Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
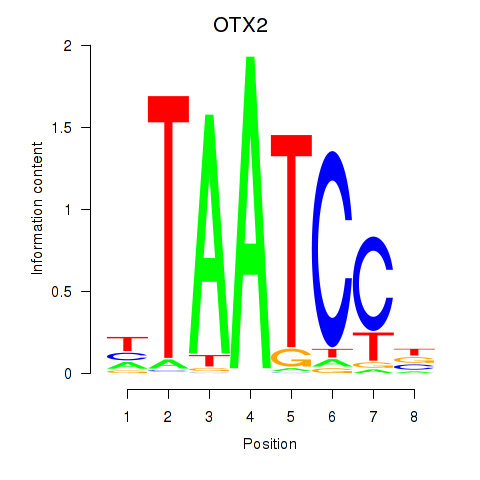
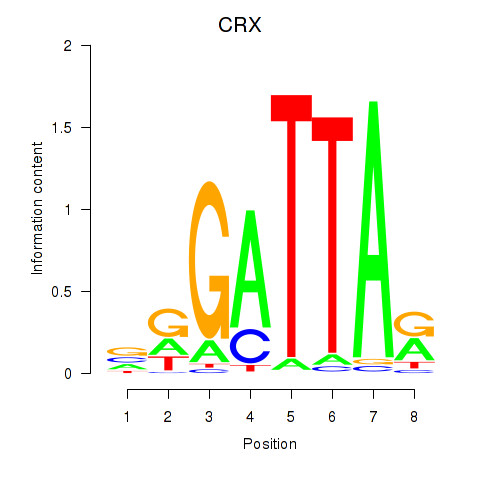
Results for OTX2_CRX
Z-value: 0.62
Transcription factors associated with OTX2_CRX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
OTX2
|
ENSG00000165588.12 | orthodenticle homeobox 2 |
|
CRX
|
ENSG00000105392.11 | cone-rod homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CRX | hg19_v2_chr19_+_48325097_48325211 | 0.53 | 4.6e-17 | Click! |
Activity profile of OTX2_CRX motif
Sorted Z-values of OTX2_CRX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_55866104 | 12.70 |
ENST00000326529.4
|
COX6B2
|
cytochrome c oxidase subunit VIb polypeptide 2 (testis) |
| chr2_-_175711133 | 8.21 |
ENST00000409597.1
ENST00000413882.1 |
CHN1
|
chimerin 1 |
| chr19_-_55866061 | 8.10 |
ENST00000588572.2
ENST00000593184.1 ENST00000589467.1 |
COX6B2
|
cytochrome c oxidase subunit VIb polypeptide 2 (testis) |
| chr5_-_88179302 | 7.33 |
ENST00000504921.2
|
MEF2C
|
myocyte enhancer factor 2C |
| chr20_-_18774614 | 6.88 |
ENST00000412553.1
|
LINC00652
|
long intergenic non-protein coding RNA 652 |
| chr22_-_38380543 | 6.55 |
ENST00000396884.2
|
SOX10
|
SRY (sex determining region Y)-box 10 |
| chr19_+_18208603 | 5.97 |
ENST00000262811.6
|
MAST3
|
microtubule associated serine/threonine kinase 3 |
| chr14_-_22005343 | 5.76 |
ENST00000327430.3
|
SALL2
|
spalt-like transcription factor 2 |
| chr6_+_150920999 | 5.45 |
ENST00000367328.1
ENST00000367326.1 |
PLEKHG1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr14_-_22005062 | 5.25 |
ENST00000317492.5
|
SALL2
|
spalt-like transcription factor 2 |
| chr6_+_111580508 | 5.23 |
ENST00000368847.4
|
KIAA1919
|
KIAA1919 |
| chr14_-_101034407 | 4.93 |
ENST00000443071.2
ENST00000557378.1 |
BEGAIN
|
brain-enriched guanylate kinase-associated |
| chr7_-_100171270 | 4.63 |
ENST00000538735.1
|
SAP25
|
Sin3A-associated protein, 25kDa |
| chr11_-_111794446 | 4.52 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr5_-_24645078 | 4.09 |
ENST00000264463.4
|
CDH10
|
cadherin 10, type 2 (T2-cadherin) |
| chr12_-_16758059 | 3.80 |
ENST00000261169.6
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr12_-_16758304 | 3.80 |
ENST00000320122.6
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr18_+_32290218 | 3.73 |
ENST00000348997.5
ENST00000588949.1 ENST00000597599.1 |
DTNA
|
dystrobrevin, alpha |
| chr17_-_48207157 | 3.68 |
ENST00000330175.4
ENST00000503131.1 |
SAMD14
|
sterile alpha motif domain containing 14 |
| chr15_+_43985084 | 3.62 |
ENST00000434505.1
ENST00000411750.1 |
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr3_-_49058479 | 3.61 |
ENST00000440857.1
|
DALRD3
|
DALR anticodon binding domain containing 3 |
| chr7_-_97881429 | 3.60 |
ENST00000420697.1
ENST00000379795.3 ENST00000415086.1 ENST00000542604.1 ENST00000447648.2 |
TECPR1
|
tectonin beta-propeller repeat containing 1 |
| chr18_-_64271363 | 3.59 |
ENST00000262150.2
|
CDH19
|
cadherin 19, type 2 |
| chr5_-_88179017 | 3.57 |
ENST00000514028.1
ENST00000514015.1 ENST00000503075.1 ENST00000437473.2 |
MEF2C
|
myocyte enhancer factor 2C |
| chr15_+_43885252 | 3.49 |
ENST00000453782.1
ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chrX_+_15767971 | 3.45 |
ENST00000479740.1
ENST00000454127.2 |
CA5B
|
carbonic anhydrase VB, mitochondrial |
| chr8_-_110656995 | 3.45 |
ENST00000276646.9
ENST00000533065.1 |
SYBU
|
syntabulin (syntaxin-interacting) |
| chr5_-_88178964 | 3.43 |
ENST00000513252.1
ENST00000508569.1 ENST00000510942.1 ENST00000506554.1 |
MEF2C
|
myocyte enhancer factor 2C |
| chr16_-_58004992 | 3.33 |
ENST00000564448.1
ENST00000251102.8 ENST00000311183.4 |
CNGB1
|
cyclic nucleotide gated channel beta 1 |
| chr3_+_129247479 | 3.33 |
ENST00000296271.3
|
RHO
|
rhodopsin |
| chr11_-_111783595 | 3.25 |
ENST00000528628.1
|
CRYAB
|
crystallin, alpha B |
| chr5_-_11588907 | 3.15 |
ENST00000513598.1
ENST00000503622.1 |
CTNND2
|
catenin (cadherin-associated protein), delta 2 |
| chr3_+_4535025 | 3.15 |
ENST00000302640.8
ENST00000354582.6 ENST00000423119.2 ENST00000357086.4 ENST00000456211.2 |
ITPR1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr15_-_27184664 | 3.11 |
ENST00000541819.2
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chrX_-_54384425 | 3.09 |
ENST00000375169.3
ENST00000354646.2 |
WNK3
|
WNK lysine deficient protein kinase 3 |
| chr11_-_62752455 | 3.01 |
ENST00000360421.4
|
SLC22A6
|
solute carrier family 22 (organic anion transporter), member 6 |
| chr5_-_11589131 | 3.00 |
ENST00000511377.1
|
CTNND2
|
catenin (cadherin-associated protein), delta 2 |
| chr11_-_62752429 | 2.96 |
ENST00000377871.3
|
SLC22A6
|
solute carrier family 22 (organic anion transporter), member 6 |
| chrX_+_69488174 | 2.96 |
ENST00000480877.2
ENST00000307959.8 |
ARR3
|
arrestin 3, retinal (X-arrestin) |
| chrX_+_41306575 | 2.90 |
ENST00000342595.2
ENST00000378220.1 |
NYX
|
nyctalopin |
| chr9_-_13279563 | 2.82 |
ENST00000541718.1
|
MPDZ
|
multiple PDZ domain protein |
| chr11_-_62752162 | 2.69 |
ENST00000458333.2
ENST00000421062.2 |
SLC22A6
|
solute carrier family 22 (organic anion transporter), member 6 |
| chr4_+_154125565 | 2.58 |
ENST00000338700.5
|
TRIM2
|
tripartite motif containing 2 |
| chrX_-_6146876 | 2.58 |
ENST00000381095.3
|
NLGN4X
|
neuroligin 4, X-linked |
| chr9_+_17135016 | 2.56 |
ENST00000425824.1
ENST00000262360.5 ENST00000380641.4 |
CNTLN
|
centlein, centrosomal protein |
| chr9_+_34179003 | 2.55 |
ENST00000545103.1
ENST00000543944.1 ENST00000536252.1 ENST00000540348.1 ENST00000297661.4 ENST00000379186.4 |
UBAP1
|
ubiquitin associated protein 1 |
| chr17_-_40264692 | 2.52 |
ENST00000591220.1
ENST00000251642.3 |
DHX58
|
DEXH (Asp-Glu-X-His) box polypeptide 58 |
| chr2_+_183943464 | 2.52 |
ENST00000354221.4
|
DUSP19
|
dual specificity phosphatase 19 |
| chr5_-_149324306 | 2.51 |
ENST00000255266.5
|
PDE6A
|
phosphodiesterase 6A, cGMP-specific, rod, alpha |
| chr9_-_13279589 | 2.50 |
ENST00000319217.7
|
MPDZ
|
multiple PDZ domain protein |
| chr9_+_17134980 | 2.49 |
ENST00000380647.3
|
CNTLN
|
centlein, centrosomal protein |
| chr19_+_56166360 | 2.49 |
ENST00000308924.4
|
U2AF2
|
U2 small nuclear RNA auxiliary factor 2 |
| chr4_-_153601136 | 2.46 |
ENST00000504064.1
ENST00000304385.3 |
TMEM154
|
transmembrane protein 154 |
| chr5_+_161494521 | 2.45 |
ENST00000356592.3
|
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr1_+_20396649 | 2.43 |
ENST00000375108.3
|
PLA2G5
|
phospholipase A2, group V |
| chr11_-_128894053 | 2.42 |
ENST00000392657.3
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr3_-_50605150 | 2.34 |
ENST00000357203.3
|
C3orf18
|
chromosome 3 open reading frame 18 |
| chr12_-_13248598 | 2.32 |
ENST00000337630.6
ENST00000545699.1 |
GSG1
|
germ cell associated 1 |
| chr1_+_18957500 | 2.32 |
ENST00000375375.3
|
PAX7
|
paired box 7 |
| chr13_+_31191920 | 2.26 |
ENST00000255304.4
|
USPL1
|
ubiquitin specific peptidase like 1 |
| chr7_-_128171123 | 2.26 |
ENST00000608477.1
|
RP11-212P7.2
|
RP11-212P7.2 |
| chr3_-_50605077 | 2.20 |
ENST00000426034.1
ENST00000441239.1 |
C3orf18
|
chromosome 3 open reading frame 18 |
| chr21_+_33245548 | 2.15 |
ENST00000270112.2
|
HUNK
|
hormonally up-regulated Neu-associated kinase |
| chr5_+_140345820 | 2.11 |
ENST00000289269.5
|
PCDHAC2
|
protocadherin alpha subfamily C, 2 |
| chr2_+_120770581 | 2.10 |
ENST00000263713.5
|
EPB41L5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr10_+_95848824 | 2.09 |
ENST00000371385.3
ENST00000371375.1 |
PLCE1
|
phospholipase C, epsilon 1 |
| chr9_+_103204553 | 2.07 |
ENST00000502978.1
ENST00000334943.6 |
MSANTD3-TMEFF1
TMEFF1
|
MSANTD3-TMEFF1 readthrough transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chr1_+_11751748 | 2.04 |
ENST00000294485.5
|
DRAXIN
|
dorsal inhibitory axon guidance protein |
| chr2_+_120770645 | 2.02 |
ENST00000443902.2
|
EPB41L5
|
erythrocyte membrane protein band 4.1 like 5 |
| chrX_+_129473859 | 1.98 |
ENST00000424447.1
|
SLC25A14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr10_-_104178857 | 1.96 |
ENST00000020673.5
|
PSD
|
pleckstrin and Sec7 domain containing |
| chr11_+_61522844 | 1.92 |
ENST00000265460.5
|
MYRF
|
myelin regulatory factor |
| chr5_+_161494770 | 1.90 |
ENST00000414552.2
ENST00000361925.4 |
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr12_+_7941989 | 1.89 |
ENST00000229307.4
|
NANOG
|
Nanog homeobox |
| chr17_-_9808887 | 1.87 |
ENST00000226193.5
|
RCVRN
|
recoverin |
| chr8_+_104892639 | 1.86 |
ENST00000436393.2
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr5_-_95768973 | 1.84 |
ENST00000311106.3
|
PCSK1
|
proprotein convertase subtilisin/kexin type 1 |
| chr3_-_98241358 | 1.79 |
ENST00000503004.1
ENST00000506575.1 ENST00000513452.1 ENST00000515620.1 |
CLDND1
|
claudin domain containing 1 |
| chr12_+_133757995 | 1.79 |
ENST00000536435.2
ENST00000228289.5 ENST00000541211.2 ENST00000500625.3 ENST00000539248.2 ENST00000542711.2 ENST00000536899.2 ENST00000542986.2 ENST00000537565.1 ENST00000541975.2 |
ZNF268
|
zinc finger protein 268 |
| chr12_-_13248562 | 1.76 |
ENST00000457134.2
ENST00000537302.1 |
GSG1
|
germ cell associated 1 |
| chr19_+_48325097 | 1.74 |
ENST00000221996.7
ENST00000539067.1 |
CRX
|
cone-rod homeobox |
| chr6_+_31554636 | 1.72 |
ENST00000433492.1
|
LST1
|
leukocyte specific transcript 1 |
| chr18_-_21891460 | 1.71 |
ENST00000357041.4
|
OSBPL1A
|
oxysterol binding protein-like 1A |
| chr3_-_54962100 | 1.68 |
ENST00000273286.5
|
LRTM1
|
leucine-rich repeats and transmembrane domains 1 |
| chr17_-_71640227 | 1.64 |
ENST00000388726.3
ENST00000392650.3 |
SDK2
|
sidekick cell adhesion molecule 2 |
| chr10_-_76868931 | 1.56 |
ENST00000372700.3
ENST00000473072.2 ENST00000491677.2 ENST00000607131.1 ENST00000372702.3 |
DUSP13
|
dual specificity phosphatase 13 |
| chr2_+_170366203 | 1.51 |
ENST00000284669.1
|
KLHL41
|
kelch-like family member 41 |
| chr7_-_128415844 | 1.51 |
ENST00000249389.2
|
OPN1SW
|
opsin 1 (cone pigments), short-wave-sensitive |
| chr13_-_52027134 | 1.50 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr10_+_95372289 | 1.49 |
ENST00000371447.3
|
PDE6C
|
phosphodiesterase 6C, cGMP-specific, cone, alpha prime |
| chr11_-_128712362 | 1.49 |
ENST00000392664.2
|
KCNJ1
|
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr3_-_45838011 | 1.45 |
ENST00000358525.4
ENST00000413781.1 |
SLC6A20
|
solute carrier family 6 (proline IMINO transporter), member 20 |
| chrX_-_49089771 | 1.45 |
ENST00000376251.1
ENST00000323022.5 ENST00000376265.2 |
CACNA1F
|
calcium channel, voltage-dependent, L type, alpha 1F subunit |
| chr3_+_137483579 | 1.38 |
ENST00000306087.1
|
SOX14
|
SRY (sex determining region Y)-box 14 |
| chr4_+_170541835 | 1.38 |
ENST00000504131.2
|
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chr11_-_118436606 | 1.38 |
ENST00000530872.1
|
IFT46
|
intraflagellar transport 46 homolog (Chlamydomonas) |
| chr12_-_21487829 | 1.36 |
ENST00000445053.1
ENST00000452078.1 ENST00000458504.1 ENST00000422327.1 ENST00000421294.1 |
SLCO1A2
|
solute carrier organic anion transporter family, member 1A2 |
| chrX_-_153363125 | 1.35 |
ENST00000407218.1
ENST00000453960.2 |
MECP2
|
methyl CpG binding protein 2 (Rett syndrome) |
| chr15_+_33603147 | 1.35 |
ENST00000415757.3
ENST00000389232.4 |
RYR3
|
ryanodine receptor 3 |
| chr9_+_82188077 | 1.35 |
ENST00000425506.1
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr10_-_48390973 | 1.34 |
ENST00000224600.4
|
RBP3
|
retinol binding protein 3, interstitial |
| chr8_-_91095099 | 1.34 |
ENST00000265431.3
|
CALB1
|
calbindin 1, 28kDa |
| chr12_-_13248705 | 1.33 |
ENST00000396310.2
|
GSG1
|
germ cell associated 1 |
| chr18_+_29769978 | 1.32 |
ENST00000269202.6
ENST00000581447.1 |
MEP1B
|
meprin A, beta |
| chrY_+_16634483 | 1.30 |
ENST00000382872.1
|
NLGN4Y
|
neuroligin 4, Y-linked |
| chr3_-_170626418 | 1.28 |
ENST00000474096.1
ENST00000295822.2 |
EIF5A2
|
eukaryotic translation initiation factor 5A2 |
| chr20_+_4702548 | 1.27 |
ENST00000305817.2
|
PRND
|
prion protein 2 (dublet) |
| chr21_-_19775973 | 1.26 |
ENST00000284885.3
|
TMPRSS15
|
transmembrane protease, serine 15 |
| chr8_-_145701718 | 1.23 |
ENST00000377317.4
|
FOXH1
|
forkhead box H1 |
| chr1_+_17248418 | 1.18 |
ENST00000375541.5
|
CROCC
|
ciliary rootlet coiled-coil, rootletin |
| chr2_+_33661382 | 1.17 |
ENST00000402538.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr7_+_136553824 | 1.16 |
ENST00000320658.5
ENST00000453373.1 ENST00000397608.3 ENST00000402486.3 ENST00000401861.1 |
CHRM2
|
cholinergic receptor, muscarinic 2 |
| chr16_-_325910 | 1.15 |
ENST00000359740.5
ENST00000316163.5 ENST00000431291.2 ENST00000397770.3 ENST00000397768.3 |
RGS11
|
regulator of G-protein signaling 11 |
| chr19_-_44124019 | 1.14 |
ENST00000300811.3
|
ZNF428
|
zinc finger protein 428 |
| chr6_-_42690312 | 1.13 |
ENST00000230381.5
|
PRPH2
|
peripherin 2 (retinal degeneration, slow) |
| chr2_-_30144432 | 1.11 |
ENST00000389048.3
|
ALK
|
anaplastic lymphoma receptor tyrosine kinase |
| chr6_+_31554826 | 1.11 |
ENST00000376089.2
ENST00000396112.2 |
LST1
|
leukocyte specific transcript 1 |
| chr15_-_34610962 | 1.09 |
ENST00000290209.5
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr22_-_19137796 | 1.08 |
ENST00000086933.2
|
GSC2
|
goosecoid homeobox 2 |
| chr1_-_241799232 | 1.07 |
ENST00000366553.1
|
CHML
|
choroideremia-like (Rab escort protein 2) |
| chr1_+_159141397 | 1.07 |
ENST00000368124.4
ENST00000368125.4 ENST00000416746.1 |
CADM3
|
cell adhesion molecule 3 |
| chr13_-_79980315 | 1.05 |
ENST00000438737.2
|
RBM26
|
RNA binding motif protein 26 |
| chr4_-_102267953 | 1.05 |
ENST00000523694.2
ENST00000507176.1 |
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr1_-_165414414 | 1.01 |
ENST00000359842.5
|
RXRG
|
retinoid X receptor, gamma |
| chr4_+_96012614 | 1.01 |
ENST00000264568.4
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr12_+_81101277 | 1.00 |
ENST00000228641.3
|
MYF6
|
myogenic factor 6 (herculin) |
| chr5_-_524445 | 1.00 |
ENST00000514375.1
ENST00000264938.3 |
SLC9A3
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 |
| chrX_-_101771645 | 0.99 |
ENST00000289373.4
|
TMSB15A
|
thymosin beta 15a |
| chr6_+_35310391 | 0.99 |
ENST00000337400.2
ENST00000311565.4 ENST00000540939.1 |
PPARD
|
peroxisome proliferator-activated receptor delta |
| chr6_+_42141029 | 0.98 |
ENST00000372958.1
|
GUCA1A
|
guanylate cyclase activator 1A (retina) |
| chr18_-_28681950 | 0.97 |
ENST00000251081.6
|
DSC2
|
desmocollin 2 |
| chr8_-_87755878 | 0.96 |
ENST00000320005.5
|
CNGB3
|
cyclic nucleotide gated channel beta 3 |
| chr10_-_74114714 | 0.96 |
ENST00000338820.3
ENST00000394903.2 ENST00000444643.2 |
DNAJB12
|
DnaJ (Hsp40) homolog, subfamily B, member 12 |
| chr12_+_58138800 | 0.95 |
ENST00000547992.1
ENST00000552816.1 ENST00000547472.1 |
TSPAN31
|
tetraspanin 31 |
| chr19_-_42931567 | 0.92 |
ENST00000244289.4
|
LIPE
|
lipase, hormone-sensitive |
| chr12_-_13248732 | 0.91 |
ENST00000396302.3
|
GSG1
|
germ cell associated 1 |
| chr15_-_35047166 | 0.90 |
ENST00000290374.4
|
GJD2
|
gap junction protein, delta 2, 36kDa |
| chr3_-_45837959 | 0.90 |
ENST00000353278.4
ENST00000456124.2 |
SLC6A20
|
solute carrier family 6 (proline IMINO transporter), member 20 |
| chrX_-_153363188 | 0.87 |
ENST00000303391.6
|
MECP2
|
methyl CpG binding protein 2 (Rett syndrome) |
| chr15_+_93443419 | 0.85 |
ENST00000557381.1
ENST00000420239.2 |
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr1_-_21606013 | 0.84 |
ENST00000357071.4
|
ECE1
|
endothelin converting enzyme 1 |
| chr1_-_117664317 | 0.82 |
ENST00000256649.4
ENST00000369464.3 ENST00000485032.1 |
TRIM45
|
tripartite motif containing 45 |
| chr9_-_20622478 | 0.82 |
ENST00000355930.6
ENST00000380338.4 |
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr6_-_35480640 | 0.81 |
ENST00000428978.1
ENST00000322263.4 |
TULP1
|
tubby like protein 1 |
| chr12_+_58138664 | 0.81 |
ENST00000257910.3
|
TSPAN31
|
tetraspanin 31 |
| chr1_-_53608289 | 0.78 |
ENST00000371491.4
|
SLC1A7
|
solute carrier family 1 (glutamate transporter), member 7 |
| chr6_+_31554962 | 0.78 |
ENST00000376092.3
ENST00000376086.3 ENST00000303757.8 ENST00000376093.2 ENST00000376102.3 |
LST1
|
leukocyte specific transcript 1 |
| chr1_-_13452656 | 0.77 |
ENST00000376132.3
|
PRAMEF13
|
PRAME family member 13 |
| chr12_-_56848426 | 0.77 |
ENST00000257979.4
|
MIP
|
major intrinsic protein of lens fiber |
| chr6_+_31555045 | 0.74 |
ENST00000396101.3
ENST00000490742.1 |
LST1
|
leukocyte specific transcript 1 |
| chr11_-_118436707 | 0.74 |
ENST00000264020.2
ENST00000264021.3 |
IFT46
|
intraflagellar transport 46 homolog (Chlamydomonas) |
| chr1_+_209757051 | 0.74 |
ENST00000009105.1
ENST00000423146.1 ENST00000361322.2 |
CAMK1G
|
calcium/calmodulin-dependent protein kinase IG |
| chr1_+_26644441 | 0.74 |
ENST00000374213.2
|
CD52
|
CD52 molecule |
| chr3_-_9291063 | 0.70 |
ENST00000383836.3
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3 |
| chr19_-_44123734 | 0.70 |
ENST00000598676.1
|
ZNF428
|
zinc finger protein 428 |
| chr10_+_35464513 | 0.70 |
ENST00000494479.1
ENST00000463314.1 ENST00000342105.3 ENST00000495301.1 ENST00000463960.1 |
CREM
|
cAMP responsive element modulator |
| chr17_+_68100989 | 0.69 |
ENST00000585558.1
ENST00000392670.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr12_+_85673868 | 0.67 |
ENST00000316824.3
|
ALX1
|
ALX homeobox 1 |
| chr8_-_133123406 | 0.66 |
ENST00000434736.2
|
HHLA1
|
HERV-H LTR-associating 1 |
| chr19_+_2476116 | 0.65 |
ENST00000215631.4
ENST00000587345.1 |
GADD45B
|
growth arrest and DNA-damage-inducible, beta |
| chr14_-_36789783 | 0.64 |
ENST00000605579.1
ENST00000604336.1 ENST00000359527.7 ENST00000603139.1 ENST00000318473.7 |
MBIP
|
MAP3K12 binding inhibitory protein 1 |
| chr12_+_70133152 | 0.64 |
ENST00000550536.1
ENST00000362025.5 |
RAB3IP
|
RAB3A interacting protein |
| chr3_-_32612263 | 0.64 |
ENST00000432458.2
ENST00000424991.1 ENST00000273130.4 |
DYNC1LI1
|
dynein, cytoplasmic 1, light intermediate chain 1 |
| chr19_-_10121144 | 0.62 |
ENST00000264828.3
|
COL5A3
|
collagen, type V, alpha 3 |
| chr1_-_53608249 | 0.62 |
ENST00000371494.4
|
SLC1A7
|
solute carrier family 1 (glutamate transporter), member 7 |
| chr10_+_52750930 | 0.61 |
ENST00000401604.2
|
PRKG1
|
protein kinase, cGMP-dependent, type I |
| chr14_-_36789865 | 0.60 |
ENST00000416007.4
|
MBIP
|
MAP3K12 binding inhibitory protein 1 |
| chr19_-_51538118 | 0.59 |
ENST00000529888.1
|
KLK12
|
kallikrein-related peptidase 12 |
| chr3_-_48229846 | 0.59 |
ENST00000302506.3
ENST00000351231.3 ENST00000437972.1 |
CDC25A
|
cell division cycle 25A |
| chr16_-_68269971 | 0.59 |
ENST00000565858.1
|
ESRP2
|
epithelial splicing regulatory protein 2 |
| chr4_+_68424434 | 0.59 |
ENST00000265404.2
ENST00000396225.1 |
STAP1
|
signal transducing adaptor family member 1 |
| chr11_+_128563652 | 0.56 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr21_-_34915147 | 0.54 |
ENST00000381831.3
ENST00000381839.3 |
GART
|
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase |
| chr18_-_28682374 | 0.54 |
ENST00000280904.6
|
DSC2
|
desmocollin 2 |
| chr2_-_214016314 | 0.54 |
ENST00000434687.1
ENST00000374319.4 |
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chrX_-_15619076 | 0.53 |
ENST00000252519.3
|
ACE2
|
angiotensin I converting enzyme 2 |
| chr5_-_58882219 | 0.53 |
ENST00000505453.1
ENST00000360047.5 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr12_-_67197760 | 0.51 |
ENST00000539540.1
ENST00000540433.1 ENST00000541947.1 ENST00000538373.1 |
GRIP1
|
glutamate receptor interacting protein 1 |
| chr10_-_86001210 | 0.49 |
ENST00000372105.3
|
LRIT1
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 1 |
| chr2_+_128175997 | 0.47 |
ENST00000234071.3
ENST00000429925.1 ENST00000442644.1 ENST00000453608.2 |
PROC
|
protein C (inactivator of coagulation factors Va and VIIIa) |
| chr19_-_48547294 | 0.46 |
ENST00000293255.2
|
CABP5
|
calcium binding protein 5 |
| chr3_+_173302222 | 0.43 |
ENST00000361589.4
|
NLGN1
|
neuroligin 1 |
| chr16_-_31076273 | 0.42 |
ENST00000426488.2
|
ZNF668
|
zinc finger protein 668 |
| chr8_+_61591337 | 0.42 |
ENST00000423902.2
|
CHD7
|
chromodomain helicase DNA binding protein 7 |
| chr2_-_119605253 | 0.41 |
ENST00000295206.6
|
EN1
|
engrailed homeobox 1 |
| chr22_-_30867973 | 0.39 |
ENST00000402286.1
ENST00000401751.1 ENST00000539629.1 ENST00000403066.1 ENST00000215812.4 |
SEC14L3
|
SEC14-like 3 (S. cerevisiae) |
| chr14_+_96722152 | 0.38 |
ENST00000216629.6
|
BDKRB1
|
bradykinin receptor B1 |
| chr16_-_31076332 | 0.37 |
ENST00000539836.3
ENST00000535577.1 ENST00000442862.2 |
ZNF668
|
zinc finger protein 668 |
| chr20_+_42984330 | 0.36 |
ENST00000316673.4
ENST00000609795.1 ENST00000457232.1 ENST00000609262.1 |
HNF4A
|
hepatocyte nuclear factor 4, alpha |
| chr10_+_118350468 | 0.35 |
ENST00000358834.4
ENST00000528052.1 ENST00000442761.1 |
PNLIPRP1
|
pancreatic lipase-related protein 1 |
| chr14_-_22005018 | 0.35 |
ENST00000546363.1
|
SALL2
|
spalt-like transcription factor 2 |
| chr1_+_180897269 | 0.33 |
ENST00000367587.1
|
KIAA1614
|
KIAA1614 |
| chr10_-_94003003 | 0.32 |
ENST00000412050.4
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr4_-_102268484 | 0.31 |
ENST00000394853.4
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr15_-_31453359 | 0.30 |
ENST00000542188.1
|
TRPM1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chrX_+_153409678 | 0.30 |
ENST00000369951.4
|
OPN1LW
|
opsin 1 (cone pigments), long-wave-sensitive |
| chr1_-_173174681 | 0.30 |
ENST00000367718.1
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4 |
| chr12_-_53171128 | 0.29 |
ENST00000332411.2
|
KRT76
|
keratin 76 |
| chr6_+_35310312 | 0.28 |
ENST00000448077.2
ENST00000360694.3 ENST00000418635.2 ENST00000444397.1 |
PPARD
|
peroxisome proliferator-activated receptor delta |
| chr5_-_33984741 | 0.27 |
ENST00000382102.3
ENST00000509381.1 ENST00000342059.3 ENST00000345083.5 |
SLC45A2
|
solute carrier family 45, member 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of OTX2_CRX
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 14.3 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 2.9 | 8.7 | GO:0097254 | renal tubular secretion(GO:0097254) |
| 1.6 | 6.6 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 1.4 | 4.1 | GO:0003383 | apical constriction(GO:0003383) mesoderm migration involved in gastrulation(GO:0007509) |
| 1.3 | 7.6 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 1.2 | 3.6 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.8 | 3.1 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.7 | 2.2 | GO:1900114 | positive regulation of histone H3-K9 trimethylation(GO:1900114) |
| 0.6 | 2.5 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.6 | 3.2 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.6 | 2.4 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.6 | 1.8 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.6 | 2.3 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.4 | 2.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.4 | 1.2 | GO:0007231 | osmosensory signaling pathway(GO:0007231) |
| 0.4 | 1.2 | GO:2000824 | negative regulation of androgen receptor activity(GO:2000824) |
| 0.4 | 3.6 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.4 | 7.5 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.3 | 5.6 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.3 | 1.7 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.3 | 6.2 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.3 | 2.4 | GO:0015824 | proline transport(GO:0015824) |
| 0.3 | 1.0 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.3 | 3.6 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.3 | 5.1 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.3 | 1.9 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.3 | 2.3 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.3 | 5.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.3 | 1.1 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.3 | 1.4 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.3 | 1.1 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.2 | 1.5 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.2 | 2.8 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.2 | 1.2 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.2 | 7.5 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.2 | 1.9 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.2 | 0.8 | GO:0034959 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.2 | 1.9 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.2 | 1.0 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.2 | 2.8 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.2 | 0.2 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.2 | 0.6 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) |
| 0.2 | 0.9 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.2 | 1.7 | GO:0086042 | cardiac muscle cell-cardiac muscle cell adhesion(GO:0086042) bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.2 | 1.3 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.2 | 1.6 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.2 | 1.3 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.2 | 0.5 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.2 | 2.6 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 1.3 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.1 | 0.4 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.1 | 0.4 | GO:0061743 | motor learning(GO:0061743) |
| 0.1 | 2.5 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 1.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 0.3 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.1 | 0.5 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 0.1 | 2.6 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.1 | 1.0 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.1 | 0.3 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.1 | 2.0 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 6.2 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.1 | 0.9 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.1 | 3.7 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.1 | 2.5 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.1 | 1.5 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 0.8 | GO:0007379 | segment specification(GO:0007379) positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 0.6 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.1 | 1.4 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.1 | 0.2 | GO:0021562 | vestibulocochlear nerve development(GO:0021562) |
| 0.1 | 1.0 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 0.5 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.1 | 0.3 | GO:0045726 | regulation of integrin biosynthetic process(GO:0045113) positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.1 | 3.4 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 10.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 0.3 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.1 | 1.3 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.1 | 1.0 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 11.3 | GO:0021915 | neural tube development(GO:0021915) |
| 0.1 | 0.5 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.1 | 2.1 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 0.2 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.1 | 0.3 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.1 | 10.6 | GO:0007601 | visual perception(GO:0007601) |
| 0.1 | 0.4 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 1.8 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.4 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 4.2 | GO:0032945 | negative regulation of mononuclear cell proliferation(GO:0032945) negative regulation of lymphocyte proliferation(GO:0050672) |
| 0.0 | 1.7 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.6 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 0.1 | GO:1903625 | negative regulation of DNA catabolic process(GO:1903625) |
| 0.0 | 3.2 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.0 | 0.2 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 1.3 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 1.4 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 1.7 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.2 | GO:0001711 | endodermal cell fate commitment(GO:0001711) |
| 0.0 | 0.8 | GO:0060348 | bone development(GO:0060348) |
| 0.0 | 1.3 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 2.2 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 1.0 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.7 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 1.3 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 0.4 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.6 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 | 0.9 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.6 | GO:0006612 | protein targeting to membrane(GO:0006612) |
| 0.0 | 0.7 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.6 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 2.2 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.2 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.1 | GO:0060737 | prostate epithelial cord elongation(GO:0060523) prostate gland morphogenetic growth(GO:0060737) |
| 0.0 | 1.1 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 1.8 | GO:0007155 | cell adhesion(GO:0007155) biological adhesion(GO:0022610) |
| 0.0 | 0.1 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.2 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) cell cycle G2/M phase transition(GO:0044839) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 20.8 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.5 | 2.5 | GO:0089701 | U2AF(GO:0089701) |
| 0.5 | 7.8 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.5 | 4.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.4 | 11.4 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.4 | 2.6 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.3 | 8.5 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.3 | 1.2 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.3 | 1.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.3 | 5.3 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.3 | 3.4 | GO:0097433 | dense body(GO:0097433) |
| 0.2 | 7.5 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.2 | 0.8 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.2 | 0.6 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.2 | 1.2 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.2 | 6.6 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.2 | 1.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 9.3 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 3.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 1.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 2.1 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 1.2 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 2.2 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 1.0 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 0.3 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.1 | 8.8 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 1.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 2.9 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.2 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 1.7 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 0.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 1.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 5.1 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 6.0 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 6.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.3 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 1.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 4.9 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 18.7 | GO:0098794 | postsynapse(GO:0098794) |
| 0.0 | 2.2 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.2 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.8 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.0 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 1.8 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 1.1 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 2.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.3 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 1.3 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.5 | GO:0005796 | Golgi lumen(GO:0005796) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.9 | 8.7 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.8 | 14.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.8 | 3.3 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.8 | 3.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.6 | 2.8 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.5 | 2.5 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.5 | 5.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.5 | 3.6 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.4 | 2.2 | GO:0002046 | opsin binding(GO:0002046) |
| 0.4 | 1.2 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.3 | 1.3 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.3 | 4.4 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.3 | 3.1 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.3 | 4.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.3 | 0.9 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.3 | 2.3 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.3 | 8.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.3 | 1.0 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.2 | 1.5 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.2 | 2.4 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.2 | 1.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.2 | 1.4 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.2 | 1.3 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.2 | 2.5 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.2 | 3.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.2 | 1.0 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.2 | 2.2 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.2 | 1.7 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.2 | 8.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.2 | 1.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.2 | 5.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 3.6 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 1.4 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.1 | 0.4 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 1.7 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 0.6 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 2.4 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.1 | 1.0 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.5 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.1 | 1.1 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 3.0 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 1.0 | GO:0015386 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 1.0 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 6.5 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.1 | 1.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 0.6 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.1 | 4.6 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 1.3 | GO:0016918 | retinal binding(GO:0016918) |
| 0.1 | 0.5 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 2.0 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 2.1 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 1.5 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.4 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.1 | 1.2 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 1.4 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 0.9 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 6.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 5.4 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 0.7 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 1.7 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 1.2 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.1 | 1.3 | GO:0036041 | long-chain fatty acid binding(GO:0036041) |
| 0.0 | 0.4 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 1.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.3 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 3.9 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.7 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 1.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 2.4 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.5 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.0 | 1.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.5 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.6 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.6 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 3.0 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 1.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 4.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.3 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 1.0 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 13.9 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 6.1 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 2.6 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.2 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 5.0 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.9 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.6 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 2.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.6 | GO:0043394 | proteoglycan binding(GO:0043394) |
| 0.0 | 3.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.6 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 1.6 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 1.9 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 1.3 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.0 | 1.1 | GO:0008233 | peptidase activity(GO:0008233) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 9.7 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.2 | 14.3 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 8.2 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 1.1 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 2.3 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 3.3 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 2.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.6 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 2.4 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.0 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 1.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.0 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.6 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 1.0 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.4 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.4 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.3 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 1.7 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 5.1 | REACTOME OPSINS | Genes involved in Opsins |
| 0.4 | 8.7 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.4 | 14.3 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.2 | 7.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 3.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.2 | 2.4 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 4.1 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 2.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 1.8 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 1.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 3.1 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 2.5 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 1.4 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.9 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 1.7 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 1.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 7.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.6 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 1.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.9 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 2.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.0 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 1.2 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 7.1 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 2.1 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.3 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.2 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |