Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
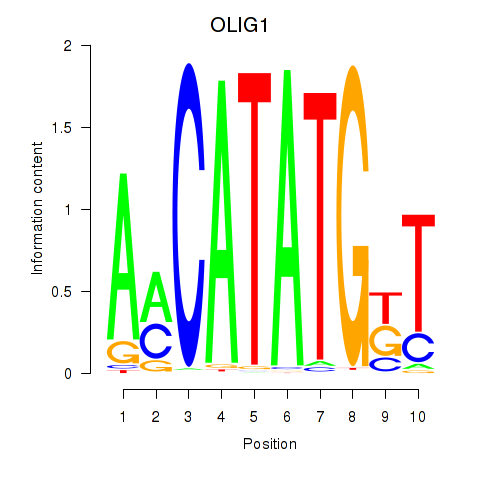
Results for OLIG1
Z-value: 0.58
Transcription factors associated with OLIG1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
OLIG1
|
ENSG00000184221.8 | oligodendrocyte transcription factor 1 |
Activity profile of OLIG1 motif
Sorted Z-values of OLIG1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_13835147 | 13.60 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr19_+_18208603 | 13.06 |
ENST00000262811.6
|
MAST3
|
microtubule associated serine/threonine kinase 3 |
| chr5_-_11588907 | 10.02 |
ENST00000513598.1
ENST00000503622.1 |
CTNND2
|
catenin (cadherin-associated protein), delta 2 |
| chr3_-_18480260 | 9.47 |
ENST00000454909.2
|
SATB1
|
SATB homeobox 1 |
| chr5_-_11589131 | 8.24 |
ENST00000511377.1
|
CTNND2
|
catenin (cadherin-associated protein), delta 2 |
| chr4_-_153456153 | 7.96 |
ENST00000603548.1
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr15_-_45670924 | 7.84 |
ENST00000396659.3
|
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr3_-_98241358 | 7.75 |
ENST00000503004.1
ENST00000506575.1 ENST00000513452.1 ENST00000515620.1 |
CLDND1
|
claudin domain containing 1 |
| chrX_-_13835461 | 7.61 |
ENST00000316715.4
ENST00000356942.5 |
GPM6B
|
glycoprotein M6B |
| chr1_+_220863187 | 7.56 |
ENST00000294889.5
|
C1orf115
|
chromosome 1 open reading frame 115 |
| chr3_-_39322728 | 7.54 |
ENST00000541347.1
ENST00000412814.1 |
CX3CR1
|
chemokine (C-X3-C motif) receptor 1 |
| chr6_+_69942298 | 7.34 |
ENST00000238918.8
|
BAI3
|
brain-specific angiogenesis inhibitor 3 |
| chr7_-_37024665 | 6.82 |
ENST00000396040.2
|
ELMO1
|
engulfment and cell motility 1 |
| chr1_+_50569575 | 6.74 |
ENST00000371827.1
|
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr2_+_54342574 | 5.99 |
ENST00000303536.4
ENST00000394666.3 |
ACYP2
|
acylphosphatase 2, muscle type |
| chr4_-_174320687 | 5.50 |
ENST00000296506.3
|
SCRG1
|
stimulator of chondrogenesis 1 |
| chr14_-_23822080 | 5.44 |
ENST00000397267.1
ENST00000354772.3 |
SLC22A17
|
solute carrier family 22, member 17 |
| chr12_-_16761007 | 5.28 |
ENST00000354662.1
ENST00000441439.2 |
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr8_+_21915368 | 5.25 |
ENST00000265800.5
ENST00000517418.1 |
DMTN
|
dematin actin binding protein |
| chr1_+_244214577 | 4.89 |
ENST00000358704.4
|
ZBTB18
|
zinc finger and BTB domain containing 18 |
| chr4_+_114214125 | 4.87 |
ENST00000509550.1
|
ANK2
|
ankyrin 2, neuronal |
| chr3_-_33700933 | 4.80 |
ENST00000480013.1
|
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr10_-_69597915 | 4.71 |
ENST00000225171.2
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr3_-_33700589 | 4.58 |
ENST00000461133.3
ENST00000496954.2 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr18_-_47813940 | 4.39 |
ENST00000586837.1
ENST00000412036.2 ENST00000589940.1 |
CXXC1
|
CXXC finger protein 1 |
| chr2_+_89986318 | 4.39 |
ENST00000491977.1
|
IGKV2D-29
|
immunoglobulin kappa variable 2D-29 |
| chr7_-_44530479 | 4.35 |
ENST00000355451.7
|
NUDCD3
|
NudC domain containing 3 |
| chr19_-_39390350 | 4.29 |
ENST00000447739.1
ENST00000358931.5 ENST00000407552.1 |
SIRT2
|
sirtuin 2 |
| chr2_-_166930131 | 4.28 |
ENST00000303395.4
ENST00000409050.1 ENST00000423058.2 ENST00000375405.3 |
SCN1A
|
sodium channel, voltage-gated, type I, alpha subunit |
| chr3_-_195310802 | 4.14 |
ENST00000421243.1
ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr14_+_24540046 | 3.93 |
ENST00000397016.2
ENST00000537691.1 ENST00000560356.1 ENST00000558450.1 |
CPNE6
|
copine VI (neuronal) |
| chr19_+_58095501 | 3.82 |
ENST00000536878.2
ENST00000597850.1 ENST00000597219.1 ENST00000598689.1 ENST00000599456.1 ENST00000307468.4 |
ZIK1
|
zinc finger protein interacting with K protein 1 |
| chr10_-_69597810 | 3.73 |
ENST00000483798.2
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr6_-_52859046 | 3.60 |
ENST00000457564.1
ENST00000541324.1 ENST00000370960.1 |
GSTA4
|
glutathione S-transferase alpha 4 |
| chr10_+_180987 | 3.35 |
ENST00000381591.1
|
ZMYND11
|
zinc finger, MYND-type containing 11 |
| chr3_+_62304648 | 3.26 |
ENST00000462069.1
ENST00000232519.5 ENST00000465142.1 |
C3orf14
|
chromosome 3 open reading frame 14 |
| chr20_-_32031680 | 3.16 |
ENST00000217381.2
|
SNTA1
|
syntrophin, alpha 1 |
| chr2_+_166428839 | 3.12 |
ENST00000342316.4
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3 |
| chr7_+_110731062 | 3.03 |
ENST00000308478.5
ENST00000451085.1 ENST00000422987.3 ENST00000421101.1 |
LRRN3
|
leucine rich repeat neuronal 3 |
| chr6_+_31553978 | 2.99 |
ENST00000376096.1
ENST00000376099.1 ENST00000376110.3 |
LST1
|
leukocyte specific transcript 1 |
| chr5_+_140739537 | 2.84 |
ENST00000522605.1
|
PCDHGB2
|
protocadherin gamma subfamily B, 2 |
| chr19_+_12035913 | 2.82 |
ENST00000591944.1
|
ZNF763
|
Uncharacterized protein; Zinc finger protein 763 |
| chr2_+_191792376 | 2.76 |
ENST00000409428.1
ENST00000409215.1 |
GLS
|
glutaminase |
| chr6_+_31553901 | 2.70 |
ENST00000418507.2
ENST00000438075.2 ENST00000376100.3 ENST00000376111.4 |
LST1
|
leukocyte specific transcript 1 |
| chr3_-_121379739 | 2.68 |
ENST00000428394.2
ENST00000314583.3 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr10_-_15902449 | 2.68 |
ENST00000277632.3
|
FAM188A
|
family with sequence similarity 188, member A |
| chr4_-_120243545 | 2.65 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr19_-_51472222 | 2.60 |
ENST00000376851.3
|
KLK6
|
kallikrein-related peptidase 6 |
| chr2_+_172543967 | 2.47 |
ENST00000534253.2
ENST00000263811.4 ENST00000397119.3 ENST00000410079.3 ENST00000438879.1 |
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr1_-_52831796 | 2.46 |
ENST00000284376.3
ENST00000438831.1 ENST00000371586.2 |
CC2D1B
|
coiled-coil and C2 domain containing 1B |
| chrX_+_120181457 | 2.43 |
ENST00000328078.1
|
GLUD2
|
glutamate dehydrogenase 2 |
| chr2_+_172544011 | 2.34 |
ENST00000508530.1
|
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr19_-_51472823 | 2.33 |
ENST00000310157.2
|
KLK6
|
kallikrein-related peptidase 6 |
| chr20_+_48429233 | 2.32 |
ENST00000417961.1
|
SLC9A8
|
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr8_-_145701718 | 2.24 |
ENST00000377317.4
|
FOXH1
|
forkhead box H1 |
| chr3_-_49466686 | 2.24 |
ENST00000273598.3
ENST00000436744.2 |
NICN1
|
nicolin 1 |
| chr5_-_115872142 | 2.23 |
ENST00000510263.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr19_-_39368887 | 2.16 |
ENST00000340740.3
ENST00000591812.1 |
RINL
|
Ras and Rab interactor-like |
| chr4_+_71600144 | 2.15 |
ENST00000502653.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr1_-_21377383 | 2.15 |
ENST00000374935.3
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr12_-_123450986 | 2.14 |
ENST00000344275.7
ENST00000442833.2 ENST00000280560.8 ENST00000540285.1 ENST00000346530.5 |
ABCB9
|
ATP-binding cassette, sub-family B (MDR/TAP), member 9 |
| chr3_+_42897512 | 2.14 |
ENST00000493193.1
|
ACKR2
|
atypical chemokine receptor 2 |
| chr10_+_51565108 | 2.10 |
ENST00000438493.1
ENST00000452682.1 |
NCOA4
|
nuclear receptor coactivator 4 |
| chr1_+_104293028 | 2.07 |
ENST00000370079.3
|
AMY1C
|
amylase, alpha 1C (salivary) |
| chr6_+_73076432 | 1.98 |
ENST00000414192.2
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr11_+_65405556 | 1.96 |
ENST00000534313.1
ENST00000533361.1 ENST00000526137.1 |
SIPA1
|
signal-induced proliferation-associated 1 |
| chr2_-_183106641 | 1.94 |
ENST00000346717.4
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr4_+_160188889 | 1.94 |
ENST00000264431.4
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr2_-_70418032 | 1.91 |
ENST00000425268.1
ENST00000428751.1 ENST00000417203.1 ENST00000417865.1 ENST00000428010.1 ENST00000447804.1 ENST00000264434.2 |
C2orf42
|
chromosome 2 open reading frame 42 |
| chr19_-_1021113 | 1.90 |
ENST00000333175.5
ENST00000356663.3 |
TMEM259
|
transmembrane protein 259 |
| chr18_-_49557 | 1.89 |
ENST00000308911.6
|
RP11-683L23.1
|
Tubulin beta-8 chain-like protein LOC260334 |
| chr12_+_6898638 | 1.82 |
ENST00000011653.4
|
CD4
|
CD4 molecule |
| chr2_+_210444748 | 1.82 |
ENST00000392194.1
|
MAP2
|
microtubule-associated protein 2 |
| chr2_+_172543919 | 1.81 |
ENST00000452242.1
ENST00000340296.4 |
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr19_-_54876558 | 1.80 |
ENST00000391742.2
ENST00000434277.2 |
LAIR1
|
leukocyte-associated immunoglobulin-like receptor 1 |
| chr2_-_183387064 | 1.80 |
ENST00000536095.1
ENST00000331935.6 ENST00000358139.2 ENST00000456212.1 |
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr14_+_69865401 | 1.80 |
ENST00000556605.1
ENST00000336643.5 ENST00000031146.4 |
SLC39A9
|
solute carrier family 39, member 9 |
| chr17_-_7145475 | 1.79 |
ENST00000571129.1
ENST00000571253.1 ENST00000573928.1 |
GABARAP
|
GABA(A) receptor-associated protein |
| chr10_+_180405 | 1.78 |
ENST00000439456.1
ENST00000397962.3 ENST00000309776.4 ENST00000381602.4 |
ZMYND11
|
zinc finger, MYND-type containing 11 |
| chr14_+_95047725 | 1.76 |
ENST00000554760.1
ENST00000554866.1 ENST00000329597.7 ENST00000556775.1 |
SERPINA5
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5 |
| chr2_-_183387283 | 1.74 |
ENST00000435564.1
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr1_-_111970353 | 1.71 |
ENST00000369732.3
|
OVGP1
|
oviductal glycoprotein 1, 120kDa |
| chr1_+_196743943 | 1.69 |
ENST00000471440.2
ENST00000391985.3 |
CFHR3
|
complement factor H-related 3 |
| chr5_-_171433579 | 1.68 |
ENST00000265094.5
ENST00000393802.2 |
FBXW11
|
F-box and WD repeat domain containing 11 |
| chr14_+_95047744 | 1.67 |
ENST00000553511.1
ENST00000554633.1 ENST00000555681.1 ENST00000554276.1 |
SERPINA5
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5 |
| chr4_-_121843985 | 1.64 |
ENST00000264808.3
ENST00000428209.2 ENST00000515109.1 ENST00000394435.2 |
PRDM5
|
PR domain containing 5 |
| chr5_-_111312622 | 1.62 |
ENST00000395634.3
|
NREP
|
neuronal regeneration related protein |
| chr1_+_24969755 | 1.60 |
ENST00000447431.2
ENST00000374389.4 |
SRRM1
|
serine/arginine repetitive matrix 1 |
| chr17_+_11924129 | 1.59 |
ENST00000353533.5
ENST00000415385.3 |
MAP2K4
|
mitogen-activated protein kinase kinase 4 |
| chr2_-_152830441 | 1.59 |
ENST00000534999.1
ENST00000397327.2 |
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr11_-_44972476 | 1.57 |
ENST00000527685.1
ENST00000308212.5 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr4_+_154622652 | 1.56 |
ENST00000260010.6
|
TLR2
|
toll-like receptor 2 |
| chr10_+_26727125 | 1.54 |
ENST00000376236.4
|
APBB1IP
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr15_+_85144217 | 1.54 |
ENST00000540936.1
ENST00000448803.2 ENST00000546275.1 ENST00000546148.1 ENST00000442073.3 ENST00000334141.3 ENST00000358472.3 ENST00000502939.2 ENST00000379358.3 ENST00000327179.6 |
ZSCAN2
|
zinc finger and SCAN domain containing 2 |
| chr3_-_48936272 | 1.53 |
ENST00000544097.1
ENST00000430379.1 ENST00000319017.4 |
SLC25A20
|
solute carrier family 25 (carnitine/acylcarnitine translocase), member 20 |
| chr1_-_21377447 | 1.49 |
ENST00000374937.3
ENST00000264211.8 |
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr2_+_130737223 | 1.49 |
ENST00000410061.2
|
RAB6C
|
RAB6C, member RAS oncogene family |
| chr14_+_21249200 | 1.49 |
ENST00000304677.2
|
RNASE6
|
ribonuclease, RNase A family, k6 |
| chr7_-_55930443 | 1.49 |
ENST00000388975.3
|
SEPT14
|
septin 14 |
| chr13_+_49551020 | 1.47 |
ENST00000541916.1
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr6_+_17110726 | 1.39 |
ENST00000354384.5
|
STMND1
|
stathmin domain containing 1 |
| chr9_-_132404374 | 1.39 |
ENST00000277459.4
ENST00000450050.2 ENST00000277458.4 |
ASB6
|
ankyrin repeat and SOCS box containing 6 |
| chr21_+_42792442 | 1.39 |
ENST00000398600.2
|
MX1
|
myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) |
| chr4_+_154178520 | 1.38 |
ENST00000433687.1
|
TRIM2
|
tripartite motif containing 2 |
| chr1_-_146054494 | 1.35 |
ENST00000401009.2
|
NBPF11
|
neuroblastoma breakpoint family, member 11 |
| chr2_-_175462934 | 1.35 |
ENST00000392546.2
ENST00000436221.1 |
WIPF1
|
WAS/WASL interacting protein family, member 1 |
| chr1_+_159272111 | 1.34 |
ENST00000368114.1
|
FCER1A
|
Fc fragment of IgE, high affinity I, receptor for; alpha polypeptide |
| chr8_+_48173514 | 1.32 |
ENST00000518074.1
ENST00000541342.1 |
SPIDR
|
scaffolding protein involved in DNA repair |
| chr8_-_102181718 | 1.29 |
ENST00000565617.1
|
KB-1460A1.5
|
KB-1460A1.5 |
| chr5_-_169725231 | 1.29 |
ENST00000046794.5
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
| chr9_-_125675576 | 1.28 |
ENST00000373659.3
|
ZBTB6
|
zinc finger and BTB domain containing 6 |
| chr8_-_23315190 | 1.27 |
ENST00000356206.6
ENST00000358689.4 ENST00000417069.2 |
ENTPD4
|
ectonucleoside triphosphate diphosphohydrolase 4 |
| chr10_+_51565188 | 1.27 |
ENST00000430396.2
ENST00000374087.4 ENST00000414907.2 |
NCOA4
|
nuclear receptor coactivator 4 |
| chr7_+_4721885 | 1.24 |
ENST00000328914.4
|
FOXK1
|
forkhead box K1 |
| chr22_-_42526802 | 1.20 |
ENST00000359033.4
ENST00000389970.3 ENST00000360608.5 |
CYP2D6
|
cytochrome P450, family 2, subfamily D, polypeptide 6 |
| chr4_-_76912070 | 1.18 |
ENST00000395711.4
ENST00000356260.5 |
SDAD1
|
SDA1 domain containing 1 |
| chr18_-_50240 | 1.17 |
ENST00000573909.1
|
RP11-683L23.1
|
Tubulin beta-8 chain-like protein LOC260334 |
| chr5_+_156607829 | 1.11 |
ENST00000422843.3
|
ITK
|
IL2-inducible T-cell kinase |
| chr5_-_171433819 | 1.11 |
ENST00000296933.6
|
FBXW11
|
F-box and WD repeat domain containing 11 |
| chr5_-_131132658 | 1.11 |
ENST00000514667.1
ENST00000511848.1 ENST00000510461.1 |
CTC-432M15.3
FNIP1
|
Folliculin-interacting protein 1 folliculin interacting protein 1 |
| chr1_+_203274639 | 1.08 |
ENST00000290551.4
|
BTG2
|
BTG family, member 2 |
| chr6_+_292051 | 1.07 |
ENST00000344450.5
|
DUSP22
|
dual specificity phosphatase 22 |
| chr14_+_88471468 | 1.06 |
ENST00000267549.3
|
GPR65
|
G protein-coupled receptor 65 |
| chr11_+_118175596 | 1.04 |
ENST00000528600.1
|
CD3E
|
CD3e molecule, epsilon (CD3-TCR complex) |
| chr22_+_37678424 | 1.03 |
ENST00000248901.6
|
CYTH4
|
cytohesin 4 |
| chr8_+_119294456 | 1.03 |
ENST00000366457.2
|
AC023590.1
|
Uncharacterized protein |
| chr12_+_12223867 | 1.02 |
ENST00000308721.5
|
BCL2L14
|
BCL2-like 14 (apoptosis facilitator) |
| chr2_-_183387430 | 1.01 |
ENST00000410103.1
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr2_-_158345462 | 1.00 |
ENST00000439355.1
ENST00000540637.1 |
CYTIP
|
cytohesin 1 interacting protein |
| chr1_+_158149737 | 0.98 |
ENST00000368171.3
|
CD1D
|
CD1d molecule |
| chr11_-_62359027 | 0.98 |
ENST00000494385.1
ENST00000308436.7 |
TUT1
|
terminal uridylyl transferase 1, U6 snRNA-specific |
| chr3_+_113251143 | 0.94 |
ENST00000264852.4
ENST00000393830.3 |
SIDT1
|
SID1 transmembrane family, member 1 |
| chr15_-_58306295 | 0.91 |
ENST00000559517.1
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr4_-_159094194 | 0.89 |
ENST00000592057.1
ENST00000585682.1 ENST00000393807.5 |
FAM198B
|
family with sequence similarity 198, member B |
| chr11_-_44972418 | 0.84 |
ENST00000525680.1
ENST00000528290.1 ENST00000530035.1 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr15_-_89755034 | 0.83 |
ENST00000563254.1
|
RLBP1
|
retinaldehyde binding protein 1 |
| chr16_-_20364122 | 0.81 |
ENST00000396138.4
ENST00000577168.1 |
UMOD
|
uromodulin |
| chr1_+_104198377 | 0.81 |
ENST00000370083.4
|
AMY1A
|
amylase, alpha 1A (salivary) |
| chrX_+_101975643 | 0.80 |
ENST00000361229.4
|
BHLHB9
|
basic helix-loop-helix domain containing, class B, 9 |
| chr1_+_17559776 | 0.80 |
ENST00000537499.1
ENST00000413717.2 ENST00000536552.1 |
PADI1
|
peptidyl arginine deiminase, type I |
| chrX_+_101975619 | 0.79 |
ENST00000457056.1
|
BHLHB9
|
basic helix-loop-helix domain containing, class B, 9 |
| chr4_+_2470664 | 0.76 |
ENST00000314289.8
ENST00000541204.1 ENST00000502316.1 ENST00000507247.1 ENST00000509258.1 ENST00000511859.1 |
RNF4
|
ring finger protein 4 |
| chr5_+_118812237 | 0.76 |
ENST00000513628.1
|
HSD17B4
|
hydroxysteroid (17-beta) dehydrogenase 4 |
| chr11_+_118175132 | 0.75 |
ENST00000361763.4
|
CD3E
|
CD3e molecule, epsilon (CD3-TCR complex) |
| chr19_+_45542773 | 0.75 |
ENST00000544944.2
|
CLASRP
|
CLK4-associating serine/arginine rich protein |
| chr20_+_57226841 | 0.73 |
ENST00000358029.4
ENST00000361830.3 |
STX16
|
syntaxin 16 |
| chr22_+_47070490 | 0.73 |
ENST00000408031.1
|
GRAMD4
|
GRAM domain containing 4 |
| chr6_+_135502501 | 0.73 |
ENST00000527615.1
ENST00000420123.2 ENST00000525369.1 ENST00000528774.1 ENST00000534121.1 ENST00000534044.1 ENST00000533624.1 |
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr4_-_89744365 | 0.72 |
ENST00000513837.1
ENST00000503556.1 |
FAM13A
|
family with sequence similarity 13, member A |
| chr19_+_55417499 | 0.72 |
ENST00000291890.4
ENST00000447255.1 ENST00000598576.1 ENST00000594765.1 |
NCR1
|
natural cytotoxicity triggering receptor 1 |
| chr17_-_7017559 | 0.71 |
ENST00000446679.2
|
ASGR2
|
asialoglycoprotein receptor 2 |
| chr1_+_12524965 | 0.70 |
ENST00000471923.1
|
VPS13D
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr11_+_89764274 | 0.70 |
ENST00000448984.1
ENST00000432771.1 |
TRIM49C
|
tripartite motif containing 49C |
| chr7_+_140396756 | 0.70 |
ENST00000460088.1
ENST00000472695.1 |
NDUFB2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa |
| chrX_+_107288239 | 0.70 |
ENST00000217957.5
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr18_-_44561988 | 0.69 |
ENST00000332567.4
|
TCEB3B
|
transcription elongation factor B polypeptide 3B (elongin A2) |
| chr15_-_50557863 | 0.68 |
ENST00000543581.1
|
HDC
|
histidine decarboxylase |
| chr19_+_39390320 | 0.64 |
ENST00000576510.1
|
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr2_+_241807870 | 0.63 |
ENST00000307503.3
|
AGXT
|
alanine-glyoxylate aminotransferase |
| chr15_-_80189380 | 0.63 |
ENST00000258874.3
|
MTHFS
|
5,10-methenyltetrahydrofolate synthetase (5-formyltetrahydrofolate cyclo-ligase) |
| chrX_+_76709648 | 0.62 |
ENST00000439435.1
|
FGF16
|
fibroblast growth factor 16 |
| chr11_-_134123142 | 0.61 |
ENST00000392595.2
ENST00000341541.3 ENST00000352327.5 ENST00000392594.3 |
THYN1
|
thymocyte nuclear protein 1 |
| chr8_-_38239732 | 0.61 |
ENST00000534155.1
ENST00000433384.2 ENST00000317025.8 ENST00000316985.3 |
WHSC1L1
|
Wolf-Hirschhorn syndrome candidate 1-like 1 |
| chr16_-_4896205 | 0.60 |
ENST00000589389.1
|
GLYR1
|
glyoxylate reductase 1 homolog (Arabidopsis) |
| chr4_+_88720698 | 0.59 |
ENST00000226284.5
|
IBSP
|
integrin-binding sialoprotein |
| chrX_+_107288197 | 0.59 |
ENST00000415430.3
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr3_-_46069223 | 0.58 |
ENST00000309285.3
|
XCR1
|
chemokine (C motif) receptor 1 |
| chr10_-_5446786 | 0.58 |
ENST00000479328.1
ENST00000380419.3 |
TUBAL3
|
tubulin, alpha-like 3 |
| chr6_-_31648150 | 0.57 |
ENST00000375858.3
ENST00000383237.4 |
LY6G5C
|
lymphocyte antigen 6 complex, locus G5C |
| chr17_-_34344991 | 0.56 |
ENST00000591423.1
|
CCL23
|
chemokine (C-C motif) ligand 23 |
| chr4_-_89744314 | 0.56 |
ENST00000508369.1
|
FAM13A
|
family with sequence similarity 13, member A |
| chr1_+_146714291 | 0.55 |
ENST00000431239.1
ENST00000369259.3 ENST00000369258.4 ENST00000361293.5 |
CHD1L
|
chromodomain helicase DNA binding protein 1-like |
| chr12_-_40013553 | 0.55 |
ENST00000308666.3
|
ABCD2
|
ATP-binding cassette, sub-family D (ALD), member 2 |
| chr6_-_25874440 | 0.54 |
ENST00000361703.6
ENST00000397060.4 |
SLC17A3
|
solute carrier family 17 (organic anion transporter), member 3 |
| chr3_-_46608010 | 0.54 |
ENST00000395905.3
|
LRRC2
|
leucine rich repeat containing 2 |
| chr2_+_177053307 | 0.53 |
ENST00000331462.4
|
HOXD1
|
homeobox D1 |
| chr3_-_101232019 | 0.53 |
ENST00000394095.2
ENST00000394091.1 ENST00000394094.2 ENST00000358203.3 ENST00000348610.3 ENST00000314261.7 |
SENP7
|
SUMO1/sentrin specific peptidase 7 |
| chr2_-_97760576 | 0.52 |
ENST00000414820.1
ENST00000272610.3 |
FAHD2B
|
fumarylacetoacetate hydrolase domain containing 2B |
| chr12_+_4699244 | 0.50 |
ENST00000540757.2
|
DYRK4
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr3_+_122103014 | 0.50 |
ENST00000232125.5
ENST00000477892.1 ENST00000469967.1 |
FAM162A
|
family with sequence similarity 162, member A |
| chr1_-_8000872 | 0.49 |
ENST00000377507.3
|
TNFRSF9
|
tumor necrosis factor receptor superfamily, member 9 |
| chr1_-_160549235 | 0.49 |
ENST00000368054.3
ENST00000368048.3 ENST00000311224.4 ENST00000368051.3 ENST00000534968.1 |
CD84
|
CD84 molecule |
| chr1_+_196743912 | 0.48 |
ENST00000367425.4
|
CFHR3
|
complement factor H-related 3 |
| chr18_+_32173276 | 0.47 |
ENST00000591816.1
ENST00000588125.1 ENST00000598334.1 ENST00000588684.1 ENST00000554864.3 ENST00000399121.5 ENST00000595022.1 ENST00000269190.7 ENST00000399097.3 |
DTNA
|
dystrobrevin, alpha |
| chr3_+_119298429 | 0.47 |
ENST00000478927.1
|
ADPRH
|
ADP-ribosylarginine hydrolase |
| chr5_+_118812294 | 0.47 |
ENST00000509514.1
|
HSD17B4
|
hydroxysteroid (17-beta) dehydrogenase 4 |
| chr22_+_24999114 | 0.46 |
ENST00000412658.1
ENST00000445029.1 ENST00000419133.1 ENST00000400382.1 ENST00000438643.2 ENST00000452551.1 ENST00000400383.1 ENST00000412898.1 ENST00000400380.1 ENST00000455483.1 ENST00000430289.1 |
GGT1
|
gamma-glutamyltransferase 1 |
| chr3_-_53916202 | 0.45 |
ENST00000335754.3
|
ACTR8
|
ARP8 actin-related protein 8 homolog (yeast) |
| chr2_-_79313973 | 0.45 |
ENST00000454188.1
|
REG1B
|
regenerating islet-derived 1 beta |
| chr19_-_37019562 | 0.45 |
ENST00000523638.1
|
ZNF260
|
zinc finger protein 260 |
| chr11_+_59824060 | 0.43 |
ENST00000395032.2
ENST00000358152.2 |
MS4A3
|
membrane-spanning 4-domains, subfamily A, member 3 (hematopoietic cell-specific) |
| chr19_-_20150252 | 0.42 |
ENST00000595736.1
ENST00000593468.1 ENST00000596019.1 |
ZNF682
|
zinc finger protein 682 |
| chr6_+_163148161 | 0.42 |
ENST00000337019.3
ENST00000366889.2 |
PACRG
|
PARK2 co-regulated |
| chr19_-_20150301 | 0.41 |
ENST00000397165.2
|
ZNF682
|
zinc finger protein 682 |
| chr7_+_64838786 | 0.40 |
ENST00000450302.2
|
ZNF92
|
zinc finger protein 92 |
| chr5_-_75919253 | 0.38 |
ENST00000296641.4
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2 |
| chr11_-_88796803 | 0.36 |
ENST00000418177.2
ENST00000455756.2 |
GRM5
|
glutamate receptor, metabotropic 5 |
| chr5_+_180336564 | 0.34 |
ENST00000505126.1
ENST00000533815.2 |
BTNL8
|
butyrophilin-like 8 |
| chr16_-_20364030 | 0.34 |
ENST00000396134.2
ENST00000573567.1 ENST00000570757.1 ENST00000424589.1 ENST00000302509.4 ENST00000571174.1 ENST00000576688.1 |
UMOD
|
uromodulin |
| chr3_+_118905564 | 0.33 |
ENST00000460625.1
|
UPK1B
|
uroplakin 1B |
Network of associatons between targets according to the STRING database.
First level regulatory network of OLIG1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.5 | GO:0002881 | negative regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002881) |
| 2.1 | 21.2 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 1.6 | 4.9 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 1.6 | 8.0 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 1.4 | 4.3 | GO:2000777 | positive regulation of oocyte maturation(GO:1900195) negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 1.4 | 4.1 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 1.2 | 9.4 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 1.1 | 5.4 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.9 | 5.3 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.9 | 5.3 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.7 | 2.2 | GO:2000824 | negative regulation of androgen receptor activity(GO:2000824) |
| 0.7 | 5.2 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.7 | 7.8 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.7 | 3.4 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.6 | 1.9 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.6 | 6.6 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.6 | 9.4 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.6 | 1.7 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.5 | 1.6 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.5 | 1.6 | GO:0032289 | central nervous system myelin formation(GO:0032289) detection of bacterial lipoprotein(GO:0042494) detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.4 | 1.3 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) |
| 0.4 | 2.0 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.4 | 4.3 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.4 | 6.5 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.4 | 1.8 | GO:0035723 | interleukin-15-mediated signaling pathway(GO:0035723) cellular response to interleukin-15(GO:0071350) |
| 0.4 | 1.8 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.4 | 2.1 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.3 | 1.2 | GO:0033076 | alkaloid catabolic process(GO:0009822) isoquinoline alkaloid metabolic process(GO:0033076) cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 0.3 | 1.1 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.3 | 19.9 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.2 | 0.5 | GO:2001183 | negative regulation of interleukin-10 secretion(GO:2001180) negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.2 | 4.9 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.2 | 0.7 | GO:1990922 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.2 | 0.7 | GO:0001694 | histamine biosynthetic process(GO:0001694) |
| 0.2 | 2.2 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.2 | 1.3 | GO:0072757 | cellular response to camptothecin(GO:0072757) |
| 0.2 | 1.3 | GO:0009191 | ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.2 | 0.6 | GO:0009078 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) |
| 0.2 | 1.2 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 0.2 | 1.4 | GO:0003374 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 0.2 | 3.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.2 | 1.0 | GO:0016045 | detection of bacterium(GO:0016045) |
| 0.2 | 0.8 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.2 | 6.8 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.2 | 5.9 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.2 | 5.1 | GO:0034243 | regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.2 | 0.5 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 0.2 | 0.8 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.2 | 2.7 | GO:0045651 | positive regulation of granulocyte differentiation(GO:0030854) positive regulation of macrophage differentiation(GO:0045651) positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.2 | 0.6 | GO:0015942 | folic acid-containing compound catabolic process(GO:0009397) formate metabolic process(GO:0015942) pteridine-containing compound catabolic process(GO:0042560) |
| 0.2 | 0.5 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 1.2 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.1 | 1.7 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.1 | 2.3 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.4 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.1 | 2.8 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.1 | 1.1 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.1 | 1.5 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.1 | 3.6 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.1 | 1.5 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.1 | 1.6 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 3.4 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 1.0 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.6 | GO:2000546 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.1 | 0.9 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.1 | 1.9 | GO:1904294 | positive regulation of ERAD pathway(GO:1904294) |
| 0.1 | 2.2 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.1 | 0.4 | GO:0072023 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.1 | 0.2 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.1 | 3.6 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.1 | 0.5 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.1 | 0.5 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.2 | GO:0019474 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.1 | 1.5 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.1 | 1.2 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 1.8 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.1 | 1.6 | GO:0051567 | histone H3-K9 methylation(GO:0051567) |
| 0.1 | 1.1 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.1 | 0.5 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.1 | 1.1 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.1 | 0.2 | GO:0018153 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.1 | 1.0 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 2.7 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 1.6 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 | 3.6 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 3.0 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.6 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.7 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.2 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 3.1 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 3.6 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 1.5 | GO:0010824 | regulation of centrosome duplication(GO:0010824) |
| 0.0 | 0.6 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 1.6 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 3.0 | GO:0050672 | negative regulation of mononuclear cell proliferation(GO:0032945) negative regulation of lymphocyte proliferation(GO:0050672) |
| 0.0 | 0.2 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.3 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.0 | 1.8 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 10.5 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 1.2 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 1.5 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 1.4 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 | 0.5 | GO:0018108 | peptidyl-tyrosine phosphorylation(GO:0018108) |
| 0.0 | 0.2 | GO:0030213 | negative regulation of fibroblast migration(GO:0010764) hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.6 | GO:0006295 | nucleotide-excision repair, preincision complex stabilization(GO:0006293) nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 1.6 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 1.0 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 1.3 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.5 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 0.5 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 4.3 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 0.7 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 1.0 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.3 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.5 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.4 | GO:0097180 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 1.1 | 8.0 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.9 | 9.4 | GO:0045180 | basal cortex(GO:0045180) |
| 0.6 | 6.8 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.5 | 1.6 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.5 | 4.3 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.4 | 2.2 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.4 | 11.0 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.4 | 3.2 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.3 | 1.3 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) |
| 0.3 | 5.3 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.3 | 1.8 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.3 | 3.6 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.3 | 7.5 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.3 | 1.3 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.3 | 4.3 | GO:0043194 | voltage-gated sodium channel complex(GO:0001518) axon initial segment(GO:0043194) |
| 0.2 | 3.4 | GO:0044754 | autolysosome(GO:0044754) |
| 0.2 | 9.5 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.2 | 25.9 | GO:0043204 | perikaryon(GO:0043204) |
| 0.2 | 3.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.2 | 1.8 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.2 | 3.4 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 2.2 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 4.9 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 7.8 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 1.0 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 2.2 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.1 | 0.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 2.7 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 5.5 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.0 | 2.0 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 1.0 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.2 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 1.4 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 10.5 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 9.0 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.5 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 1.8 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 2.5 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.0 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.7 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 3.1 | GO:0031301 | integral component of organelle membrane(GO:0031301) |
| 0.0 | 0.8 | GO:0044297 | cell body(GO:0044297) |
| 0.0 | 1.6 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 1.6 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.9 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 9.9 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.4 | GO:0042734 | presynaptic membrane(GO:0042734) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.5 | GO:0016495 | C-X3-C chemokine receptor activity(GO:0016495) |
| 1.5 | 4.4 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 1.4 | 4.3 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 1.0 | 6.0 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 1.0 | 8.0 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.9 | 6.5 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.6 | 2.4 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.6 | 2.8 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.5 | 2.1 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.5 | 1.5 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.5 | 11.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.4 | 1.9 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.4 | 3.4 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.4 | 1.8 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.3 | 8.5 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.3 | 6.6 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.3 | 0.6 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.3 | 1.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.3 | 0.8 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.3 | 1.3 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.2 | 1.0 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.2 | 2.3 | GO:0015386 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.2 | 10.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 3.2 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.2 | 0.8 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.2 | 1.2 | GO:0033989 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.2 | 0.8 | GO:0016160 | amylase activity(GO:0016160) |
| 0.2 | 0.6 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.2 | 4.3 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.2 | 1.6 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.2 | 1.3 | GO:0019863 | immunoglobulin receptor activity(GO:0019763) IgE binding(GO:0019863) |
| 0.2 | 3.6 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.2 | 2.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.2 | 0.8 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.2 | 0.9 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.2 | 18.2 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.2 | 1.7 | GO:0004568 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 0.1 | 1.8 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.1 | 4.2 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 1.0 | GO:0030884 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.1 | 1.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 0.7 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.1 | 0.5 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 1.8 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 4.1 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 0.9 | GO:0016918 | retinal binding(GO:0016918) |
| 0.1 | 3.9 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 0.6 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 1.6 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.5 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 5.6 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 0.5 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 1.6 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 1.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 3.3 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 1.2 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 0.7 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 4.3 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 0.3 | GO:0004771 | sterol esterase activity(GO:0004771) methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.1 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 9.5 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 2.7 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 13.0 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.4 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 1.0 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 10.0 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.9 | GO:0004950 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.0 | 0.2 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.3 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.5 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.2 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.2 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.0 | 0.6 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 1.0 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 2.5 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.4 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.5 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 1.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.7 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 0.6 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 1.0 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 1.0 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 3.9 | GO:0030695 | GTPase regulator activity(GO:0030695) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 7.2 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 6.8 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 2.9 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 4.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 9.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 6.5 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 4.9 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 3.6 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 2.8 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 1.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.5 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 3.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 4.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.7 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 1.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.6 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.1 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.3 | 2.9 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.2 | 8.0 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.2 | 4.7 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.2 | 1.6 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.1 | 8.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 5.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 5.9 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.1 | 4.1 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 9.5 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 1.8 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 2.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 1.5 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 2.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 2.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 2.4 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 1.6 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.1 | 6.6 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 1.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 4.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 1.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 2.0 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 1.5 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 5.0 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 1.6 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 0.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.6 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.0 | 1.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 2.4 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 4.1 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.5 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 1.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.7 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |