Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for NR6A1
Z-value: 1.00
Transcription factors associated with NR6A1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR6A1
|
ENSG00000148200.12 | nuclear receptor subfamily 6 group A member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR6A1 | hg19_v2_chr9_-_127533582_127533615 | 0.45 | 2.6e-12 | Click! |
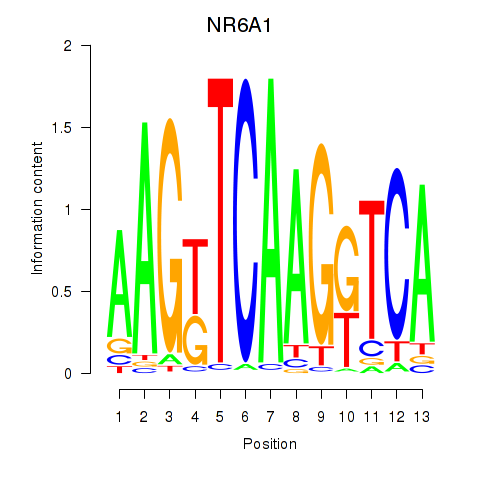
Activity profile of NR6A1 motif
Sorted Z-values of NR6A1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_130554803 | 13.68 |
ENST00000535487.1
|
RP11-474D1.2
|
RP11-474D1.2 |
| chr14_-_106539557 | 13.14 |
ENST00000390599.2
|
IGHV1-8
|
immunoglobulin heavy variable 1-8 |
| chr14_-_106642049 | 12.25 |
ENST00000390605.2
|
IGHV1-18
|
immunoglobulin heavy variable 1-18 |
| chr21_+_10862622 | 11.69 |
ENST00000302092.5
ENST00000559480.1 |
IGHV1OR21-1
|
immunoglobulin heavy variable 1/OR21-1 (non-functional) |
| chr14_-_106963409 | 11.31 |
ENST00000390621.2
|
IGHV1-45
|
immunoglobulin heavy variable 1-45 |
| chr19_-_11669960 | 10.83 |
ENST00000589171.1
ENST00000590700.1 ENST00000586683.1 ENST00000593077.1 ENST00000252445.3 |
ELOF1
|
elongation factor 1 homolog (S. cerevisiae) |
| chr14_-_96180435 | 10.44 |
ENST00000556450.1
ENST00000555202.1 ENST00000554012.1 ENST00000402399.1 |
TCL1A
|
T-cell leukemia/lymphoma 1A |
| chr22_+_23229960 | 10.25 |
ENST00000526893.1
ENST00000532223.2 ENST00000531372.1 |
IGLL5
|
immunoglobulin lambda-like polypeptide 5 |
| chr17_-_39093672 | 10.20 |
ENST00000209718.3
ENST00000436344.3 ENST00000485751.1 |
KRT23
|
keratin 23 (histone deacetylase inducible) |
| chr16_-_75498308 | 9.53 |
ENST00000569540.1
|
TMEM170A
|
transmembrane protein 170A |
| chr5_-_138725560 | 9.06 |
ENST00000412103.2
ENST00000457570.2 |
MZB1
|
marginal zone B and B1 cell-specific protein |
| chr19_+_42381173 | 9.02 |
ENST00000221972.3
|
CD79A
|
CD79a molecule, immunoglobulin-associated alpha |
| chr5_-_138725594 | 9.00 |
ENST00000302125.8
|
MZB1
|
marginal zone B and B1 cell-specific protein |
| chr15_-_20170354 | 8.90 |
ENST00000338912.5
|
IGHV1OR15-9
|
immunoglobulin heavy variable 1/OR15-9 (non-functional) |
| chr16_-_75498553 | 8.73 |
ENST00000569276.1
ENST00000357613.4 ENST00000561878.1 ENST00000566980.1 ENST00000567194.1 |
TMEM170A
RP11-77K12.1
|
transmembrane protein 170A Uncharacterized protein |
| chr19_+_42381337 | 8.55 |
ENST00000597454.1
ENST00000444740.2 |
CD79A
|
CD79a molecule, immunoglobulin-associated alpha |
| chr12_-_91574142 | 8.45 |
ENST00000547937.1
|
DCN
|
decorin |
| chr19_-_43586820 | 7.92 |
ENST00000406487.1
|
PSG2
|
pregnancy specific beta-1-glycoprotein 2 |
| chr14_-_106733624 | 7.60 |
ENST00000390610.2
|
IGHV1-24
|
immunoglobulin heavy variable 1-24 |
| chr15_-_22448819 | 7.59 |
ENST00000604066.1
|
IGHV1OR15-1
|
immunoglobulin heavy variable 1/OR15-1 (non-functional) |
| chr19_-_43383789 | 7.35 |
ENST00000595356.1
|
PSG1
|
pregnancy specific beta-1-glycoprotein 1 |
| chr2_-_60780702 | 7.30 |
ENST00000359629.5
|
BCL11A
|
B-cell CLL/lymphoma 11A (zinc finger protein) |
| chr2_-_60780607 | 7.27 |
ENST00000537768.1
ENST00000335712.6 ENST00000356842.4 |
BCL11A
|
B-cell CLL/lymphoma 11A (zinc finger protein) |
| chr19_-_43383850 | 7.16 |
ENST00000436291.2
ENST00000595124.1 ENST00000244296.2 |
PSG1
|
pregnancy specific beta-1-glycoprotein 1 |
| chr11_-_68518910 | 6.80 |
ENST00000544963.1
ENST00000443940.2 |
MTL5
|
metallothionein-like 5, testis-specific (tesmin) |
| chr1_+_92417716 | 6.50 |
ENST00000402388.1
|
BRDT
|
bromodomain, testis-specific |
| chr8_+_103563792 | 6.48 |
ENST00000285402.3
|
ODF1
|
outer dense fiber of sperm tails 1 |
| chr2_-_60780536 | 6.44 |
ENST00000538214.1
|
BCL11A
|
B-cell CLL/lymphoma 11A (zinc finger protein) |
| chr19_-_43708378 | 6.33 |
ENST00000599746.1
|
PSG4
|
pregnancy specific beta-1-glycoprotein 4 |
| chr7_+_150521691 | 6.30 |
ENST00000493429.1
|
AOC1
|
amine oxidase, copper containing 1 |
| chr8_+_9046503 | 6.26 |
ENST00000512942.2
|
RP11-10A14.5
|
RP11-10A14.5 |
| chrX_-_70329118 | 6.16 |
ENST00000374188.3
|
IL2RG
|
interleukin 2 receptor, gamma |
| chr19_-_43269809 | 6.13 |
ENST00000406636.3
ENST00000404209.4 ENST00000306511.4 |
PSG8
|
pregnancy specific beta-1-glycoprotein 8 |
| chr14_-_106453155 | 6.07 |
ENST00000390594.2
|
IGHV1-2
|
immunoglobulin heavy variable 1-2 |
| chr19_-_43383819 | 5.98 |
ENST00000312439.6
ENST00000403380.3 |
PSG1
|
pregnancy specific beta-1-glycoprotein 1 |
| chr5_-_130970723 | 5.51 |
ENST00000308008.6
ENST00000296859.6 ENST00000507093.1 ENST00000510071.1 ENST00000509018.1 ENST00000307984.5 |
RAPGEF6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr6_+_31620191 | 5.44 |
ENST00000375918.2
ENST00000375920.4 |
APOM
|
apolipoprotein M |
| chr19_-_43709703 | 5.33 |
ENST00000599391.1
ENST00000244295.9 |
PSG4
|
pregnancy specific beta-1-glycoprotein 4 |
| chr2_-_25564750 | 5.31 |
ENST00000321117.5
|
DNMT3A
|
DNA (cytosine-5-)-methyltransferase 3 alpha |
| chr19_-_43690642 | 5.25 |
ENST00000407356.1
ENST00000407568.1 ENST00000404580.1 ENST00000599812.1 |
PSG5
|
pregnancy specific beta-1-glycoprotein 5 |
| chr16_+_20817761 | 5.23 |
ENST00000568046.1
ENST00000261377.6 |
AC004381.6
|
Putative RNA exonuclease NEF-sp |
| chr1_+_151739131 | 5.19 |
ENST00000400999.1
|
OAZ3
|
ornithine decarboxylase antizyme 3 |
| chr11_-_64512273 | 5.15 |
ENST00000377497.3
ENST00000377487.1 ENST00000430645.1 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr16_-_3306587 | 5.10 |
ENST00000541159.1
ENST00000536379.1 ENST00000219596.1 ENST00000339854.4 |
MEFV
|
Mediterranean fever |
| chr21_-_46330545 | 5.06 |
ENST00000320216.6
ENST00000397852.1 |
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr19_-_43709772 | 5.05 |
ENST00000596907.1
ENST00000451895.1 |
PSG4
|
pregnancy specific beta-1-glycoprotein 4 |
| chr17_-_37308824 | 5.00 |
ENST00000415163.1
ENST00000441877.1 ENST00000444911.2 |
PLXDC1
|
plexin domain containing 1 |
| chr19_-_6720686 | 4.97 |
ENST00000245907.6
|
C3
|
complement component 3 |
| chr19_-_43382142 | 4.96 |
ENST00000597058.1
|
PSG1
|
pregnancy specific beta-1-glycoprotein 1 |
| chr17_+_45810594 | 4.96 |
ENST00000177694.1
|
TBX21
|
T-box 21 |
| chr19_-_43244610 | 4.93 |
ENST00000595140.1
ENST00000327495.5 |
PSG3
|
pregnancy specific beta-1-glycoprotein 3 |
| chr11_+_113185292 | 4.89 |
ENST00000429951.1
ENST00000442859.1 ENST00000531164.1 ENST00000529850.1 ENST00000314756.3 ENST00000525965.1 |
TTC12
|
tetratricopeptide repeat domain 12 |
| chr19_-_43690674 | 4.87 |
ENST00000342951.6
ENST00000366175.3 |
PSG5
|
pregnancy specific beta-1-glycoprotein 5 |
| chr4_+_7045042 | 4.80 |
ENST00000310074.7
ENST00000512388.1 |
TADA2B
|
transcriptional adaptor 2B |
| chr13_-_114898016 | 4.75 |
ENST00000542651.1
ENST00000334062.7 |
RASA3
|
RAS p21 protein activator 3 |
| chr14_-_107095662 | 4.75 |
ENST00000390630.2
|
IGHV4-61
|
immunoglobulin heavy variable 4-61 |
| chr11_+_113185533 | 4.61 |
ENST00000393020.1
|
TTC12
|
tetratricopeptide repeat domain 12 |
| chr1_+_111415757 | 4.48 |
ENST00000429072.2
ENST00000271324.5 |
CD53
|
CD53 molecule |
| chr17_+_7531281 | 4.47 |
ENST00000575729.1
ENST00000340624.5 |
SHBG
|
sex hormone-binding globulin |
| chr22_+_40390930 | 4.44 |
ENST00000333407.6
|
FAM83F
|
family with sequence similarity 83, member F |
| chr7_+_156742399 | 4.42 |
ENST00000275820.3
|
NOM1
|
nucleolar protein with MIF4G domain 1 |
| chr8_-_145701718 | 4.39 |
ENST00000377317.4
|
FOXH1
|
forkhead box H1 |
| chr6_+_106534192 | 4.38 |
ENST00000369091.2
ENST00000369096.4 |
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr1_+_1260147 | 4.38 |
ENST00000343938.4
|
GLTPD1
|
glycolipid transfer protein domain containing 1 |
| chr14_-_106471723 | 4.32 |
ENST00000390595.2
|
IGHV1-3
|
immunoglobulin heavy variable 1-3 |
| chr4_+_668348 | 4.31 |
ENST00000511290.1
|
MYL5
|
myosin, light chain 5, regulatory |
| chr1_-_207224307 | 4.23 |
ENST00000315927.4
|
YOD1
|
YOD1 deubiquitinase |
| chr22_+_22936998 | 4.20 |
ENST00000390303.2
|
IGLV3-32
|
immunoglobulin lambda variable 3-32 (non-functional) |
| chr7_-_100844193 | 4.19 |
ENST00000440203.2
ENST00000379423.3 ENST00000223114.4 |
MOGAT3
|
monoacylglycerol O-acyltransferase 3 |
| chr6_+_129204337 | 4.18 |
ENST00000421865.2
|
LAMA2
|
laminin, alpha 2 |
| chr19_+_49999631 | 4.18 |
ENST00000270625.2
ENST00000596873.1 ENST00000594493.1 ENST00000599561.1 |
RPS11
|
ribosomal protein S11 |
| chr11_-_104480019 | 4.18 |
ENST00000536529.1
ENST00000545630.1 ENST00000538641.1 |
RP11-886D15.1
|
RP11-886D15.1 |
| chr21_+_43639211 | 4.18 |
ENST00000450121.1
ENST00000398449.3 ENST00000361802.2 |
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr19_-_43709817 | 4.13 |
ENST00000433626.2
ENST00000405312.3 |
PSG4
|
pregnancy specific beta-1-glycoprotein 4 |
| chr19_-_43773589 | 4.12 |
ENST00000291752.5
ENST00000244293.7 ENST00000596730.1 ENST00000593948.1 ENST00000270077.3 ENST00000443718.3 ENST00000418820.2 |
PSG9
|
pregnancy specific beta-1-glycoprotein 9 |
| chr2_+_69001913 | 4.11 |
ENST00000409030.3
ENST00000409220.1 |
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr19_-_43530643 | 4.04 |
ENST00000320078.7
|
PSG11
|
pregnancy specific beta-1-glycoprotein 11 |
| chr14_-_47120956 | 4.00 |
ENST00000298283.3
|
RPL10L
|
ribosomal protein L10-like |
| chr7_+_94536898 | 3.98 |
ENST00000433360.1
ENST00000340694.4 ENST00000424654.1 |
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A |
| chr11_+_113185251 | 3.93 |
ENST00000529221.1
|
TTC12
|
tetratricopeptide repeat domain 12 |
| chr5_-_41213607 | 3.92 |
ENST00000337836.5
ENST00000433294.1 |
C6
|
complement component 6 |
| chr4_+_159593271 | 3.91 |
ENST00000512251.1
ENST00000511912.1 |
ETFDH
|
electron-transferring-flavoprotein dehydrogenase |
| chr16_+_30759563 | 3.90 |
ENST00000563588.1
ENST00000565924.1 ENST00000424889.3 |
PHKG2
|
phosphorylase kinase, gamma 2 (testis) |
| chr3_-_49158218 | 3.88 |
ENST00000417901.1
ENST00000306026.5 ENST00000434032.2 |
USP19
|
ubiquitin specific peptidase 19 |
| chr1_-_20834586 | 3.85 |
ENST00000264198.3
|
MUL1
|
mitochondrial E3 ubiquitin protein ligase 1 |
| chr19_-_43422019 | 3.84 |
ENST00000402603.4
ENST00000594375.1 |
PSG6
|
pregnancy specific beta-1-glycoprotein 6 |
| chr19_-_43530600 | 3.79 |
ENST00000598133.1
ENST00000403486.1 ENST00000306322.7 ENST00000401740.1 |
PSG11
|
pregnancy specific beta-1-glycoprotein 11 |
| chr3_-_49158312 | 3.77 |
ENST00000398892.3
ENST00000453664.1 ENST00000398888.2 |
USP19
|
ubiquitin specific peptidase 19 |
| chrX_-_118827333 | 3.71 |
ENST00000360156.7
ENST00000354228.4 ENST00000489216.1 ENST00000354416.3 ENST00000394610.1 ENST00000343984.5 |
SEPT6
|
septin 6 |
| chr17_-_37309480 | 3.62 |
ENST00000539608.1
|
PLXDC1
|
plexin domain containing 1 |
| chr6_-_32339671 | 3.60 |
ENST00000442822.2
ENST00000375015.4 ENST00000533191.1 |
C6orf10
|
chromosome 6 open reading frame 10 |
| chrX_-_6453159 | 3.53 |
ENST00000381089.3
ENST00000398729.1 |
VCX3A
|
variable charge, X-linked 3A |
| chr11_-_5271122 | 3.48 |
ENST00000330597.3
|
HBG1
|
hemoglobin, gamma A |
| chr3_+_37284824 | 3.46 |
ENST00000431105.1
|
GOLGA4
|
golgin A4 |
| chr9_+_131217459 | 3.45 |
ENST00000497812.2
ENST00000393533.2 |
ODF2
|
outer dense fiber of sperm tails 2 |
| chrX_+_8432871 | 3.42 |
ENST00000381032.1
ENST00000453306.1 ENST00000444481.1 |
VCX3B
|
variable charge, X-linked 3B |
| chr20_+_43803517 | 3.41 |
ENST00000243924.3
|
PI3
|
peptidase inhibitor 3, skin-derived |
| chr9_-_130679257 | 3.39 |
ENST00000361444.3
ENST00000335791.5 ENST00000343609.2 |
ST6GALNAC4
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 |
| chr6_+_28109703 | 3.36 |
ENST00000457389.2
ENST00000330236.6 |
ZKSCAN8
|
zinc finger with KRAB and SCAN domains 8 |
| chr10_+_114710516 | 3.36 |
ENST00000542695.1
ENST00000346198.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr6_-_32339649 | 3.32 |
ENST00000527965.1
ENST00000532023.1 ENST00000447241.2 ENST00000375007.4 ENST00000534588.1 |
C6orf10
|
chromosome 6 open reading frame 10 |
| chr13_+_23755099 | 3.29 |
ENST00000537476.1
|
SGCG
|
sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein) |
| chr9_-_111619239 | 3.26 |
ENST00000374667.3
|
ACTL7B
|
actin-like 7B |
| chr2_+_68592305 | 3.25 |
ENST00000234313.7
|
PLEK
|
pleckstrin |
| chr16_+_30759700 | 3.24 |
ENST00000328273.7
|
PHKG2
|
phosphorylase kinase, gamma 2 (testis) |
| chr16_+_31128978 | 3.22 |
ENST00000448516.2
ENST00000219797.4 |
KAT8
|
K(lysine) acetyltransferase 8 |
| chr8_-_27472198 | 3.19 |
ENST00000519472.1
ENST00000523589.1 ENST00000522413.1 ENST00000523396.1 ENST00000560366.1 |
CLU
|
clusterin |
| chr2_-_220108309 | 3.18 |
ENST00000409640.1
|
GLB1L
|
galactosidase, beta 1-like |
| chr19_-_10679697 | 3.15 |
ENST00000335766.2
|
CDKN2D
|
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) |
| chr1_-_202679535 | 3.14 |
ENST00000367268.4
|
SYT2
|
synaptotagmin II |
| chr2_+_219575543 | 3.11 |
ENST00000457313.1
ENST00000415717.1 ENST00000392102.1 |
TTLL4
|
tubulin tyrosine ligase-like family, member 4 |
| chr4_+_39184024 | 3.09 |
ENST00000399820.3
ENST00000509560.1 ENST00000512112.1 ENST00000288634.7 ENST00000506503.1 |
WDR19
|
WD repeat domain 19 |
| chr1_-_207095212 | 3.08 |
ENST00000420007.2
|
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr11_-_119599794 | 3.03 |
ENST00000264025.3
|
PVRL1
|
poliovirus receptor-related 1 (herpesvirus entry mediator C) |
| chr1_+_156338993 | 3.02 |
ENST00000368249.1
ENST00000368246.2 ENST00000537040.1 ENST00000400992.2 ENST00000255013.3 ENST00000451864.2 |
RHBG
|
Rh family, B glycoprotein (gene/pseudogene) |
| chr13_+_23755054 | 3.00 |
ENST00000218867.3
|
SGCG
|
sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein) |
| chr12_+_2921788 | 2.99 |
ENST00000228799.2
ENST00000419778.2 ENST00000542548.1 |
ITFG2
|
integrin alpha FG-GAP repeat containing 2 |
| chr11_-_64511575 | 2.99 |
ENST00000431822.1
ENST00000377486.3 ENST00000394432.3 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr4_+_128702969 | 2.96 |
ENST00000508776.1
ENST00000439123.2 |
HSPA4L
|
heat shock 70kDa protein 4-like |
| chr16_+_3451184 | 2.91 |
ENST00000575752.1
ENST00000571936.1 ENST00000344823.5 |
ZNF174
|
zinc finger protein 174 |
| chr7_+_99933730 | 2.90 |
ENST00000610247.1
|
PILRB
|
paired immunoglobin-like type 2 receptor beta |
| chr1_-_226111929 | 2.89 |
ENST00000343818.6
ENST00000432920.2 |
PYCR2
RP4-559A3.7
|
pyrroline-5-carboxylate reductase family, member 2 Uncharacterized protein |
| chr3_+_46618727 | 2.88 |
ENST00000296145.5
|
TDGF1
|
teratocarcinoma-derived growth factor 1 |
| chr16_-_3450963 | 2.88 |
ENST00000573327.1
ENST00000571906.1 ENST00000573830.1 ENST00000439568.2 ENST00000422427.2 ENST00000304926.3 ENST00000396852.4 |
ZSCAN32
|
zinc finger and SCAN domain containing 32 |
| chr19_-_10679644 | 2.82 |
ENST00000393599.2
|
CDKN2D
|
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) |
| chr10_+_70480963 | 2.81 |
ENST00000265872.6
ENST00000535016.1 ENST00000538031.1 ENST00000543719.1 ENST00000539539.1 ENST00000543225.1 ENST00000536012.1 ENST00000494903.2 |
CCAR1
|
cell division cycle and apoptosis regulator 1 |
| chr1_+_176432298 | 2.78 |
ENST00000367661.3
ENST00000367662.3 |
PAPPA2
|
pappalysin 2 |
| chr19_-_344786 | 2.74 |
ENST00000264819.4
|
MIER2
|
mesoderm induction early response 1, family member 2 |
| chr11_+_118754475 | 2.71 |
ENST00000292174.4
|
CXCR5
|
chemokine (C-X-C motif) receptor 5 |
| chr7_+_102004322 | 2.70 |
ENST00000496391.1
|
PRKRIP1
|
PRKR interacting protein 1 (IL11 inducible) |
| chrY_-_21906623 | 2.69 |
ENST00000382806.2
|
KDM5D
|
lysine (K)-specific demethylase 5D |
| chr1_-_161102421 | 2.69 |
ENST00000490843.2
ENST00000368006.3 ENST00000392188.1 ENST00000545495.1 |
DEDD
|
death effector domain containing |
| chr12_-_53594227 | 2.67 |
ENST00000550743.2
|
ITGB7
|
integrin, beta 7 |
| chr19_+_49990811 | 2.67 |
ENST00000391857.4
ENST00000467825.2 |
RPL13A
|
ribosomal protein L13a |
| chr2_+_207024306 | 2.66 |
ENST00000236957.5
ENST00000392221.1 ENST00000392222.2 ENST00000445505.1 |
EEF1B2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr1_+_119957554 | 2.60 |
ENST00000543831.1
ENST00000433745.1 ENST00000369416.3 |
HSD3B2
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 |
| chr6_-_31620403 | 2.55 |
ENST00000451898.1
ENST00000439687.2 ENST00000362049.6 ENST00000424480.1 |
BAG6
|
BCL2-associated athanogene 6 |
| chr1_+_29213584 | 2.55 |
ENST00000343067.4
ENST00000356093.2 ENST00000398863.2 ENST00000373800.3 ENST00000349460.4 |
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
| chr19_-_7293942 | 2.53 |
ENST00000341500.5
ENST00000302850.5 |
INSR
|
insulin receptor |
| chr6_-_31620455 | 2.52 |
ENST00000437771.1
ENST00000404765.2 ENST00000375964.6 ENST00000211379.5 |
BAG6
|
BCL2-associated athanogene 6 |
| chr10_-_70092671 | 2.50 |
ENST00000358769.2
ENST00000432941.1 ENST00000495025.2 |
PBLD
|
phenazine biosynthesis-like protein domain containing |
| chr16_+_31085714 | 2.47 |
ENST00000300850.5
ENST00000564189.1 ENST00000428260.1 |
ZNF646
|
zinc finger protein 646 |
| chr2_-_175547571 | 2.38 |
ENST00000409415.3
ENST00000359761.3 ENST00000272746.5 |
WIPF1
|
WAS/WASL interacting protein family, member 1 |
| chr11_+_3876859 | 2.37 |
ENST00000300737.4
|
STIM1
|
stromal interaction molecule 1 |
| chr3_-_194072019 | 2.35 |
ENST00000429275.1
ENST00000323830.3 |
CPN2
|
carboxypeptidase N, polypeptide 2 |
| chrX_+_30233668 | 2.27 |
ENST00000378988.4
|
MAGEB2
|
melanoma antigen family B, 2 |
| chr11_-_133826852 | 2.26 |
ENST00000533871.2
ENST00000321016.8 |
IGSF9B
|
immunoglobulin superfamily, member 9B |
| chr11_-_108464321 | 2.26 |
ENST00000265843.4
|
EXPH5
|
exophilin 5 |
| chr3_+_137906154 | 2.26 |
ENST00000466749.1
ENST00000358441.2 ENST00000489213.1 |
ARMC8
|
armadillo repeat containing 8 |
| chr17_-_2614927 | 2.24 |
ENST00000435359.1
|
CLUH
|
clustered mitochondria (cluA/CLU1) homolog |
| chr22_+_18593507 | 2.23 |
ENST00000330423.3
|
TUBA8
|
tubulin, alpha 8 |
| chr7_-_75115548 | 2.21 |
ENST00000453279.2
|
POM121C
|
POM121 transmembrane nucleoporin C |
| chr17_-_71410794 | 2.19 |
ENST00000424778.1
|
SDK2
|
sidekick cell adhesion molecule 2 |
| chr9_-_137809718 | 2.19 |
ENST00000371806.3
|
FCN1
|
ficolin (collagen/fibrinogen domain containing) 1 |
| chr7_+_72349920 | 2.17 |
ENST00000395270.1
ENST00000446813.1 ENST00000257622.4 |
POM121
|
POM121 transmembrane nucleoporin |
| chr1_-_48866517 | 2.17 |
ENST00000371841.1
|
SPATA6
|
spermatogenesis associated 6 |
| chr3_+_121903181 | 2.17 |
ENST00000498619.1
|
CASR
|
calcium-sensing receptor |
| chr2_+_219575653 | 2.15 |
ENST00000442769.1
ENST00000424644.1 |
TTLL4
|
tubulin tyrosine ligase-like family, member 4 |
| chr7_-_87104963 | 2.14 |
ENST00000359206.3
ENST00000358400.3 ENST00000265723.4 |
ABCB4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr3_+_171561127 | 2.12 |
ENST00000334567.5
ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr7_+_142985308 | 2.11 |
ENST00000310447.5
|
CASP2
|
caspase 2, apoptosis-related cysteine peptidase |
| chr6_-_31620149 | 2.09 |
ENST00000435080.1
ENST00000375976.4 ENST00000441054.1 |
BAG6
|
BCL2-associated athanogene 6 |
| chr7_-_95225768 | 2.07 |
ENST00000005178.5
|
PDK4
|
pyruvate dehydrogenase kinase, isozyme 4 |
| chr19_+_3933085 | 2.04 |
ENST00000168977.2
ENST00000599576.1 |
NMRK2
|
nicotinamide riboside kinase 2 |
| chr12_+_9980069 | 2.02 |
ENST00000354855.3
ENST00000324214.4 ENST00000279544.3 |
KLRF1
|
killer cell lectin-like receptor subfamily F, member 1 |
| chr16_+_31366455 | 2.00 |
ENST00000268296.4
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr12_+_69186125 | 1.99 |
ENST00000399333.3
|
AC124890.1
|
HCG1774533, isoform CRA_a; PRO2268; Uncharacterized protein |
| chr9_-_21351377 | 1.98 |
ENST00000380210.1
|
IFNA6
|
interferon, alpha 6 |
| chrX_-_8139308 | 1.97 |
ENST00000317103.4
|
VCX2
|
variable charge, X-linked 2 |
| chr12_+_10366016 | 1.91 |
ENST00000546017.1
ENST00000535576.1 ENST00000539170.1 |
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr21_+_34697209 | 1.88 |
ENST00000270139.3
|
IFNAR1
|
interferon (alpha, beta and omega) receptor 1 |
| chr9_+_123970052 | 1.87 |
ENST00000373823.3
|
GSN
|
gelsolin |
| chr4_+_113152881 | 1.85 |
ENST00000274000.5
|
AP1AR
|
adaptor-related protein complex 1 associated regulatory protein |
| chr10_+_114135004 | 1.82 |
ENST00000393081.1
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5 |
| chr1_+_78511586 | 1.80 |
ENST00000370759.3
|
GIPC2
|
GIPC PDZ domain containing family, member 2 |
| chr19_-_10446449 | 1.79 |
ENST00000592439.1
|
ICAM3
|
intercellular adhesion molecule 3 |
| chr12_+_53399942 | 1.79 |
ENST00000262056.9
|
EIF4B
|
eukaryotic translation initiation factor 4B |
| chr12_+_60058458 | 1.78 |
ENST00000548610.1
|
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr1_-_161102367 | 1.76 |
ENST00000464113.1
|
DEDD
|
death effector domain containing |
| chr1_-_153517473 | 1.75 |
ENST00000368715.1
|
S100A4
|
S100 calcium binding protein A4 |
| chr11_+_65190245 | 1.75 |
ENST00000499732.1
ENST00000501122.2 ENST00000601801.1 |
NEAT1
|
nuclear paraspeckle assembly transcript 1 (non-protein coding) |
| chr19_-_5839703 | 1.74 |
ENST00000524754.1
|
FUT6
|
fucosyltransferase 6 (alpha (1,3) fucosyltransferase) |
| chr19_-_5839721 | 1.74 |
ENST00000286955.5
ENST00000318336.4 |
FUT6
|
fucosyltransferase 6 (alpha (1,3) fucosyltransferase) |
| chr21_-_37838739 | 1.73 |
ENST00000399139.1
|
CLDN14
|
claudin 14 |
| chr4_-_170948361 | 1.73 |
ENST00000393702.3
|
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr2_-_231084820 | 1.72 |
ENST00000258382.5
ENST00000338556.3 |
SP110
|
SP110 nuclear body protein |
| chr7_-_6048702 | 1.71 |
ENST00000265849.7
|
PMS2
|
PMS2 postmeiotic segregation increased 2 (S. cerevisiae) |
| chr1_-_203198790 | 1.71 |
ENST00000367229.1
ENST00000255427.3 ENST00000535569.1 |
CHIT1
|
chitinase 1 (chitotriosidase) |
| chr20_+_3052264 | 1.71 |
ENST00000217386.2
|
OXT
|
oxytocin/neurophysin I prepropeptide |
| chr15_+_64752927 | 1.70 |
ENST00000416172.1
|
ZNF609
|
zinc finger protein 609 |
| chr19_-_22193731 | 1.68 |
ENST00000601773.1
ENST00000397126.4 ENST00000601993.1 ENST00000599916.1 |
ZNF208
|
zinc finger protein 208 |
| chr11_-_18270182 | 1.67 |
ENST00000528349.1
ENST00000526900.1 ENST00000529528.1 ENST00000414546.2 ENST00000256733.4 |
SAA2
|
serum amyloid A2 |
| chr1_-_154928562 | 1.65 |
ENST00000368463.3
ENST00000539880.1 ENST00000542459.1 ENST00000368460.3 ENST00000368465.1 |
PBXIP1
|
pre-B-cell leukemia homeobox interacting protein 1 |
| chr16_-_21170762 | 1.63 |
ENST00000261383.3
ENST00000415178.1 |
DNAH3
|
dynein, axonemal, heavy chain 3 |
| chr5_+_156607829 | 1.56 |
ENST00000422843.3
|
ITK
|
IL2-inducible T-cell kinase |
| chr16_+_31366536 | 1.50 |
ENST00000562522.1
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr2_+_24163281 | 1.48 |
ENST00000309033.4
|
UBXN2A
|
UBX domain protein 2A |
| chr6_+_31637944 | 1.48 |
ENST00000375864.4
|
LY6G5B
|
lymphocyte antigen 6 complex, locus G5B |
| chr3_-_114790179 | 1.47 |
ENST00000462705.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr19_+_3933579 | 1.47 |
ENST00000593949.1
|
NMRK2
|
nicotinamide riboside kinase 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR6A1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.0 | 21.0 | GO:1903860 | negative regulation of dendrite extension(GO:1903860) regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 3.6 | 18.1 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 2.2 | 8.9 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 2.2 | 6.5 | GO:0051037 | histone displacement(GO:0001207) regulation of transcription involved in meiotic cell cycle(GO:0051037) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 1.8 | 7.2 | GO:1904379 | maintenance of unfolded protein(GO:0036506) protein localization to cytosolic proteasome complex(GO:1904327) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 1.7 | 5.1 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 1.5 | 4.4 | GO:2000824 | negative regulation of androgen receptor activity(GO:2000824) |
| 1.4 | 4.2 | GO:1990168 | protein K29-linked deubiquitination(GO:0035523) protein K33-linked deubiquitination(GO:1990168) |
| 1.4 | 4.2 | GO:0009726 | detection of endogenous stimulus(GO:0009726) |
| 1.4 | 5.4 | GO:0034445 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 1.2 | 7.1 | GO:0045819 | positive regulation of glycogen catabolic process(GO:0045819) |
| 1.1 | 18.3 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 1.1 | 2.1 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 1.1 | 6.3 | GO:0071504 | response to heparin(GO:0071503) cellular response to heparin(GO:0071504) |
| 1.0 | 10.4 | GO:0010918 | positive regulation of mitochondrial membrane potential(GO:0010918) |
| 1.0 | 5.2 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 1.0 | 3.1 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 1.0 | 3.1 | GO:0061055 | myotome development(GO:0061055) |
| 1.0 | 3.0 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 1.0 | 4.0 | GO:1904048 | regulation of spontaneous neurotransmitter secretion(GO:1904048) |
| 1.0 | 69.1 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.9 | 5.3 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.9 | 6.2 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.9 | 5.3 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.9 | 4.4 | GO:0033078 | extrathymic T cell differentiation(GO:0033078) |
| 0.8 | 2.4 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.8 | 3.8 | GO:0060339 | negative regulation of mitochondrial fusion(GO:0010637) negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.7 | 3.0 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.7 | 4.2 | GO:1902847 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.7 | 8.5 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.7 | 2.7 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.6 | 2.5 | GO:1990535 | transformation of host cell by virus(GO:0019087) neuron projection maintenance(GO:1990535) |
| 0.6 | 2.5 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.6 | 6.0 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.6 | 1.7 | GO:0002125 | maternal aggressive behavior(GO:0002125) positive regulation of female receptivity(GO:0045925) |
| 0.5 | 3.3 | GO:0033625 | positive regulation of integrin activation(GO:0033625) protein secretion by platelet(GO:0070560) |
| 0.5 | 1.5 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.4 | 1.8 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.4 | 1.3 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.4 | 4.2 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.4 | 1.2 | GO:0051106 | positive regulation of DNA ligation(GO:0051106) |
| 0.4 | 2.7 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.4 | 7.6 | GO:1900037 | negative regulation of skeletal muscle tissue development(GO:0048642) regulation of cellular response to hypoxia(GO:1900037) |
| 0.4 | 5.0 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.4 | 2.7 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.4 | 1.9 | GO:0044856 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.4 | 3.4 | GO:0048625 | maintenance of DNA repeat elements(GO:0043570) myoblast fate commitment(GO:0048625) |
| 0.4 | 1.1 | GO:0050894 | determination of affect(GO:0050894) |
| 0.4 | 2.1 | GO:0001554 | luteolysis(GO:0001554) |
| 0.4 | 1.1 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.3 | 2.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.3 | 5.5 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.3 | 4.5 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.3 | 3.5 | GO:0019317 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.3 | 75.7 | GO:0007565 | female pregnancy(GO:0007565) |
| 0.3 | 5.9 | GO:0097242 | beta-amyloid clearance(GO:0097242) |
| 0.3 | 4.2 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.3 | 1.6 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.3 | 1.0 | GO:1990418 | response to insulin-like growth factor stimulus(GO:1990418) |
| 0.2 | 1.0 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.2 | 1.7 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.2 | 1.5 | GO:0045144 | meiotic sister chromatid segregation(GO:0045144) |
| 0.2 | 17.6 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.2 | 1.0 | GO:0044821 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.2 | 3.5 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.2 | 3.4 | GO:0007620 | copulation(GO:0007620) |
| 0.2 | 1.8 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.2 | 2.4 | GO:0002566 | somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.2 | 3.2 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.2 | 4.5 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.2 | 2.5 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.2 | 2.7 | GO:0048535 | lymph node development(GO:0048535) |
| 0.2 | 0.6 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 0.2 | 4.4 | GO:0048820 | hair follicle maturation(GO:0048820) |
| 0.2 | 2.6 | GO:0006702 | androgen biosynthetic process(GO:0006702) |
| 0.2 | 1.0 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.2 | 2.9 | GO:0007351 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.2 | 0.6 | GO:0010725 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) |
| 0.2 | 2.6 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.2 | 0.9 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.2 | 0.4 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.2 | 1.9 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.2 | 0.5 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.2 | 0.7 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.2 | 2.3 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.2 | 1.7 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.2 | 0.9 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.2 | 3.5 | GO:0009435 | NAD biosynthetic process(GO:0009435) negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 3.1 | GO:1903859 | regulation of dendrite extension(GO:1903859) |
| 0.1 | 1.6 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.1 | 2.9 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.1 | 0.9 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 1.0 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.1 | 2.2 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.1 | 2.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 3.4 | GO:0097503 | sialylation(GO:0097503) |
| 0.1 | 1.8 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.1 | 0.4 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.1 | 0.6 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.1 | 0.5 | GO:0035962 | response to interleukin-13(GO:0035962) |
| 0.1 | 0.5 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.4 | GO:1903365 | regulation of fear response(GO:1903365) regulation of behavioral fear response(GO:2000822) |
| 0.1 | 2.7 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 2.1 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.1 | 2.2 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 0.3 | GO:0042361 | menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 0.1 | 1.0 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 | 4.0 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 8.6 | GO:0021510 | spinal cord development(GO:0021510) |
| 0.1 | 1.3 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.1 | 1.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.1 | 4.4 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.1 | 3.1 | GO:2000758 | positive regulation of histone acetylation(GO:0035066) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.1 | 0.8 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 0.4 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 0.9 | GO:0032754 | positive regulation of interleukin-5 production(GO:0032754) |
| 0.1 | 1.6 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.1 | 9.4 | GO:0070268 | cornification(GO:0070268) |
| 0.1 | 1.8 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.1 | 0.2 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 2.2 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.1 | 0.5 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 6.3 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 1.7 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 0.3 | GO:0045915 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.1 | 2.3 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.1 | 0.7 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.1 | 1.0 | GO:0032435 | negative regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032435) |
| 0.1 | 5.0 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.1 | 0.5 | GO:0007606 | sensory perception of chemical stimulus(GO:0007606) |
| 0.1 | 1.5 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.1 | 0.6 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.9 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 0.5 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.3 | GO:0070857 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) regulation of bile acid biosynthetic process(GO:0070857) |
| 0.0 | 0.2 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.8 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 2.2 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.6 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 | 1.5 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.6 | GO:0002507 | tolerance induction(GO:0002507) |
| 0.0 | 3.5 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 1.4 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) |
| 0.0 | 2.7 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 4.1 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 1.2 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.9 | GO:0042269 | regulation of natural killer cell mediated cytotoxicity(GO:0042269) |
| 0.0 | 1.4 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 | 8.7 | GO:0048232 | spermatogenesis(GO:0007283) male gamete generation(GO:0048232) |
| 0.0 | 0.9 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 1.7 | GO:0050918 | positive chemotaxis(GO:0050918) |
| 0.0 | 2.4 | GO:0038096 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.0 | 0.3 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.0 | 0.3 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.0 | 0.4 | GO:0097237 | cellular response to toxic substance(GO:0097237) |
| 0.0 | 1.7 | GO:0003014 | renal system process(GO:0003014) |
| 0.0 | 1.8 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 1.0 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 1.8 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.0 | 1.8 | GO:0001837 | epithelial to mesenchymal transition(GO:0001837) |
| 0.0 | 1.1 | GO:0060349 | bone morphogenesis(GO:0060349) |
| 0.0 | 0.8 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.5 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.0 | 1.0 | GO:0060041 | retina development in camera-type eye(GO:0060041) |
| 0.0 | 1.1 | GO:1990823 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.7 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.3 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 2.3 | GO:0002250 | adaptive immune response(GO:0002250) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 17.6 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 1.8 | 5.4 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 1.8 | 7.2 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 1.4 | 9.9 | GO:0001520 | outer dense fiber(GO:0001520) |
| 1.3 | 67.0 | GO:0042571 | immunoglobulin complex(GO:0019814) immunoglobulin complex, circulating(GO:0042571) |
| 1.0 | 5.1 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 1.0 | 6.0 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.9 | 7.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.8 | 15.7 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.7 | 4.4 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.7 | 6.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.7 | 2.7 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.6 | 8.5 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.6 | 4.0 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.5 | 2.2 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.5 | 7.4 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.5 | 3.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.5 | 2.5 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.5 | 1.5 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) condensed chromosome inner kinetochore(GO:0000939) |
| 0.5 | 3.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.4 | 1.3 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.4 | 3.9 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.4 | 2.4 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.4 | 2.7 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.4 | 4.4 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.4 | 2.2 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.4 | 5.3 | GO:0001741 | XY body(GO:0001741) |
| 0.4 | 1.1 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.3 | 4.8 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.3 | 2.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.3 | 2.4 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.3 | 4.8 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.3 | 2.6 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.3 | 3.7 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.3 | 2.7 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.3 | 5.6 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.2 | 3.5 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.2 | 1.3 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.2 | 2.2 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.2 | 2.3 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 8.5 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 0.7 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 5.2 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 1.4 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 1.9 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 1.0 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 16.7 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 1.6 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.1 | 4.2 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 1.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 3.1 | GO:0005770 | late endosome(GO:0005770) |
| 0.1 | 19.8 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 1.5 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 12.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 1.8 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 3.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 9.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 0.5 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 1.7 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 2.1 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 1.9 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 0.8 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 4.1 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 13.7 | GO:0099572 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.1 | 1.0 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 1.2 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.6 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.9 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.9 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 4.2 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.5 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 1.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 1.3 | GO:0030054 | cell junction(GO:0030054) |
| 0.0 | 0.9 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 1.7 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 3.2 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 5.4 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 0.8 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.4 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.7 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.0 | 7.1 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 3.5 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 7.8 | GO:0042175 | nuclear outer membrane-endoplasmic reticulum membrane network(GO:0042175) |
| 0.0 | 1.0 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.9 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 5.6 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 1.2 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.9 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 2.7 | GO:0016324 | apical plasma membrane(GO:0016324) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.2 | GO:0004913 | interleukin-4 receptor activity(GO:0004913) interleukin-7 receptor activity(GO:0004917) |
| 1.4 | 4.2 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 1.4 | 4.2 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 1.4 | 4.2 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 1.2 | 67.0 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 1.1 | 3.4 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 1.1 | 5.3 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 1.0 | 5.2 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 1.0 | 7.6 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.9 | 5.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.9 | 3.5 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.9 | 2.6 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) steroid delta-isomerase activity(GO:0004769) |
| 0.8 | 5.0 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.8 | 6.6 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.7 | 2.9 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.7 | 5.1 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.7 | 3.5 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.7 | 6.3 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.6 | 4.5 | GO:0005497 | androgen binding(GO:0005497) |
| 0.6 | 1.9 | GO:0019961 | interferon binding(GO:0019961) |
| 0.5 | 2.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.5 | 1.5 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.4 | 3.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.4 | 7.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.4 | 1.8 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.4 | 1.8 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.4 | 2.1 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.4 | 3.2 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.4 | 7.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.4 | 2.7 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.4 | 1.1 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 0.4 | 10.8 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.4 | 2.2 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.3 | 2.4 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.3 | 6.0 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.3 | 2.5 | GO:0043559 | insulin binding(GO:0043559) |
| 0.3 | 2.2 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.3 | 3.2 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.3 | 3.5 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.3 | 10.4 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.3 | 1.8 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.3 | 0.8 | GO:0017057 | glucose-6-phosphate dehydrogenase activity(GO:0004345) 6-phosphogluconolactonase activity(GO:0017057) |
| 0.2 | 2.7 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.2 | 3.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.2 | 1.7 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.2 | 4.5 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.2 | 2.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.2 | 4.8 | GO:0015278 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.2 | 4.4 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.2 | 0.6 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.2 | 0.6 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.2 | 1.9 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.2 | 3.8 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.2 | 3.4 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.2 | 6.5 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.2 | 28.5 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.2 | 1.1 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.2 | 2.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.2 | 1.3 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.2 | 0.6 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.2 | 2.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.2 | 1.9 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.2 | 1.7 | GO:0008061 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 0.2 | 4.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 4.2 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 4.0 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 2.4 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 2.9 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 0.5 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.1 | 1.2 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 0.3 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.1 | 0.7 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 3.0 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 19.2 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 1.8 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 2.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 7.1 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 1.6 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) dynein intermediate chain binding(GO:0045505) |
| 0.1 | 1.3 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.1 | 0.9 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.1 | 1.8 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.1 | 1.0 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.1 | 2.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 2.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 1.5 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 0.5 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 1.0 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.1 | 7.9 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 0.7 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 1.2 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 4.2 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 3.1 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 2.7 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.1 | 3.3 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 0.3 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.9 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 1.7 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 2.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 1.7 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 2.7 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 4.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.5 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.9 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.6 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 1.0 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.9 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.6 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 4.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.7 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 3.6 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.0 | 1.3 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.6 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.3 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 2.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.2 | GO:0004075 | biotin carboxylase activity(GO:0004075) biotin binding(GO:0009374) |
| 0.0 | 1.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 5.4 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 1.4 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 0.3 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 1.4 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 3.4 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 1.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 15.6 | GO:0004888 | transmembrane signaling receptor activity(GO:0004888) |
| 0.0 | 2.8 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.4 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 2.8 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 1.4 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.5 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.0 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.5 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.3 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.4 | GO:0008235 | metalloexopeptidase activity(GO:0008235) |
| 0.0 | 1.0 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 1.1 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 15.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 2.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.2 | 9.6 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.2 | 9.1 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 5.0 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.2 | 18.9 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.2 | 4.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 1.9 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.2 | 8.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 4.5 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 2.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 1.9 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.1 | 2.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 12.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 4.5 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.1 | 5.0 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 7.7 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 7.7 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 2.9 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 2.4 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.1 | 1.6 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.1 | 0.6 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 3.5 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 2.1 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 8.1 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.0 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 2.7 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 3.4 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.8 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.7 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.5 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.9 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 1.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.8 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.6 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.9 | PID E2F PATHWAY | E2F transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 19.9 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.4 | 6.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.3 | 8.5 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.3 | 6.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.3 | 5.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.2 | 7.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 4.5 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 5.0 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.2 | 2.5 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.2 | 4.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.2 | 9.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.2 | 2.6 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 15.8 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 3.9 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 1.1 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.1 | 2.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 2.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 1.8 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 2.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 3.1 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 1.8 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 6.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 4.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.1 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 2.2 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.1 | 1.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 4.1 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 1.0 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 2.1 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 1.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 1.0 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.7 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.5 | REACTOME SIGNALING BY FGFR MUTANTS | Genes involved in Signaling by FGFR mutants |
| 0.0 | 1.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 5.3 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 3.6 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 1.6 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.5 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.2 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 1.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 3.1 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.3 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.7 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 3.2 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 0.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |