO:2000811) |
| 0.2 | 3.2 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.2 | 39.3 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.2 | 1.5 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.2 | 1.0 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.2 | 3.1 | GO:0006972 | hyperosmotic response(GO:0006972) |
| 0.2 | 0.8 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.2 | 1.5 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.2 | 9.0 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.2 | 5.1 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.2 | 3.9 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.2 | 1.2 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.1 | 3.0 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.1 | 7.4 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 2.4 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 0.9 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.1 | 12.2 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.1 | 3.3 | GO:0043691 | reverse cholesterol transport(GO:0043691) |
| 0.1 | 3.1 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 3.1 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.1 | 102.6 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.1 | 2.1 | GO:0030220 | platelet formation(GO:0030220) |
| 0.1 | 4.3 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.1 | 2.5 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.1 | 0.9 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) atrial cardiac muscle tissue morphogenesis(GO:0055009) |
| 0.1 | 1.3 | GO:0036315 | cellular response to sterol(GO:0036315) cellular response to cholesterol(GO:0071397) |
| 0.1 | 3.2 | GO:0006625 | protein targeting to peroxisome(GO:0006625) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.1 | 0.1 | GO:1902732 | positive regulation of chondrocyte proliferation(GO:1902732) |
| 0.1 | 1.8 | GO:0032754 | positive regulation of interleukin-5 production(GO:0032754) |
| 0.1 | 4.3 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 0.9 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.1 | 1.8 | GO:0033015 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.1 | 3.3 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 14.5 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.1 | 3.0 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.1 | 4.4 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 1.6 | GO:2000758 | positive regulation of histone acetylation(GO:0035066) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.1 | 2.5 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 6.1 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 4.8 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 0.4 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.1 | 1.0 | GO:0000963 | mitochondrial RNA processing(GO:0000963) |
| 0.1 | 4.2 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.1 | 5.0 | GO:1905114 | Wnt signaling pathway(GO:0016055) cell-cell signaling by wnt(GO:0198738) cell surface receptor signaling pathway involved in cell-cell signaling(GO:1905114) |
| 0.1 | 1.8 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.1 | 4.3 | GO:0032092 | positive regulation of protein binding(GO:0032092) |
| 0.1 | 9.1 | GO:0006096 | glycolytic process(GO:0006096) ATP generation from ADP(GO:0006757) |
| 0.1 | 2.3 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.1 | 3.1 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.1 | 0.2 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.1 | 10.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 9.9 | GO:0051480 | regulation of cytosolic calcium ion concentration(GO:0051480) |
| 0.1 | 1.6 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.1 | 2.5 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 0.4 | GO:0016050 | vesicle organization(GO:0016050) |
| 0.1 | 0.9 | GO:2000398 | regulation of T cell differentiation in thymus(GO:0033081) regulation of thymocyte aggregation(GO:2000398) |
| 0.1 | 2.5 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 0.5 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.1 | 1.6 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.1 | 2.3 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 0.2 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.1 | 1.5 | GO:0097503 | sialylation(GO:0097503) |
| 0.1 | 0.7 | GO:0001696 | gastric acid secretion(GO:0001696) |
| 0.0 | 1.5 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.0 | 4.5 | GO:0006633 | fatty acid biosynthetic process(GO:0006633) |
| 0.0 | 2.3 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.9 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.4 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 1.5 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.1 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 1.7 | GO:0034113 | heterotypic cell-cell adhesion(GO:0034113) |
| 0.0 | 0.3 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.0 | 1.0 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 0.6 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 1.6 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.9 | GO:0030819 | positive regulation of cAMP biosynthetic process(GO:0030819) |
| 0.0 | 0.1 | GO:0044266 | angiotensin catabolic process in blood(GO:0002005) multicellular organismal protein catabolic process(GO:0044254) protein digestion(GO:0044256) multicellular organismal macromolecule catabolic process(GO:0044266) multicellular organismal protein metabolic process(GO:0044268) |
| 0.0 | 0.4 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.8 | GO:0030218 | erythrocyte differentiation(GO:0030218) |
| 0.0 | 0.1 | GO:0010644 | cell communication by electrical coupling(GO:0010644) |
| 0.0 | 0.8 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.9 | GO:0042472 | inner ear morphogenesis(GO:0042472) |
| 0.0 | 2.4 | GO:0001654 | eye development(GO:0001654) |
| 0.0 | 0.3 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 0.3 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.0 | GO:0042415 | norepinephrine metabolic process(GO:0042415) surfactant homeostasis(GO:0043129) |
| 0.0 | 0.1 | GO:0097237 | cellular response to toxic substance(GO:0097237) |
| 0.0 | 1.1 | GO:0006334 | nucleosome assembly(GO:0006334) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.1 | 28.5 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 7.0 | 27.9 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 3.8 | 49.9 | GO:0097433 | dense body(GO:0097433) |
| 3.6 | 36.0 | GO:0097449 | astrocyte projection(GO:0097449) |
| 3.5 | 21.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 3.0 | 51.3 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 3.0 | 41.6 | GO:0097227 | sperm annulus(GO:0097227) |
| 2.7 | 13.4 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 2.6 | 41.7 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 2.3 | 16.4 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 2.1 | 36.4 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 2.0 | 8.1 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 2.0 | 11.8 | GO:1990130 | Iml1 complex(GO:1990130) |
| 1.7 | 11.8 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 1.7 | 6.7 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 1.6 | 18.6 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 1.5 | 10.5 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 1.4 | 5.7 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 1.4 | 11.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 1.4 | 5.5 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 1.3 | 18.1 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 1.2 | 9.3 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 1.1 | 219.3 | GO:0030426 | growth cone(GO:0030426) |
| 1.0 | 7.0 | GO:0097452 | GAIT complex(GO:0097452) |
| 1.0 | 44.1 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.9 | 2.7 | GO:0098536 | deuterosome(GO:0098536) |
| 0.9 | 3.5 | GO:0043293 | apoptosome(GO:0043293) |
| 0.8 | 7.5 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.8 | 5.8 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.7 | 4.3 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.7 | 2.8 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.7 | 3.3 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.6 | 11.5 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.5 | 2.7 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.5 | 4.3 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.5 | 28.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.5 | 8.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.5 | 39.8 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.5 | 8.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.4 | 4.7 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.4 | 9.5 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.4 | 10.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.4 | 22.9 | GO:0000786 | nucleosome(GO:0000786) |
| 0.4 | 2.3 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.4 | 6.2 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.4 | 7.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.4 | 3.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.3 | 11.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.3 | 5.3 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.3 | 3.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.3 | 1.9 | GO:0032420 | stereocilium(GO:0032420) |
| 0.3 | 0.6 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.3 | 1.8 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.3 | 5.5 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.3 | 7.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.3 | 2.2 | GO:0032797 | SMN complex(GO:0032797) Gemini of coiled bodies(GO:0097504) |
| 0.3 | 1.0 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.3 | 2.5 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.3 | 2.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.2 | 26.8 | GO:0016234 | inclusion body(GO:0016234) |
| 0.2 | 3.0 | GO:0036038 | MKS complex(GO:0036038) |
| 0.2 | 1.7 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.2 | 6.6 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.2 | 6.1 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.2 | 1.0 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.2 | 19.7 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.2 | 3.0 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.2 | 2.9 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.2 | 0.2 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.2 | 3.4 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.2 | 12.2 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.2 | 1.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.2 | 4.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.2 | 1.2 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.2 | 1.6 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.2 | 2.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.2 | 39.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.2 | 24.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.2 | 23.3 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.2 | 4.9 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.1 | 4.0 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 3.8 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 5.3 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 4.0 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 2.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 2.1 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 1.8 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 4.2 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 0.9 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 11.5 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 4.1 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 1.5 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 5.1 | GO:0043679 | axon terminus(GO:0043679) |
| 0.1 | 1.1 | GO:0001741 | XY body(GO:0001741) |
| 0.1 | 2.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 2.3 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 1.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 14.3 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.1 | 1.2 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 6.8 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 1.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 4.3 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 6.6 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 7.8 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.1 | 5.4 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.1 | 0.6 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 3.7 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 1.0 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.1 | 3.0 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 7.2 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 96.6 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 6.1 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 2.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 3.3 | GO:0005657 | replication fork(GO:0005657) |
| 0.1 | 5.2 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.8 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 2.7 | GO:0044297 | cell body(GO:0044297) |
| 0.0 | 3.1 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.9 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 2.8 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.6 | GO:0042827 | platelet dense granule(GO:0042827) |
| 0.0 | 3.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 59.1 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 1.0 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 0.7 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 3.7 | GO:0043005 | neuron projection(GO:0043005) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.9 | 70.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 5.0 | 15.0 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 4.4 | 22.2 | GO:0047820 | D-glutamate cyclase activity(GO:0047820) |
| 4.4 | 22.0 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 4.3 | 77.0 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 3.6 | 14.4 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 3.6 | 28.5 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 3.5 | 14.1 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 3.0 | 11.8 | GO:0008431 | vitamin E binding(GO:0008431) |
| 2.7 | 35.6 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 2.7 | 13.5 | GO:0052845 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 2.6 | 7.9 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 2.6 | 36.8 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 2.5 | 12.5 | GO:0004359 | glutaminase activity(GO:0004359) |
| 2.5 | 76.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 2.4 | 7.1 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 2.4 | 11.8 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 2.4 | 9.4 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 2.3 | 29.7 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 2.2 | 13.4 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 2.2 | 6.7 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 2.1 | 6.4 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 1.9 | 11.3 | GO:0008443 | phosphofructokinase activity(GO:0008443) |
| 1.8 | 7.4 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 1.8 | 5.3 | GO:0019961 | interferon binding(GO:0019961) |
| 1.8 | 21.1 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 1.8 | 24.5 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 1.7 | 7.0 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) aspartic-type endopeptidase inhibitor activity(GO:0019828) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 1.7 | 5.0 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 1.6 | 17.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 1.5 | 44.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 1.4 | 4.3 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 1.4 | 41.0 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 1.4 | 11.0 | GO:0004064 | arylesterase activity(GO:0004064) |
| 1.4 | 5.5 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 1.4 | 5.5 | GO:0047298 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 1.3 | 4.0 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 1.3 | 6.4 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 1.3 | 3.8 | GO:0052870 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 1.2 | 7.5 | GO:0043532 | angiostatin binding(GO:0043532) |
| 1.2 | 9.3 | GO:0030172 | troponin C binding(GO:0030172) |
| 1.2 | 5.8 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 1.1 | 6.6 | GO:0003796 | lysozyme activity(GO:0003796) |
| 1.1 | 13.1 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 1.1 | 33.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 1.0 | 9.3 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 1.0 | 10.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 1.0 | 18.2 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 1.0 | 16.4 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.9 | 8.2 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.9 | 2.7 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.9 | 2.6 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.9 | 2.6 | GO:0004507 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 0.9 | 4.3 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.8 | 8.2 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.8 | 3.2 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.8 | 2.3 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.7 | 4.5 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.7 | 19.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.7 | 4.4 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.7 | 13.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.7 | 3.6 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.7 | 11.5 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.7 | 3.6 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.7 | 4.3 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.7 | 46.2 | GO:0043621 | protein self-association(GO:0043621) |
| 0.7 | 7.5 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.7 | 6.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.7 | 5.3 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.7 | 2.6 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.6 | 3.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.6 | 1.8 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.6 | 6.1 | GO:0008061 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 0.6 | 10.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.6 | 3.0 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.6 | 4.1 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.6 | 2.3 | GO:0035473 | lipase binding(GO:0035473) |
| 0.6 | 4.0 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.6 | 5.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.6 | 4.4 | GO:0016406 | carnitine O-acyltransferase activity(GO:0016406) |
| 0.5 | 2.2 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.5 | 10.8 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.5 | 7.5 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.5 | 6.4 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.5 | 1.6 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.5 | 1.6 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.5 | 5.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.5 | 3.6 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.5 | 15.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.5 | 2.0 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.5 | 3.5 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.5 | 3.0 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.5 | 3.9 | GO:0005497 | androgen binding(GO:0005497) |
| 0.5 | 4.3 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.5 | 10.6 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.5 | 3.8 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.4 | 1.8 | GO:0019862 | IgA binding(GO:0019862) |
| 0.4 | 4.9 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.4 | 1.3 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.4 | 10.7 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.4 | 7.2 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.4 | 3.3 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.4 | 3.7 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.4 | 3.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.4 | 4.4 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.4 | 1.6 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.4 | 1.6 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.4 | 10.4 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.4 | 1.8 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.4 | 3.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.4 | 3.6 | GO:0009374 | biotin carboxylase activity(GO:0004075) biotin binding(GO:0009374) |
| 0.4 | 5.0 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.4 | 3.6 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.4 | 4.2 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.3 | 5.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.3 | 2.7 | GO:0043559 | insulin binding(GO:0043559) |
| 0.3 | 5.3 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.3 | 15.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.3 | 8.4 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.3 | 5.3 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.3 | 1.9 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.3 | 69.7 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.3 | 8.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.3 | 6.9 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.3 | 6.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.3 | 4.8 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.3 | 9.3 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.3 | 0.8 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.3 | 4.7 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.3 | 36.3 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.3 | 3.8 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.3 | 4.8 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.3 | 5.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.3 | 6.1 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.3 | 23.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.3 | 1.0 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.2 | 1.5 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.2 | 4.6 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.2 | 3.3 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.2 | 3.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.2 | 1.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.2 | 6.2 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.2 | 1.4 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.2 | 6.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.2 | 9.6 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.2 | 4.2 | GO:0016722 | oxidoreductase activity, oxidizing metal ions(GO:0016722) |
| 0.2 | 2.5 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.2 | 2.3 | GO:0048185 | activin binding(GO:0048185) |
| 0.2 | 1.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.2 | 3.0 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.2 | 3.3 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.2 | 0.8 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.2 | 2.0 | GO:0031432 | titin binding(GO:0031432) |
| 0.2 | 2.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.9 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.1 | 8.2 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 2.8 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 3.8 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 0.9 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.1 | 2.6 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 0.9 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.1 | 1.0 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 2.3 | GO:0070016 | gamma-catenin binding(GO:0045295) armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.7 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 3.3 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 12.7 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 0.3 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 33.5 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 1.9 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 3.0 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 11.1 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.1 | 2.8 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 1.7 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.2 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.1 | 3.1 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.8 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 3.2 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 26.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 4.8 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 5.4 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.1 | 1.0 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 2.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.4 | GO:0099583 | postsynaptic neurotransmitter receptor activity(GO:0098960) neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.1 | 0.3 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.1 | 2.0 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 2.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 1.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 5.9 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 0.3 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 0.7 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 1.9 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 3.0 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 1.5 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 1.7 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 1.8 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 0.6 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.1 | 1.0 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 1.3 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 1.3 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.1 | 5.4 | GO:0005230 | extracellular ligand-gated ion channel activity(GO:0005230) |
| 0.1 | 2.7 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 0.8 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 3.1 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 3.3 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.0 | 10.0 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 1.1 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.9 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 2.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 6.2 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 1.5 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.5 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.3 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 5.8 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 2.2 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.0 | 1.2 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 2.0 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 1.4 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 2.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.7 | GO:0001159 | core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 5.6 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 1.8 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 1.5 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 1.5 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 6.6 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 0.1 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.0 | 5.8 | GO:0004888 | transmembrane signaling receptor activity(GO:0004888) |
| 0.0 | 1.5 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 0.1 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.0 | 0.6 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.7 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 71.9 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 1.3 | 190.8 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.9 | 15.0 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.9 | 37.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.7 | 3.6 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.6 | 33.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.5 | 2.7 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.5 | 1.0 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.4 | 8.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.4 | 19.0 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.4 | 14.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.4 | 2.0 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.4 | 15.1 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.4 | 5.3 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.4 | 7.5 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.4 | 5.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.4 | 12.7 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.4 | 11.2 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.4 | 14.0 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.4 | 3.6 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.3 | 5.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.3 | 3.9 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.3 | 9.7 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.3 | 1.3 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.3 | 4.5 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.3 | 6.9 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.3 | 5.8 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.3 | 13.8 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.3 | 6.7 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.2 | 12.9 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.2 | 10.9 | PID ATM PATHWAY | ATM pathway |
| 0.2 | 12.7 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.2 | 2.2 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.2 | 4.2 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.2 | 7.8 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.2 | 3.5 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.2 | 6.8 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 7.8 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.2 | 5.3 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.2 | 11.8 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.2 | 5.1 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.2 | 7.2 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.2 | 4.4 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.2 | 7.1 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.2 | 7.7 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 5.3 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 3.9 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 7.0 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 2.6 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 8.7 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 3.9 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 3.3 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 2.2 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 2.9 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 0.6 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 14.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 1.5 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 1.5 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 6.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.2 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.5 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.5 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 44.1 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 1.6 | 27.9 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 1.0 | 12.5 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 1.0 | 20.7 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.9 | 11.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.9 | 13.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.8 | 10.9 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.7 | 11.0 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.7 | 8.9 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.6 | 29.0 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.6 | 31.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.6 | 13.0 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.6 | 15.6 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.5 | 14.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.5 | 9.8 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.5 | 16.1 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.4 | 47.0 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.4 | 16.1 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.4 | 24.4 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.4 | 15.2 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.4 | 8.5 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.4 | 13.1 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.4 | 4.4 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.4 | 10.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.4 | 7.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.4 | 8.4 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.3 | 5.2 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.3 | 5.5 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.3 | 10.7 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.3 | 5.4 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.3 | 6.6 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.3 | 9.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.3 | 3.8 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.3 | 4.4 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.3 | 3.6 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.3 | 4.1 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.3 | 2.7 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.3 | 3.5 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.3 | 6.1 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.3 | 8.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.3 | 6.1 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.3 | 6.0 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.2 | 5.5 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.2 | 4.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.2 | 3.3 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.2 | 3.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.2 | 3.8 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.2 | 4.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.2 | 4.6 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.2 | 32.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.2 | 5.3 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.2 | 3.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.2 | 6.0 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.2 | 11.4 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.2 | 1.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 3.0 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 2.6 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 3.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 2.0 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.1 | 10.2 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 34.9 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 2.3 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.1 | 4.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 3.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 2.1 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 1.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 4.8 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 1.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 2.8 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 3.5 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.1 | 3.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.9 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 2.9 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 5.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 2.3 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 1.4 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.1 | 3.2 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 0.8 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 3.8 | REACTOME BIOLOGICAL OXIDATIONS | Genes involved in Biological oxidations |
| 0.0 | 1.9 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 1.6 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 2.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 2.2 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 2.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.7 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 4.8 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 1.1 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 2.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 1.4 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.5 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |