Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
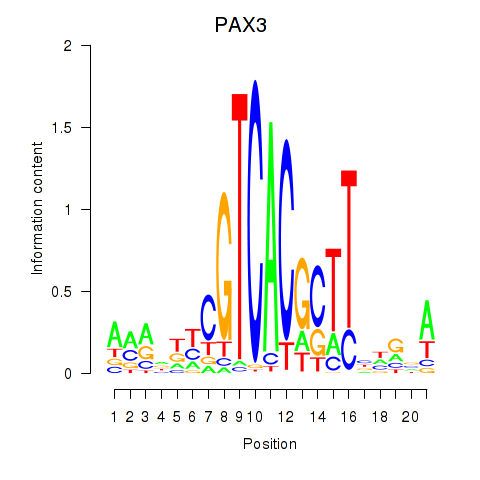
Results for PAX3
Z-value: 1.10
Transcription factors associated with PAX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PAX3
|
ENSG00000135903.14 | paired box 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PAX3 | hg19_v2_chr2_-_223163465_223163730 | 0.25 | 2.1e-04 | Click! |
Activity profile of PAX3 motif
Sorted Z-values of PAX3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_-_44258360 | 50.17 |
ENST00000330884.4
ENST00000249130.5 |
SULT4A1
|
sulfotransferase family 4A, member 1 |
| chrX_+_38420783 | 47.37 |
ENST00000422612.2
ENST00000286824.6 ENST00000545599.1 |
TSPAN7
|
tetraspanin 7 |
| chrX_+_38420623 | 41.84 |
ENST00000378482.2
|
TSPAN7
|
tetraspanin 7 |
| chr11_+_73358594 | 27.60 |
ENST00000227214.6
ENST00000398494.4 ENST00000543085.1 |
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chrX_-_13835461 | 24.28 |
ENST00000316715.4
ENST00000356942.5 |
GPM6B
|
glycoprotein M6B |
| chr1_+_50513686 | 20.66 |
ENST00000448907.2
|
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr3_-_58572760 | 19.04 |
ENST00000447756.2
|
FAM107A
|
family with sequence similarity 107, member A |
| chr15_+_99791567 | 18.34 |
ENST00000558879.1
ENST00000301981.3 ENST00000422500.2 ENST00000447360.2 ENST00000442993.2 |
LRRC28
|
leucine rich repeat containing 28 |
| chr1_+_99729813 | 15.84 |
ENST00000457765.1
|
LPPR4
|
Lipid phosphate phosphatase-related protein type 4 |
| chr9_-_91793675 | 15.38 |
ENST00000375835.4
ENST00000375830.1 |
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3 |
| chr16_+_23847339 | 13.64 |
ENST00000303531.7
|
PRKCB
|
protein kinase C, beta |
| chr15_-_52861157 | 13.10 |
ENST00000564163.1
|
ARPP19
|
cAMP-regulated phosphoprotein, 19kDa |
| chr1_+_84630053 | 12.86 |
ENST00000394838.2
ENST00000370682.3 ENST00000432111.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr11_+_31391381 | 12.07 |
ENST00000465995.1
ENST00000536040.1 |
DNAJC24
|
DnaJ (Hsp40) homolog, subfamily C, member 24 |
| chr4_+_156680153 | 12.07 |
ENST00000502959.1
ENST00000505764.1 ENST00000507146.1 ENST00000264424.8 ENST00000503520.1 |
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3 |
| chr19_+_52693259 | 10.75 |
ENST00000322088.6
ENST00000454220.2 ENST00000444322.2 ENST00000477989.1 |
PPP2R1A
|
protein phosphatase 2, regulatory subunit A, alpha |
| chr2_-_44588624 | 10.72 |
ENST00000438314.1
ENST00000409936.1 |
PREPL
|
prolyl endopeptidase-like |
| chr7_-_103629963 | 10.44 |
ENST00000428762.1
ENST00000343529.5 ENST00000424685.2 |
RELN
|
reelin |
| chr1_+_202830876 | 10.32 |
ENST00000456105.2
|
RP11-480I12.7
|
RP11-480I12.7 |
| chr18_+_23806437 | 10.17 |
ENST00000578121.1
|
TAF4B
|
TAF4b RNA polymerase II, TATA box binding protein (TBP)-associated factor, 105kDa |
| chr2_-_44588694 | 10.14 |
ENST00000409957.1
|
PREPL
|
prolyl endopeptidase-like |
| chr19_+_37407212 | 9.93 |
ENST00000427117.1
ENST00000587130.1 ENST00000333987.7 ENST00000415168.1 ENST00000444991.1 |
ZNF568
|
zinc finger protein 568 |
| chr2_-_44588679 | 9.79 |
ENST00000409411.1
|
PREPL
|
prolyl endopeptidase-like |
| chr4_+_156680143 | 9.26 |
ENST00000505154.1
|
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3 |
| chr8_+_117950422 | 9.17 |
ENST00000378279.3
|
AARD
|
alanine and arginine rich domain containing protein |
| chr15_+_51973680 | 9.15 |
ENST00000542355.2
|
SCG3
|
secretogranin III |
| chr19_-_57183114 | 9.10 |
ENST00000537055.2
ENST00000601659.1 |
ZNF835
|
zinc finger protein 835 |
| chr2_-_44588893 | 8.76 |
ENST00000409272.1
ENST00000410081.1 ENST00000541738.1 |
PREPL
|
prolyl endopeptidase-like |
| chr15_+_51973550 | 8.68 |
ENST00000220478.3
|
SCG3
|
secretogranin III |
| chr6_+_89674246 | 8.55 |
ENST00000369474.1
|
AL079342.1
|
Uncharacterized protein; cDNA FLJ27030 fis, clone SLV07741 |
| chr15_-_52861323 | 8.43 |
ENST00000569723.1
ENST00000249822.4 ENST00000567669.1 ENST00000569281.2 ENST00000563566.1 ENST00000567830.1 |
ARPP19
|
cAMP-regulated phosphoprotein, 19kDa |
| chr6_-_32908765 | 8.02 |
ENST00000416244.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr19_+_2841433 | 7.76 |
ENST00000334241.4
ENST00000585966.1 ENST00000591539.1 |
ZNF555
|
zinc finger protein 555 |
| chr15_-_42783303 | 7.58 |
ENST00000565380.1
ENST00000564754.1 |
ZNF106
|
zinc finger protein 106 |
| chr7_+_29519486 | 7.42 |
ENST00000409041.4
|
CHN2
|
chimerin 2 |
| chr5_+_177457525 | 7.27 |
ENST00000511856.1
ENST00000511189.1 |
FAM153C
|
family with sequence similarity 153, member C |
| chr14_+_24583836 | 7.07 |
ENST00000559115.1
ENST00000558215.1 ENST00000557810.1 ENST00000561375.1 ENST00000446197.3 ENST00000559796.1 ENST00000560713.1 ENST00000560901.1 ENST00000559382.1 |
DCAF11
|
DDB1 and CUL4 associated factor 11 |
| chr10_-_75226166 | 6.91 |
ENST00000544628.1
|
PPP3CB
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr6_+_127587755 | 6.88 |
ENST00000368314.1
ENST00000476956.1 ENST00000609447.1 ENST00000356799.2 ENST00000477776.1 ENST00000609944.1 |
RNF146
|
ring finger protein 146 |
| chr22_+_18593507 | 6.68 |
ENST00000330423.3
|
TUBA8
|
tubulin, alpha 8 |
| chr19_+_35168567 | 6.43 |
ENST00000457781.2
ENST00000505163.1 ENST00000505242.1 ENST00000423823.2 ENST00000507959.1 ENST00000446502.2 |
ZNF302
|
zinc finger protein 302 |
| chr10_+_96162242 | 6.32 |
ENST00000225235.4
|
TBC1D12
|
TBC1 domain family, member 12 |
| chr9_+_104161123 | 6.31 |
ENST00000374861.3
ENST00000339664.2 ENST00000259395.4 |
ZNF189
|
zinc finger protein 189 |
| chr15_+_90808919 | 6.21 |
ENST00000379095.3
|
NGRN
|
neugrin, neurite outgrowth associated |
| chrX_-_48931648 | 6.19 |
ENST00000376386.3
ENST00000376390.4 |
PRAF2
|
PRA1 domain family, member 2 |
| chr8_-_145159083 | 6.12 |
ENST00000398712.2
|
SHARPIN
|
SHANK-associated RH domain interactor |
| chr16_-_18468926 | 6.09 |
ENST00000545114.1
|
RP11-1212A22.4
|
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chrX_+_103217207 | 5.86 |
ENST00000563257.1
ENST00000540220.1 ENST00000436583.1 ENST00000567181.1 ENST00000569577.1 |
TMSB15B
|
thymosin beta 15B |
| chr16_-_21849091 | 5.67 |
ENST00000537951.1
|
NPIPB4
|
nuclear pore complex interacting protein family, member B4 |
| chr14_+_22964877 | 5.63 |
ENST00000390494.1
|
TRAJ43
|
T cell receptor alpha joining 43 |
| chr10_-_104178857 | 5.56 |
ENST00000020673.5
|
PSD
|
pleckstrin and Sec7 domain containing |
| chr19_+_35168633 | 5.44 |
ENST00000505365.2
|
ZNF302
|
zinc finger protein 302 |
| chr6_+_14117872 | 5.43 |
ENST00000379153.3
|
CD83
|
CD83 molecule |
| chr7_+_99816859 | 5.39 |
ENST00000317271.2
|
PVRIG
|
poliovirus receptor related immunoglobulin domain containing |
| chr9_-_73029540 | 5.17 |
ENST00000377126.2
|
KLF9
|
Kruppel-like factor 9 |
| chr13_+_96085847 | 5.08 |
ENST00000376873.3
|
CLDN10
|
claudin 10 |
| chr19_-_46296011 | 4.93 |
ENST00000377735.3
ENST00000270223.6 |
DMWD
|
dystrophia myotonica, WD repeat containing |
| chr10_+_91087651 | 4.85 |
ENST00000371818.4
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr14_-_24711806 | 4.81 |
ENST00000540705.1
ENST00000538777.1 ENST00000558566.1 ENST00000559019.1 |
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr6_+_127588020 | 4.76 |
ENST00000309649.3
ENST00000610162.1 ENST00000610153.1 ENST00000608991.1 ENST00000480444.1 |
RNF146
|
ring finger protein 146 |
| chr14_-_24584138 | 4.72 |
ENST00000558280.1
ENST00000561028.1 |
NRL
|
neural retina leucine zipper |
| chr16_-_21416640 | 4.71 |
ENST00000542817.1
|
NPIPB3
|
nuclear pore complex interacting protein family, member B3 |
| chr19_+_10527449 | 4.59 |
ENST00000592685.1
ENST00000380702.2 |
PDE4A
|
phosphodiesterase 4A, cAMP-specific |
| chrX_+_70364667 | 4.50 |
ENST00000536169.1
ENST00000395855.2 ENST00000374051.3 ENST00000358741.3 |
NLGN3
|
neuroligin 3 |
| chr2_-_128051670 | 4.48 |
ENST00000493187.2
|
ERCC3
|
excision repair cross-complementing rodent repair deficiency, complementation group 3 |
| chr6_-_30815936 | 4.40 |
ENST00000442852.1
|
XXbac-BPG27H4.8
|
XXbac-BPG27H4.8 |
| chr12_+_112563303 | 4.09 |
ENST00000412615.2
|
TRAFD1
|
TRAF-type zinc finger domain containing 1 |
| chr16_+_27214802 | 4.02 |
ENST00000380948.2
ENST00000286096.4 |
KDM8
|
lysine (K)-specific demethylase 8 |
| chr16_-_71523236 | 4.01 |
ENST00000288177.5
ENST00000569072.1 |
ZNF19
|
zinc finger protein 19 |
| chr2_-_11810284 | 3.85 |
ENST00000306928.5
|
NTSR2
|
neurotensin receptor 2 |
| chr15_-_52861394 | 3.76 |
ENST00000563277.1
ENST00000566423.1 |
ARPP19
|
cAMP-regulated phosphoprotein, 19kDa |
| chr12_-_53594227 | 3.65 |
ENST00000550743.2
|
ITGB7
|
integrin, beta 7 |
| chr2_-_128051708 | 3.61 |
ENST00000285398.2
|
ERCC3
|
excision repair cross-complementing rodent repair deficiency, complementation group 3 |
| chr5_-_180688105 | 3.60 |
ENST00000327767.4
|
TRIM52
|
tripartite motif containing 52 |
| chr16_-_75018968 | 3.48 |
ENST00000262144.6
|
WDR59
|
WD repeat domain 59 |
| chr12_+_112563335 | 3.42 |
ENST00000549358.1
ENST00000257604.5 ENST00000548092.1 ENST00000552896.1 |
TRAFD1
|
TRAF-type zinc finger domain containing 1 |
| chr14_+_24584508 | 3.42 |
ENST00000559354.1
ENST00000560459.1 ENST00000559593.1 ENST00000396941.4 ENST00000396936.1 |
DCAF11
|
DDB1 and CUL4 associated factor 11 |
| chr12_-_89919965 | 3.41 |
ENST00000548729.1
|
POC1B-GALNT4
|
POC1B-GALNT4 readthrough |
| chr19_+_7968728 | 3.33 |
ENST00000397981.3
ENST00000545011.1 ENST00000397983.3 ENST00000397979.3 |
MAP2K7
|
mitogen-activated protein kinase kinase 7 |
| chr19_-_2783363 | 3.30 |
ENST00000221566.2
|
SGTA
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha |
| chr17_-_3461092 | 3.29 |
ENST00000301365.4
ENST00000572519.1 |
TRPV3
|
transient receptor potential cation channel, subfamily V, member 3 |
| chr18_-_59854203 | 3.27 |
ENST00000589339.1
ENST00000357637.5 ENST00000585458.1 ENST00000400334.3 ENST00000587134.1 ENST00000585923.1 ENST00000590765.1 ENST00000589720.1 ENST00000588571.1 ENST00000585344.1 |
PIGN
|
phosphatidylinositol glycan anchor biosynthesis, class N |
| chr1_-_7913089 | 3.18 |
ENST00000361696.5
|
UTS2
|
urotensin 2 |
| chr5_+_49961727 | 3.18 |
ENST00000505697.2
ENST00000503750.2 ENST00000514342.2 |
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr12_-_89920030 | 2.93 |
ENST00000413530.1
ENST00000547474.1 |
GALNT4
POC1B-GALNT4
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4) POC1B-GALNT4 readthrough |
| chr2_+_171034646 | 2.92 |
ENST00000409044.3
ENST00000408978.4 |
MYO3B
|
myosin IIIB |
| chr10_-_102289611 | 2.90 |
ENST00000299166.4
ENST00000370320.4 ENST00000531258.1 ENST00000370322.1 ENST00000535773.1 |
NDUFB8
SEC31B
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 8, 19kDa SEC31 homolog B (S. cerevisiae) |
| chr1_+_171283331 | 2.77 |
ENST00000367749.3
|
FMO4
|
flavin containing monooxygenase 4 |
| chr10_+_118305435 | 2.63 |
ENST00000369221.2
|
PNLIP
|
pancreatic lipase |
| chr3_-_119379427 | 2.54 |
ENST00000264231.3
ENST00000468801.1 ENST00000538678.1 |
POPDC2
|
popeye domain containing 2 |
| chr2_+_44589036 | 2.53 |
ENST00000402247.1
ENST00000407131.1 ENST00000403853.3 ENST00000378494.3 |
CAMKMT
|
calmodulin-lysine N-methyltransferase |
| chr19_-_54784353 | 2.53 |
ENST00000391746.1
|
LILRB2
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 2 |
| chr12_+_10460417 | 2.50 |
ENST00000381908.3
ENST00000336164.4 ENST00000350274.5 |
KLRD1
|
killer cell lectin-like receptor subfamily D, member 1 |
| chr1_+_1222489 | 2.46 |
ENST00000379099.3
|
SCNN1D
|
sodium channel, non-voltage-gated 1, delta subunit |
| chr19_-_40596828 | 2.44 |
ENST00000414720.2
ENST00000455521.1 ENST00000340963.5 ENST00000595773.1 |
ZNF780A
|
zinc finger protein 780A |
| chr9_-_21239978 | 2.36 |
ENST00000380222.2
|
IFNA14
|
interferon, alpha 14 |
| chr2_-_114514181 | 2.30 |
ENST00000409342.1
|
SLC35F5
|
solute carrier family 35, member F5 |
| chr9_+_131549483 | 2.26 |
ENST00000372648.5
ENST00000539497.1 |
TBC1D13
|
TBC1 domain family, member 13 |
| chr9_-_35650900 | 2.20 |
ENST00000259608.3
|
SIT1
|
signaling threshold regulating transmembrane adaptor 1 |
| chr8_+_107670064 | 2.18 |
ENST00000312046.6
|
OXR1
|
oxidation resistance 1 |
| chr21_-_30365136 | 2.09 |
ENST00000361371.5
ENST00000389194.2 ENST00000389195.2 |
LTN1
|
listerin E3 ubiquitin protein ligase 1 |
| chrX_+_102883620 | 2.08 |
ENST00000372626.3
|
TCEAL1
|
transcription elongation factor A (SII)-like 1 |
| chrX_-_118699325 | 2.04 |
ENST00000320339.4
ENST00000371594.4 ENST00000536133.1 |
CXorf56
|
chromosome X open reading frame 56 |
| chr17_+_3118915 | 2.01 |
ENST00000304094.1
|
OR1A1
|
olfactory receptor, family 1, subfamily A, member 1 |
| chr19_-_41388657 | 1.99 |
ENST00000301146.4
ENST00000291764.3 |
CYP2A7
|
cytochrome P450, family 2, subfamily A, polypeptide 7 |
| chr1_-_205180664 | 1.94 |
ENST00000367161.3
ENST00000367162.3 ENST00000367160.4 |
DSTYK
|
dual serine/threonine and tyrosine protein kinase |
| chrX_-_15872914 | 1.80 |
ENST00000380291.1
ENST00000545766.1 ENST00000421527.2 ENST00000329235.2 |
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr11_-_104972158 | 1.77 |
ENST00000598974.1
ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1
CARD16
CARD17
|
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr2_-_219524193 | 1.76 |
ENST00000450560.1
ENST00000449707.1 ENST00000432460.1 ENST00000411696.2 |
ZNF142
|
zinc finger protein 142 |
| chr9_-_13175823 | 1.70 |
ENST00000545857.1
|
MPDZ
|
multiple PDZ domain protein |
| chr1_-_52344471 | 1.66 |
ENST00000352171.7
ENST00000354831.7 |
NRD1
|
nardilysin (N-arginine dibasic convertase) |
| chr6_+_35227449 | 1.62 |
ENST00000373953.3
ENST00000440666.2 ENST00000339411.5 |
ZNF76
|
zinc finger protein 76 |
| chr20_-_23807358 | 1.61 |
ENST00000304725.2
|
CST2
|
cystatin SA |
| chr15_+_81591757 | 1.56 |
ENST00000558332.1
|
IL16
|
interleukin 16 |
| chr4_-_168155730 | 1.56 |
ENST00000502330.1
ENST00000357154.3 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr4_-_170924888 | 1.52 |
ENST00000502832.1
ENST00000393704.3 |
MFAP3L
|
microfibrillar-associated protein 3-like |
| chrX_+_102883887 | 1.40 |
ENST00000372625.3
ENST00000372624.3 |
TCEAL1
|
transcription elongation factor A (SII)-like 1 |
| chr5_-_157079428 | 1.37 |
ENST00000265007.6
|
SOX30
|
SRY (sex determining region Y)-box 30 |
| chr11_+_112038088 | 1.35 |
ENST00000530752.1
ENST00000280358.4 |
TEX12
|
testis expressed 12 |
| chr12_-_57400227 | 1.29 |
ENST00000300101.2
|
ZBTB39
|
zinc finger and BTB domain containing 39 |
| chr12_+_10460549 | 1.29 |
ENST00000543420.1
ENST00000543777.1 |
KLRD1
|
killer cell lectin-like receptor subfamily D, member 1 |
| chr4_-_168155700 | 1.26 |
ENST00000357545.4
ENST00000512648.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr17_-_59668550 | 1.18 |
ENST00000521764.1
|
NACA2
|
nascent polypeptide-associated complex alpha subunit 2 |
| chr12_+_9142131 | 1.15 |
ENST00000356986.3
ENST00000266551.4 |
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1 |
| chr3_+_49726932 | 1.15 |
ENST00000327697.6
ENST00000432042.1 ENST00000454491.1 |
RNF123
|
ring finger protein 123 |
| chr6_-_169654139 | 1.13 |
ENST00000366787.3
|
THBS2
|
thrombospondin 2 |
| chr9_+_131549610 | 1.08 |
ENST00000223865.8
|
TBC1D13
|
TBC1 domain family, member 13 |
| chr19_+_55105085 | 1.06 |
ENST00000251372.3
ENST00000453777.1 |
LILRA1
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 1 |
| chr15_-_43622736 | 1.01 |
ENST00000544735.1
ENST00000567039.1 ENST00000305641.5 |
LCMT2
|
leucine carboxyl methyltransferase 2 |
| chrX_+_37850026 | 0.89 |
ENST00000341016.3
|
CXorf27
|
chromosome X open reading frame 27 |
| chr2_-_68479614 | 0.63 |
ENST00000234310.3
|
PPP3R1
|
protein phosphatase 3, regulatory subunit B, alpha |
| chr1_+_158323486 | 0.57 |
ENST00000444681.2
ENST00000368167.3 |
CD1E
|
CD1e molecule |
| chr1_+_158323244 | 0.57 |
ENST00000434258.1
|
CD1E
|
CD1e molecule |
| chrX_+_135230712 | 0.56 |
ENST00000535737.1
|
FHL1
|
four and a half LIM domains 1 |
| chr4_-_168155577 | 0.50 |
ENST00000541354.1
ENST00000509854.1 ENST00000512681.1 ENST00000421836.2 ENST00000510741.1 ENST00000510403.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr20_-_17511962 | 0.44 |
ENST00000377873.3
|
BFSP1
|
beaded filament structural protein 1, filensin |
| chr11_-_615942 | 0.40 |
ENST00000397562.3
ENST00000330243.5 ENST00000397570.1 ENST00000397574.2 |
IRF7
|
interferon regulatory factor 7 |
| chr15_-_80263506 | 0.31 |
ENST00000335661.6
|
BCL2A1
|
BCL2-related protein A1 |
| chr6_-_32908792 | 0.29 |
ENST00000418107.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chrX_+_47077632 | 0.26 |
ENST00000457458.2
|
CDK16
|
cyclin-dependent kinase 16 |
| chrX_+_1734051 | 0.24 |
ENST00000381229.4
ENST00000381233.3 |
ASMT
|
acetylserotonin O-methyltransferase |
| chr1_+_154244987 | 0.20 |
ENST00000328703.7
ENST00000457918.2 ENST00000483970.2 ENST00000435087.1 ENST00000532105.1 |
HAX1
|
HCLS1 associated protein X-1 |
| chrX_+_68725084 | 0.06 |
ENST00000252338.4
|
FAM155B
|
family with sequence similarity 155, member B |
| chr12_-_57352103 | 0.05 |
ENST00000398138.3
|
RDH16
|
retinol dehydrogenase 16 (all-trans) |
Network of associatons between targets according to the STRING database.
First level regulatory network of PAX3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.1 | 21.3 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 4.5 | 13.6 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 3.7 | 14.9 | GO:2000969 | positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 2.8 | 8.3 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 2.7 | 10.8 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 2.6 | 12.9 | GO:0097338 | response to clozapine(GO:0097338) |
| 2.4 | 24.3 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 2.1 | 25.3 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 1.6 | 3.2 | GO:0010841 | positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) |
| 1.5 | 12.1 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 1.4 | 50.2 | GO:0034035 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 1.2 | 4.7 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 1.1 | 3.3 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 1.0 | 6.1 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.9 | 3.6 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.9 | 39.4 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.8 | 4.8 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.7 | 5.2 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.6 | 2.5 | GO:0002774 | Fc receptor mediated inhibitory signaling pathway(GO:0002774) |
| 0.6 | 5.4 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.6 | 6.9 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.5 | 4.0 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.5 | 3.3 | GO:0042636 | negative regulation of hair cycle(GO:0042636) |
| 0.4 | 1.8 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.4 | 3.3 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.4 | 30.1 | GO:0007602 | phototransduction(GO:0007602) |
| 0.4 | 8.1 | GO:0009650 | UV protection(GO:0009650) |
| 0.4 | 4.8 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.3 | 1.9 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.3 | 5.6 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 13.7 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.2 | 6.3 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.2 | 6.2 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.2 | 10.2 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.2 | 5.1 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.2 | 2.8 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.2 | 15.4 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.2 | 7.5 | GO:0045824 | negative regulation of innate immune response(GO:0045824) |
| 0.2 | 2.6 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.2 | 3.3 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.1 | 4.6 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.1 | 2.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 1.8 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.1 | 2.4 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.1 | 2.0 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 3.5 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.1 | 2.0 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 1.1 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.1 | 0.2 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.1 | 3.6 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.1 | 1.4 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.1 | 4.1 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 1.6 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.1 | 0.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 2.2 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.1 | 2.9 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 2.9 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 3.3 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 7.6 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 1.1 | GO:2000181 | negative regulation of angiogenesis(GO:0016525) negative regulation of vasculature development(GO:1901343) negative regulation of blood vessel morphogenesis(GO:2000181) |
| 0.0 | 3.8 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 9.2 | GO:0030324 | lung development(GO:0030324) |
| 0.0 | 2.0 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 6.1 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 7.3 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.0 | 0.4 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 18.4 | GO:0001558 | regulation of cell growth(GO:0001558) |
| 0.0 | 0.3 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 4.0 | GO:0030335 | positive regulation of cell migration(GO:0030335) |
| 0.0 | 1.7 | GO:0022610 | cell adhesion(GO:0007155) biological adhesion(GO:0022610) |
| 0.0 | 7.4 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 2.9 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 59.1 | GO:0007166 | cell surface receptor signaling pathway(GO:0007166) |
| 0.0 | 2.4 | GO:0002027 | regulation of heart rate(GO:0002027) |
| 0.0 | 9.6 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 3.4 | GO:0006486 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 6.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 1.0 | 4.8 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.9 | 3.6 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.8 | 7.5 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.8 | 8.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.7 | 17.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.6 | 12.9 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.5 | 3.5 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.4 | 8.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.4 | 1.8 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.3 | 10.5 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.3 | 10.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.3 | 1.4 | GO:0000801 | central element(GO:0000801) |
| 0.2 | 10.2 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.2 | 1.2 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.1 | 20.7 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.1 | 1.7 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.1 | 5.6 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 6.3 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 23.8 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 15.3 | GO:0005856 | cytoskeleton(GO:0005856) |
| 0.0 | 95.8 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 5.1 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 4.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 2.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 24.8 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 9.6 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 1.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 35.7 | GO:0005829 | cytosol(GO:0005829) |
| 0.0 | 51.0 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 0.2 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 13.6 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 3.5 | 10.4 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 2.2 | 50.2 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 1.7 | 39.4 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 1.3 | 8.1 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 1.1 | 3.3 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 1.1 | 3.3 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.8 | 12.9 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.8 | 3.8 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.7 | 10.2 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.7 | 17.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.7 | 6.9 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.6 | 25.3 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.6 | 20.7 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.5 | 4.0 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.3 | 4.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.3 | 12.1 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.3 | 8.3 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.3 | 2.5 | GO:0032396 | inhibitory MHC class I receptor activity(GO:0032396) |
| 0.3 | 2.8 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.3 | 4.7 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.2 | 5.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 2.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.2 | 15.4 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.2 | 2.5 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.2 | 2.4 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.2 | 0.6 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.1 | 1.8 | GO:0089720 | caspase binding(GO:0089720) |
| 0.1 | 10.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 5.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 4.4 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 2.2 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 3.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 1.1 | GO:0030882 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.1 | 2.0 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 3.8 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 4.8 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 4.7 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 2.6 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 3.2 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 1.6 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 3.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 2.0 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 7.6 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 7.4 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 9.7 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 13.7 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 2.9 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 1.9 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 7.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 2.0 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 1.0 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 3.6 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 44.0 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 0.3 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 3.4 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 2.5 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 1.6 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 5.4 | GO:0060089 | receptor activity(GO:0004872) molecular transducer activity(GO:0060089) |
| 0.0 | 3.2 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 4.6 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.2 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 15.2 | GO:0004871 | signal transducer activity(GO:0004871) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 13.6 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.3 | 13.8 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.3 | 12.9 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.2 | 10.8 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.2 | 15.4 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.2 | 7.5 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 7.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 3.6 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.1 | 10.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 1.8 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.1 | 2.4 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 4.8 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 50.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.8 | 10.8 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.7 | 15.4 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.4 | 12.9 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.4 | 21.2 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.4 | 10.6 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.3 | 18.3 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.2 | 7.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 3.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 5.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 1.8 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 4.8 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 1.8 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 1.9 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.1 | 8.3 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 2.6 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 5.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 7.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 16.1 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 2.0 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 1.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 2.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.4 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 6.5 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 1.1 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |